Supplemental Digital Content is available in the text
Keywords: Bayesian false discovery probability, male infertility, meta-analysis, Methylenetetrahydrofolate reductase, polymorphism
Abstract
Background:
18 previous meta-analyses have been published on the methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms with male infertility risk. However, results of the previous meta-analyses were still inconsistent. Moreover, their meta-analyses did not assess false-positive report probabilities except one study. Furthermore, many new studies have been published, and therefore an updated meta-analysis and re-analysis of systematic previous meta-analyses were performed to further explore these issues.
Objectives:
To determine the association between MTHFR C677T and A1298C polymorphisms and male infertility risk.
Methods:
Crude odds ratios and their 95% confidence intervals were used to assess the association between MTHFR C677T and A1298C polymorphisms and male infertility risk. We used the Bayesian false discovery probability (BFDP) to assess the credibility of statistically significant associations.
Results:
Fifty-nine studies were included concerning the MTHFR C677T and 28 studies were found on the MTHFR A1298C with male infertility risk. Overall, the MTHFR C677T was associated with increased male infertility risk in overall populations, Africans, East Asians, West Asians, South Asians, azoospermia, and Oligoasthenoteratozoospermia (OAT). In further sensitivity analysis and BFDP test, the positive results were only considered as “noteworthy” in the overall population (TT vs CC: BFDP = 0.294, CT + TT vs CC: BFDP = 0.300, T vs C: BFDP = 0.336), East Asians (TT vs CC: BFDP = 0.089, TT vs CT + CC: BFDP = 0.020, T vs C: BFDP < 0.001), West Asians (TT vs CC: BFDP = 0.584), hospital-based studies (TT vs CC: BFDP = 0.726, TT vs CT + CC: BFDP = 0.126), and OAT (TT vs CT + CC: BFDP = 0.494) for MTHFR C677T. In addition, a significantly increased male infertility risk was found in East Asians and population-based studies for MTHFR A1298C. However, we did not find that the positive results were considered as “noteworthy” in the overall and all subgroup analyses for MTHFR A1298C.
Conclusions:
In summary, this study indicates that the MTHFR C677T is associated with increased male infertility risk in East Asians, West Asians, and OAT. No significant association was observed on the MTHFR A1298C with male infertility risk.
1. Introduction
Infertility, defined as the inability to conceive after one year of regular unprotected sexual intercourse by the World Health Organization, has been a major health problem which is multifactorial in nature and affected approximately 15% to 20% of all couples trying for pregnancy.[1–3] Male factors infertility accounts for 40% to 50% about the cases of infertility.[4–5] The etiological factors of male infertility are multifactorial syndrome with a very complex pathogenesis, involving lifestyle, organic diseases, genetic factors, environmental risk factors, and their interactions.[6–8]
Folate play much essential roles for the maintenance of genome integrity in Deoxyribonucleic acid synthesis, repair and methylation.[9] Methylenetetrahydrofolate reductase (MTHFR) gene has the chromosomal locus 1p36.6 and is 2.2 kb in length with a total of 11 exons, which is involved in folate and homocysteine metabolism. A change of C to T at nucleotide 677 in MTHFR C677T (Ala222Val, rs1801133) results in an amino acid substance change of an alanine to valine, and this substance is associated with reduced enzyme activity that leads to reduced plasma folate levels.[10,11] The MTHFR A1298C polymorphism, marked as rs1801131 in the NCBI database, is located at exon 7 and results in a 1298A-C mutation resulting in a glu429-to-ala (E429A) substitution at codon 429,[12] is also associated with decreased enzyme activity.[13,14]
To date, sixty-six studies have been published on the MTHFR C677T and A1298C polymorphisms with male infertility risk. However, the results of these studies were still contradictory. In addition, 15 previous meta-analyses[15,17,19–29,31,32] have been reported on the MTHFR C677T polymorphism with male infertility risk (as shown in Table 1). Among these publications, two studies[32] investigated this issue in Caucasians, two studies[22,27] in Asians, one study[25] in Chinese population, and 11 studies[15,17,19–21,23,24,26,28,29,31] in overall populations. Moreover, ten previous meta-analyses[15,16,18,20,24–27,30,32] have also been published on the MTHFR A1298C polymorphism with male infertility risk (as shown in Table 2). However, the previous meta-analysis results still inconsistent. Moreover, their meta-analyses did not assess false-positive report probabilities except Liu et al[26] by using the Benjamini-Hochberg methods, which control for false discovery rate, furthermore, many new studies have been published, and therefore an updated and high quality meta-analysis were performed to further explore the issues. For all we know, this is the first meta-analysis to further investigate the positive result using a Bayesian method.
Table 1.
Results of previous meta-analysis between MTHFR C677T polymorphism with male infertility risk.
| CT vs. CC | TT vs. CC | (CT + TT) vs. CC | TT vs. (CC + CT) | T vs. C | ||||||||||
| First author/year | Variable | n (Cases/Controls) | OR (95% CI) | Ph/I2 (%) | OR (95% CI) | Ph/I2 (%) | OR (95% CI) | Ph/I2 (%) | OR (95% CI) | Ph/I2 (%) | OR (95% CI) | Ph/I2 (%) | Whether performed assessment of literature quality | Whether performed P adjust |
| Ullah[32] 2019 | Low income | 8 (NA) | NA | NA | NA | NA | 1.87 (0.96, 3.64) | NA | NA | NA | NA | NA | No | No |
| Middle income | 13 (NA) | NA | NA | NA | NA | 1.38 (1.02, 1.88) | NA | NA | NA | NA | NA | |||
| High income | 9 (NA) | NA | NA | NA | NA | 1.26 (0.92, 1.71) | NA | NA | NA | NA | NA | |||
| Shi[27] 2019 | Asian | 20 (4734/3967) | 1.35 (1.22, 1.49) | NA/38 | 2.08 (1.79, 2.44) | NA/44 | 1.49 (1.35, 1.64) | NA/50 | 1.67 (1.49, 1.89) | NA/27 | 1.43 (1.33, 1.52) | NA/49 | Yes | No |
| East Asian | 13 (3013/2571) | 1.45 (1.28, 1.67) | NA/20 | 2.13 (1.82, 2.50) | NA/17 | 1.61 (1.43, 1.75) | NA/28 | 1.67 (1.45, 1.89) | NA/3 | 1.45 (1.35, 1.56) | NA/20 | |||
| South/West Asia | 7 (1721/1396) | 1.22 (1.03, 1.43) | NA/53 | 1.89 (1.35, 2.63) | NA/68 | 1.32 (1.12, 1.54) | NA/65 | 1.78 (1.29, 2.14) | NA/57 | 1.33 (1.16, 1.52) | NA/71 | |||
| Hong[19] 2017 | Overall | 15 (3853/3613) | 1.34 (1.03, 1.74) | <.001/80 | 1.86 (1.36, 2.54) | 0.009/55 | 1.46 (1.05, 2.04) | <.001/89 | 1.42 (1.19, 1.70) | .03/49 | 1.38 (1.18, 1.63) | .0007/66 | Yes | No |
| Caucasian | 2 (NA) | NA | NA | NA | NA | NA | NA | NA | NA | 1.23 (0.85, 1.70) | .10/63 | |||
| East-asian | 5 (NA) | NA | NA | NA | NA | NA | NA | NA | NA | 1.39 (1.20, 1.61) | .23/29 | |||
| Middle-estern | 2 (NA) | NA | NA | NA | NA | NA | NA | NA | NA | 1.30 (1.05, 1.63) | .78/0 | |||
| Indian | 3 (NA) | NA | NA | NA | NA | NA | NA | NA | NA | 1.25 (0.74, 2.13) | .0003/88 | |||
| Mixed-race | 1 (NA) | NA | NA | NA | NA | NA | NA | NA | NA | 1.96 (1.35, 2.85) | .001/63 | |||
| Rai[22] 2017 | Asian | 17 (4392/3667) | 1.40 (1.18, 1.62) | .005/52.7 | 2.10 (1.61, 2.61) | .02/47.4 | 1.53 (1.30, 1.77) | .005/53.7 | 1.70 (1.38, 2.10) | .03/43.7 | 1.99 (1.58, 2.51) | <.001/89.4 | No | No |
| Ren[25] 2017 | Chinese | 9 (1713/1104) | NA | NA | 2.08 (1.68, 2.58) | NA/35 | 1.51 (1.30, 1.77) | NA/29 | 1.58 (1.31, 1.90) | NA/0.0 | 1.47 (1.32, 1.63) | NA/42 | Yes | No |
| Yang et al.[15] 2016 | Overall | 21 (4505/4024) | 1.21 (1.04, 1.41) | .001/54.7 | 1.63 (1.22, 2.18) | <.001/69.4 | 1.29 (1.09, 1.54) | <.001/68.6 | 1.46 (1.16, 1.85) | <.001/60.3 | 1.26 (1.10, 1.46) | <.001/76.1 | No | No |
| Caucasian | 13 (NA) | 1.13 (0.90, 1.42) | NA | 1.38 (0.84, 2.27) | NA | 1.17 (0.90, 1.51) | NA | 1.30 (0.86, 1.98) | NA | 1.16 (0.92, 1.45) | NA | |||
| Asian | 8 (NA) | 1.32 (1.14–1.53) | NA | 1.90 (1.54, 2.35) | NA | 1.47 (1.25, 1.73) | NA | 1.63 (1.36, 1.96) | NA | 1.40 (1.24, 1.59) | NA | |||
| Zhu[17] 2016 | Overall | 26 (5659/5528) | 1.09 (1.00, 1.19) | .008/45 | 1.83 (1.48, 2.26) | <.001/74 | 1.19 (1.04, 1.36) | .0002/57 | 1.54 (1.27, 1.88) | .002/51 | 1.23 (1.10,1.37) | <.001/66 | No | No |
| Asian | 16 (NA) | 1.22 (1.10, 1.36) | .08/36 | 2.43 (2.08, 2.83) | .13/30 | 1.36 (1.19, 1.56) | .04/42 | 1.81 (1.47, 2.23) | .08/36 | 1.37 (1.27, 1.47) | .01/51 | |||
| Caucasian | 10 (NA) | 0.89 (0.77, 1.03) | .28/17 | 1.08 (0.83, 1.41) | .04/49 | 0.93 (0.81, 1.06) | .15/32 | 1.14 (0.92, 1.41) | .05/48 | 0.99 (0.89, 1.09) | .03/51 | |||
| Azoo∗ | 12 (NA) | 1.67 (1.36, 2.06) | .14/31 | 1.17 (1.02, 1.35) | .10/36 | 1.26 (1.10, 1.44) | .04/42 | 1.50 (1.25, 1.82) | .3/15 | 1.25 (1.14, 1.38) | .03/49 | |||
| OAT† | 14 (NA) | 1.01 (0.90, 1.14) | .05/41 | 1.41 (1.00, 1.99) | <.001/69 | 1.10 (0.91, 1.33) | .15/32 | 1.43 (1.04, 1.98) | <.001/69 | 1.17 (0.98, 1.40) | <.001/76 | |||
| Gong[23] 2015 | Overall | 26 (5575/5447) | NA | NA | 1.76 (1.53, 2.01) | NA/54.0 | 1.34 (1.23, 1.46) | NA/68.2 | 1.60 (1.41, 1.81) | NA/36.9 | 1.32 (1.24, 1.41) | NA/71.5 | No | No |
| Asian | 10 (NA) | NA | NA | NA | NA | 1.54 (1.35, 1.76) | NA/0.0 | NA | NA | NA | NA | |||
| Caucasian | 11 (NA) | NA | NA | NA | NA | 1.19 (1.05, 1.36) | NA/48.1 | NA | NA | NA | NA | |||
| Azoo∗ | 1412/3532 | NA | NA | NA | NA | 1.36 (1.18, 1.55) | NA/49.1 | NA | NA | NA | NA | |||
| OAT† | 615/1865 | NA | NA | NA | NA | 1.35 (1.11, 1.64) | NA/44.7 | NA | NA | NA | NA | |||
| Liu[26] 2015 | Overall | 32 (NA) | 1.17 (1.03, 1.33) | <.001/NA | 1.62 (1.29, 2.04) | <.001/NA | 1.26 (1.10, 1.45) | <.001/NA | 1.47 (1.23, 1.77) | .002/NA | 1.25 (1.12, 1.40) | <.001/NA | Yes | Yes (FDR) |
| Asian | 17 (NA) | 1.28 (1.13, 1.46) | .153/NA | 2.15 (1.67, 2.75) | .036/NA | 1.44 (1.24, 1.66) | .019/NA | 1.79 (1.48, 2.16) | .193/NA | 1.42 (1.27, 1.60) | .005/NA | |||
| Azoo∗ | 14 (NA) | 1.21 (1.04, 1.41) | .298/NA | 1.64 (1.12, 2.42) | .002/NA | 1.31 (1.06, 1.61) | .015/NA | 1.49 (1.07, 2.06) | .011/NA | 1.27 (1.05, 1.54) | <.001/NA | |||
| OAT† | 16 (NA) | 1.17 (0.96, 1.44) | .001/NA | 1.52 (1.12, 2.06) | .008/NA | 1.25 (1.01, 1.55) | <.001/NA | 1.43 (1.13, 1.82) | .098/NA | 1.24 (1.05, 1.47) | <.001/NA | |||
| Nikzad[28] 2015 | Overall | 23 (5174/5253) | NA | NA | 1.44 (1.09, 1.89) | <.001/66 | 1.21 (1.06, 1.39) | <.001/60 | 1.38 (1.14, 1.68) | .017/43 | 1.21 (1.08, 1.36) | < .001/67 | No | No |
| Weiner[29] 2014 | Overall | 17 (2972/3436) | NA | NA | NA | NA | 1.05 (0.99, 1.11) | NA | NA | NA | NA | NA | No | No |
| Azoo∗ | 6 (NA) | NA | NA | NA | NA | 1.18 (0.92, 1.51) | NA | NA | NA | NA | NA | |||
| Gupta[31] 2013 | Overall | 13 (3094/2877) | NA | NA | NA | NA | 1.31 (1.17, 1.46) | NA | NA | NA | 1.30 (1.20, 1.41) | NA | No | No |
| Azoo∗ | NA | NA | NA | NA | NA | 1.65 (1.36, 1.99) | NA | NA | NA | 1.52 (1.32, 1.17) | NA | |||
| OAT† | NA | NA | NA | NA | NA | 1.08 (0.88, 1.34) | NA | NA | NA | 1.43 (1.18, 1.73) | NA | |||
| Wu[21] 2012 | Overall | 10 (2275/1958) | 1.11 (0.86, 1.43) | .004/NA | 1.39 (0.93, 2.07) | <.001/NA | 1.15 (0.89, 1.49) | <.001/NA | 1.34 (0.99, 1.81) | .012/NA | 1.17 (0.95, 1.43) | <.001/NA | No | No |
| Asian | 6 (NA) | 1.70 (0.97, 1.72) | .017/NA | 1.79 (1.08, 2.96) | .003/NA | 1.42 (1.02, 1.96) | .001/NA | 1.50 (1.21, 1.86) | .055/NA | 1.36 (1.06, 1.75) | <.001/NA | |||
| Caucasian | 3 (NA) | 0.71 (0.45, 1.12) | .932/NA | 0.71 (0.45, 1.12) | .522/NA | 0.75 (0.55, 1.01) | .774/NA | 0.81 (0.53, 1.23) | .534/NA | 0.81 (0.65, 1.01) | .534/NA | |||
| Azoo∗ | 5 (NA) | 1.45 (1.18, 1.79) | .340/NA | 1.89 (1.43, 2.51) | .308/NA | 1.55 (1.28, 1.88) | .257/NA | 1.51 (1.17, 1.95) | .333/NA | 1.38 (1.20, 1.57) | .155/NA | |||
| OAT† | 7 (NA) | 0.90 (0.74, 1.08) | .062/NA | 1.02 (0.78, 1.32) | .064/NA | 0.91 (0.69, 1.19) | .031/NA | 1.08 (0.85, 1.38) | .119/NA | 0.96 (0.78, 1.18) | .017/NA | |||
| Wei[24] 2012 | Overall | 11 (2217/2312) | 1.22 (0.96, 1.56) | .001/NA | 1.40 (0.98, 2.00) | .001/NA | 1.26 (0.97, 1.65) | <.001/NA | 1.28 (1.00, 1.64) | .09/NA | NA | NA | No | No |
| Caucasian | 5 (635/611) | 1.17 (0.67, 2.06) | .001/NA | 1.18 (0.60, 2.34) | .01/NA | 1.17 (0.65, 2.12) | <.001/NA | 1.09 (0.70, 1.71) | .15/NA | NA | NA | |||
| Asian | 6 (1582/1701) | 1.26 (0.98, 1.62) | .03/NA | 1.57 (1.05, 2.37) | .02/NA | 1.34 (1.01, 1.77) | .004/NA | 1.40 (1.05, 1.86) | .17/NA | NA | NA | |||
| Tüttelmann[20] 2007 | Overall | 8 (1843/1791) | NA | NA | NA | NA | 1.39 (1.15, 1.69) | NA | NA | NA | NA | NA | No | No |
Table 2.
Results of previous meta-analysis between MTHFR A1298C polymorphism with male infertility risk.
| AC vs. AA | CC vs. AA | (AC + CC) vs. AA | CC vs. (AA + AC) | C vs. A | ||||||||||
| First author | Variable | n (Cases/Controls) | OR (95% CI) | Ph/I2 (%) | OR (95% CI) | Ph/I2 (%) | OR (95% CI) | Ph/I2 (%) | OR (95% CI) | Ph/I2 (%) | OR (95% CI) | Ph/I2 (%) | Whether performed assessment of literature quality | Whether performed P adjust |
| Shi[27] 2019 | Asian | 12 (2673/2328) | 1.20 (1.08, 1.37) | NA/27 | 1.64 (1.08, 2.56) | NA/ 58 | 1.27 (1.14, 1.43) | NA/46 | 1.61 (1.27, 2.04) | NA/50 | 1.22 (1.05, 1.41) | NA/57 | Yes | No |
| East Asian | 7 (1759/1586) | 1.35 (1.16, 1.56) | NA/0 | 2.17 (1.11, 4.17) | NA/ 65 | 1.43 (1.25, 1.67) | NA/38 | 2.04 (1.47, 2.86) | NA/59 | 1.37 (1.12, 1.67) | NA/56 | |||
| South/West Asia | 5 (878/742) | 0.96 (0.78, 1.19) | NA/0 | 1.14 (0.78, 1.67) | NA/0 | 1.00 (0.82,1.22) | NA/0 | 1.20 (0.84, 1.72) | NA/0 | 1.03 (0.77, 1.20) | NA/0 | |||
| Ullah[32] 2019 | Low income | 6 (NA) | NA | NA | NA | NA | NA | NA | 1.71 (1.19, 2.47) | NA | NA | NA | No | No |
| Middle income | 10 (NA) | NA | NA | NA | NA | NA | NA | 1.02 (0.81, 1.28) | NA | NA | NA | |||
| High income | 4 (NA) | NA | NA | NA | NA | NA | NA | 0.86 (0.62, 1.19) | NA | NA | NA | |||
| Zhang[16] 2017 | Overall | 20 (4293/4507) | 1.02 (0.93, 1.12) | .165/NA | 1.01 (0.85, 1.20) | .100/NA | 1.02 (0.93, 1.12) | .157/NA | 1.01 (0.86, 1.19) | .111/NA | 1.02 (0.95, 1.09) | .148/NA | Yes | No |
| Caucasian | 15 (NA) | 0.95 (0.85, 1.06) | .142/NA | 0.94 (0.78, 1.14) | .063/NA | 0.95 (0.86, 1.06) | .177/NA | 0.96 (0.80, 1.14) | .056/NA | 0.96 (0.89, 1.04) | .202/NA | |||
| Asian | 5 (NA) | 1.20 (1.01, 1.44) | .994/NA | 1.41 (0.93, 2.15) | .860/NA | 1.23 (1.04, 1.45) | .996/NA | 1.33 (0.88, 2.02) | .846/NA | 1.20 (1.04, 1.39) | .985/NA | |||
| Ren[25] 2017 | Chinese | 3 (540/457) | NA | NA | 1.34 (0.66, 2.71) | NA/0.0 | 1.27 (0.95, 1.65) | NA/0.0 | 1.44 (0.72, 2.88) | NA/9 | 1.22 (0.97, 1.53) | NA/0.0 | Yes | No |
| Yang[15] 2016 | Overall | 13 (2785/3094) | 1.02 (0.91, 1.14) | .216/22.5 | 1.29 (1.03, 1.62) | .330/11.5 | 1.06 (0.95, 1.18) | .224/21.7 | 1.29 (1.03, 1.60) | .345/10.0 | 1.08 (0.99, 1.18) | .294/15.0 | No | No |
| Caucasian | 8 (NA) | 0.90 (0.78, 1.04) | NA | 1.31 (1.00, 1.72) | NA | 0.95 (0.83, 1.09) | NA | 1.35 (1.04, 1.74) | NA | 1.02 (0.92, 1.14) | NA | |||
| Asian | 5 (NA) | 1.24 (1.03, 1.48) | NA | 1.24 (0.82, 1.88) | NA | 1.24 (1.04, 1.47) | NA | 1.16 (0.77, 1.75) | NA | 1.19 (1.03, 1.37) | NA | |||
| Liu[26] 2015 | Overall | 17 (NA) | NA | NA | NA | NA | NA | NA | 1.11 (0.87, 1.41) | NA | NA | NA | Yes | Yes (FDR) |
| Gupta[30] 2013 | Overall | 10 (2734/2737) | NA | NA | NA | NA | 1.05 (0.89, 1.23) | .058/45.4 | NA | NA | NA | NA | No | No |
| Azoo∗ | NA | NA | NA | NA | NA | 0.97 (0.79, 1.18) | .697/0.0 | NA | NA | NA | NA | |||
| OAT† | NA | NA | NA | NA | NA | 0.96 (0.74, 1.24) | .006/66.6 | NA | NA | NA | NA | |||
| Shen[18] 2012 | Overall | 7 (1633/1735) | 1.10 (0.95, 1.27) | .855/NA | 1.29 (0.97, 1.72) | .087/NA | 1.13 (0.98, 1.30) | .578/NA | 1.26 (0.95, 1.65) | .119/NA | 1.12 (1.00, 1.26) | .215/NA | No | No |
| Asian | 5 (NA) | 1.11 (0.94, 1.31) | .919/NA | 1.18 (0.84, 1.66) | .435/NA | 1.12 (0.96, 1.31) | .917/NA | 1.14 (0.82, 1.59) | .415/NA | 1.10 (0.97, 1.25) | .841/NA | |||
| Caucasian | 2 (NA) | 1.06 (0.77, 1.45) | .206/NA | 1.55 (0.42, 5.72) | .012/NA | 1.15 (0.85, 1.55) | .052/NA | 1.52 (0.49, 4.71) | .023/NA | 0.81 (0.65, 1.01) | .011/NA | |||
| Azoo∗ | 4 (NA) | 1.01 (0.78, 1.31) | .840/NA | 1.66 (1.01, 2.73) | .124/NA | 1.08 (0.85, 1.38) | .965/NA | 1.67 (1.03, 2.71) | .078/NA | 1.14 (0.94, 1.38) | .625/NA | |||
| OAT† | 5 (NA) | 1.10 (0.91, 1.34) | .401/NA | 1.15 (0.82, 1.63) | .140/NA | 1.12 (0.93, 1.34) | .177/NA | 1.12 (0.81, 1.56) | .290/NA | 1.09 (0.95, 1.26) | .079/NA | |||
| Wei[24] 2012 | Overall | 7 (1633/1735) | 1.30 (0.87, 1.95) | .09/NA | 1.10 (0.95, 1.27) | .86/NA | 1.13 (0.98, 1.30) | .58/NA | 1.26 (0.95, 1.65) | .12/N1 | NA | NA | No | No |
| Caucasian | 2 (406/346) | 1.55 (0.42, 5.72) | .01/NA | 1.06 (0.77, 1.45) | .21/NA | 1.15 (0.85, 1.55) | .05/NA | 1.54 (0.94, 2.54) | .02/N1 | NA | NA | |||
| Asian | 5 (1227/1389) | 1.16 (0.82, 1.64) | .44/NA | 1.11 (0.94, 1.31) | .92/NA | 1.12 (0.96, 1.32) | .92/NA | 1.14 (0.82, 1.59) | .42/NA | NA | NA | |||
| Tüttelmann[20] 2007 | Overall | 2 (539/525) | NA | NA | NA | NA | 0.97 (0.54, 1.74) | NA | NA | NA | NA | NA | No | No |
2. Materials and methods
2.1. Search strategy
The eligible studies were searched (the deadline was April 9, 2020) to used three databases (PubMed, CNKI, and WangFang). Retrieval strategy was designed by the following keywords (methylenetetrahydrofolate reductase OR MTHFR) AND (polymorphism OR mutation OR variant) AND (infertility OR azoospermia OR oligoasthenoteratozoospermia OR oligozoospermia OR subinfertility). Language did not be restrict in this study. We send emails to the corresponding authors if data of a few studies did not be collect by full-text. In addition, the previous meta-analyses were also carefully examined by reference lists.
2.2. Inclusion and exclusion criteria
The inclusion criteria as following:
-
(1)
human case-control or cohort studies (Infertility was defined as conception failure after at least 1 year of regular unprotected sexual intercourse among couples; Controls were healthy without a history of infertility, and had one child at least with normal sperm parameters. In addition, Cases and controls should be comparable),
-
(2)
studies on the MTHFR C677T and A1298C polymorphisms and male infertility risk,
-
(3)
If more than one study had been published using the same case series, we selected one study including the maximum sample size, and
-
(4)
the genotype data or odds ratios (ORs) and their 95% confidence intervals (CIs) provided.
The exclusion criteria as following:
-
(1)
data not listed,
-
(2)
not human case–control or cohort studies, and
-
(3)
reviews, meta-analyses, conference abstracts, letters, and editorials.
2.3. Data extraction and quality score assessment
Two authors independently extracted data from selected studies including the following information:
-
(1)
first author's name,
-
(2)
year of publication,
-
(3)
country,
-
(4)
ethnicity,
-
(5)
source of controls,
-
(6)
sample size, and
-
(7)
genotype distribution of male infertility cases and controls.
Two investigators assessed independently the quality of eligible articles. The literature quality assessment criteria was shown in supplemental Table 1. The biggest score value is eleven by the quality assessment; scoring ≥ 5 were considered as high quality studies. A third author adjudicated inconsistent scores.
2.4. Statistical analysis
We evaluated the association between the MTHFR C677T and A1298C polymorphisms and male infertility risk by pooled the crude ORs and their 95% CIs. The pooled ORs with the corresponding 95% CIs were performed by the following genetic models: a dominant model: (CT + TT) vs. CC for the MTHFR C677T polymorphism and (AC + CC) vs. AA for the MTHFR A1298C polymorphism, a recessive model: TT vs (CC + CT) for the C677T and (AC + CC) vs AA for the A1298C, a heterozygote model: CT vs. CC for the C677T and AC vs. AA for the A1298C, a homozygote model: TT vs CC for the C677T and CC vs. AA for the A1298C, and an allele model: T vs. C for the C677T and C vs. A for the A1298C. Heterogeneity among studies was checked according to the Cochran Q[94] and I2 value[95]. The P > .10 and/or I2 < 50% indicate a lack of heterogeneity among studies, hence, the pooled crude ORs was calculated using a fixed-effects model (Mantel–Haenszel method)[96]; otherwise, a random-effect model (DerSimonian and Laird method) was applied[97]. A meta-regression analysis was used to explore sources of heterogeneity[98] if heterogeneity among studies was significant. Subgroup analyses were conducted according to ethnicity, source of controls and type of male infertility. Sensitivity analyses were also performed to estimate the robustness of the pooled results. We used the following methods to perform the sensitivity analyses: excluded the studies of Hardy-Weinberg dis-equilibrium (HWD) and quality scores < 5. Hardy-Weinberg equilibrium (HWE) was calculated by chi-square goodness-of-fit test, and significant deviation was considered in control groups if the P value < .05. The publication bias was assessed to using Begg funnel[99] and Egger test.[100] Last, a Bayesian false discovery probability (BFDP: a cutoff value was set up to be a level of 0.8 and a prior probability of 0.001)[101] was used to evaluate positive results whether were noteworthy or not. All statistical analyses were conducted using STATA version 12.0 (STATA Corporation, College Station, TX).
3. Results
3.1. Study characteristics
A flowchart of study selection is listed in Figure 1. Overall, we retrieved 243 publications by several databases. Among these publications, sixty-six articles were selected after filtering titles, abstracts, and full texts. In addition, the sample size of four publications[54,75,86,92] overlapped with those of another four publications.[2,57,61,88] Therefore, sixty-two publications were involved in the final analysis. Table 3 lists the main characteristics of the selected studies. Fifty-nine studies[2,28,29,31–43,46–53,55–63,65–74,76–85,87–91] were included concerning the MTHFR C677T polymorphism (11,767 male infertility cases and 10,591 controls; two studies on Africans, thirteen on Caucasians, twenty-seven on East Asians, seven on West Asians, eight on South Asians, and two mixed populations; fifty-three hospital-based studies and six population-based studies; twenty-four azoospermia studies and thirty-seven Oligoasthenoteratozoospermia (OAT) studies) with male infertility risk. Twenty-eight studies were found on the MTHFR A1298C polymorphis[2,29,30,32,37,38,41,43–45,49,50,52,53,55–59,61,63–65,72,73,80,81,91] (5,976 male infertility cases and 5,774 controls; four studies on South Asians, 7 on West Asians, nine on East Asians, six on Caucasians, one on Africans, and one mixed populations; twenty-five hospital-based studies and three population-based studies; twelve azoospermia studies and twelve OAT studies) with male infertility risk. In addition, HWD of controls was observed in six studies[2,32,43,69,76,80] for C677T polymorphism and six studies[32,44,49,63,81,91] for A1298C polymorphism.
Figure 1.
OA_ Guidelines Flow Diagram Study selection flowchart in the current meta-analysis.
Table 3.
Characteristics of studies included in the current meta-analysis.
| Case | Control | |||||||||||||||||
| Azoospermia | OAT | Total | ||||||||||||||||
| First Author/Year | Country | Ethnicity | SC | Sample size | CC | CT | TT | CC | CT | TT | CC | CT | TT | CC | CT | TT | HWE | Quality score |
| MTHFR C667T | ||||||||||||||||||
| Bezold[33] 2001 | German | Caucasian | HB | 255/200 | – | – | – | – | – | – | 114 | 93 | 48 | 92 | 89 | 19 | 0.705 | 5 |
| Stuppia[34] 2003 | Italy | Caucasian | HB | 93/105 | 8 | 6 | 7 | 29 | 31 | 12 | 37 | 37 | 19 | 33 | 43 | 29 | 0.066 | 4 |
| Ebisch[35] 2003 | Netherlands | Caucasian | PB | 77/113 | – | – | – | – | – | – | 42 | 28 | 7 | 50 | 48 | 15 | 0.522 | 4 |
| Singh[36] 2005 | India | South Asian | PB | 151/200 | – | – | – | – | – | – | 105 | 40 | 6 | 163 | 37 | 0 | 0.149 | 6 |
| Park[37] 2005 | Korea | East Asian | HB | 373/396 | 75 | 164 | 47 | 28 | 40 | 17 | 105 | 205 | 63 | 145 | 200 | 51 | 0.161 | 5 |
| Lee[38] 2006 | Korea | East Asian | HB | 360/325 | 44 | 100 | 30 | 71 | 81 | 34 | 115 | 181 | 64 | 118 | 166 | 41 | 0.138 | 5 |
| Paracchini[39] 2006 | Italy | Caucasian | HB | 59/46 | – | – | – | – | – | – | 11 | 32 | 16 | 18 | 21 | 7 | 0.83 | 4 |
| A[40] 2007 | China | East Asian | HB | 355/252 | 83 | 97 | 48 | 47 | 63 | 17 | 130 | 160 | 65 | 128 | 95 | 29 | 0.085 | 5 |
| Dhillon[41] 2007 | India | South Asian | HB | 179/200 | – | – | – | 81 | 77 | 21 | 81 | 77 | 21 | 70 | 100 | 30 | 0.556 | 5 |
| Sun[42] 2007 | China | East Asian | NR | 182/53 | – | – | – | 22 | 75 | 52 | 27 | 86 | 69 | 15 | 28 | 10 | 0.63 | 3 |
| Zhang[73] 2007 | China | East Asian | HB | 165/132 | – | – | – | – | – | – | 41 | 93 | 31 | 48 | 60 | 24 | 0.492 | 4 |
| Ravel[43] 2009 | French | Caucasian | HB | 250/113 | 33 | 31 | 6 | 85 | 70 | 25 | 118 | 101 | 31 | 49 | 52 | 31 | 0.024 | 3 |
| Yang[72] 2010 | China | East Asian | HB | 131/293 | – | – | – | 34 | 55 | 42 | 34 | 55 | 42 | 98 | 142 | 53 | 0.901 | 4 |
| Đorđević[61] 2010 | Serbia | Caucasian | HB | 52/56 | – | – | – | – | – | – | 22 | 24 | 6 | 23 | 26 | 7 | 0.934 | 5 |
| Zhang[82] 2010 | China | East Asian | HB | 491/430 | – | – | – | – | – | – | 43 | 253 | 195 | 87 | 213 | 130 | 0.988 | 5 |
| Gava[45] 2011 | Brazil | Mixed | HB | 156/233 | 27 | 15 | 7 | 54 | 45 | 8 | 81 | 60 | 15 | 167 | 53 | 13 | 0.003 | 3 |
| Safarinejad[46] 2011 | Iran | West Asian | HB | 164/328 | – | – | – | 58 | 80 | 26 | 58 | 80 | 26 | 144 | 148 | 36 | 0.826 | 7 |
| Liu[47] 2011 | China | East Asian | HB | 75/72 | – | – | – | 27 | 38 | 10 | 27 | 38 | 10 | 40 | 28 | 4 | 0.753 | 3 |
| Qiu[48] 2011 | China | East Asian | HB | 271/180 | 42 | 66 | 50 | 33 | 46 | 34 | 75 | 112 | 84 | 63 | 85 | 32 | 0.72 | 4 |
| Murphy[64] 2011 | Swede | Caucasian | HB | 153/184 | – | – | – | – | – | – | 73 | 63 | 13 | 94 | 73 | 15 | 0.876 | 6 |
| Kumar[67] 2011 | India | South Asian | HB | 100/100 | – | – | – | – | – | – | 86 | 14 | 0 | 81 | 19 | 0 | 0.294 | 4 |
| Gupta[31] 2011 | India | South Asian | HB | 522/315 | 49 | 15 | 4 | 144 | 46 | 10 | 378 | 116 | 28 | 251 | 58 | 6 | 0.229 | 5 |
| Vani[49] 2012 | India | South Asian | HB | 206/230 | – | – | – | – | – | – | 158 | 42 | 6 | 188 | 42 | 0 | 0.128 | 4 |
| Eloualid[50] 2012 | Morocco | African | HB | 344/690 | 65 | 37 | 8 | 134 | 88 | 12 | 199 | 125 | 20 | 351 | 286 | 53 | 0.611 | 6 |
| Chellat[69] 2012 | Algeria | Mixed | HB | 74/84 | 20 | 19 | 7 | 11 | 14 | 3 | 31 | 33 | 10 | 36 | 38 | 10 | 0.995 | 3 |
| Liu[68] 2012 | China | East Asian | HB | 75/72 | – | – | – | 27 | 38 | 10 | 27 | 38 | 10 | 40 | 28 | 4 | 0.753 | 3 |
| Stangler[66] 2013 | Slovene | Caucasian | PB | 100/111 | – | – | – | – | – | – | 29 | 51 | 20 | 47 | 50 | 14 | 0.902 | 6 |
| Camprubi[71] 2013 | Spain | Caucasian | HB | 107/25 | – | – | – | 42 | 36 | 14 | 47 | 43 | 17 | 8 | 15 | 2 | 0.172 | 3 |
| Pei J[75] 2013 | China | East Asian | HB | 290/90 | – | – | – | – | – | – | 39 | 138 | 113 | 24 | 47 | 19 | 0.651 | 4 |
| Balkan[81] 2014 | Turkey | West Asian | NR | 108/125 | 57 | 40 | 11 | – | – | – | 57 | 40 | 11 | 78 | 36 | 11 | 0.032 | 3 |
| Mfady[51] 2014 | Jordan | West Asian | HB | 150/150 | – | – | – | – | – | – | 67 | 63 | 20 | 74 | 67 | 9 | 0.221 | 5 |
| Naqvi[52] 2014 | India | South Asian | HB | 637/364 | 34 | 11 | 4 | 413 | 143 | 33 | 447 | 154 | 36 | 275 | 79 | 10 | 0.145 | 7 |
| Li SS[54] 2014 | China | East Asian | HB | 82/133 | – | – | – | – | – | – | 14 | 36 | 32 | 36 | 61 | 36 | 0.34 | 4 |
| Weiner[29] 2014 | Russia | Caucasian | PB | 271/301 | 49 | 41 | 8 | 40 | 31 | 11 | 129 | 116 | 26 | 153 | 115 | 33 | 0.113 | 7 |
| Vardarli[62] 2014 | Turkey | Caucasian | HB | 100/50 | 23 | 22 | 5 | 21 | 22 | 7 | 44 | 44 | 12 | 30 | 20 | 0 | 0.077 | 4 |
| Hussein[77] 2014 | Egypt | African | HB | 107/107 | 64 | 35 | 8 | – | – | – | 64 | 35 | 8 | 62 | 32 | 13 | 0.012 | 3 |
| Ng[78] 2014 | Canada | Caucasian | NR | 39/19 | 10 | 10 | 2 | 12 | 4 | 1 | 22 | 14 | 3 | 8 | 5 | 3 | 0.219 | 2 |
| Ni W[56] 2015 | China | East Asian | PB | 296/204 | – | – | – | – | – | – | 117 | 135 | 44 | 84 | 94 | 26 | 0.97 | 7 |
| Gurkan[57] 2015 | Turkey | West Asian | HB | 137/134 | 41 | 25 | 9 | 29 | 24 | 9 | 70 | 49 | 18 | 71 | 55 | 8 | 0.533 | 5 |
| Li XY[58] 2015 | China | East Asian | HB | 162/120 | 36 | 49 | 15 | 25 | 28 | 9 | 61 | 77 | 24 | 48 | 54 | 18 | 0.661 | 5 |
| Kurzawski[59] 2015 | Poland | Caucasian | HB | 284/352 | – | – | – | – | – | – | 143 | 113 | 28 | 166 | 150 | 36 | 0.806 | 5 |
| Kim[60] 2015 | Korea | East Asian | HB | 85/246 | 30 | 44 | 11 | – | – | – | 30 | 44 | 11 | 87 | 106 | 53 | 0.057 | 4 |
| Nikzad[28] 2015 | Iran | West Asian | HB | 242/255 | 47 | 49 | 11 | 62 | 60 | 13 | 109 | 109 | 24 | 144 | 98 | 13 | 0.48 | 5 |
| Karimian[53] 2016 | Iran | West Asian | HB | 118/132 | – | – | – | 51 | 59 | 8 | 51 | 59 | 8 | 77 | 52 | 3 | 0.087 | 4 |
| Irfan[79] 2016 | Pakistan | South Asian | PB | 437/218 | 36 | 18 | 3 | 249 | 118 | 3 | 285 | 136 | 16 | 187 | 30 | 1 | 0.862 | 9 |
| Najafipour[74] 2017 | Iran | West Asian | HB | 280/120 | 25 | 34 | 11 | 88 | 89 | 33 | 113 | 123 | 44 | 66 | 43 | 11 | 0.31 | 4 |
| Ma FF[86] 2017 | China | East Asian | HB | 140/96 | 40 | 30 | 4 | 36 | 22 | 8 | 76 | 52 | 12 | 44 | 44 | 8 | 0.514 | 4 |
| Hu[70] 2017 | China | East Asian | HB | 186/131 | – | – | – | 68 | 80 | 38 | 68 | 80 | 38 | 72 | 41 | 18 | 0.005 | 3 |
| Wang Y[83] 2018 | China | East Asian | HB | 76/95 | – | – | – | 14 | 34 | 11 | 15 | 37 | 24 | 24 | 54 | 17 | 0.163 | 3 |
| Hu LL[84] 2018 | China | East Asian | HB | 145/88 | – | – | – | 23 | 60 | 62 | 23 | 60 | 62 | 11 | 48 | 29 | 0.194 | 4 |
| Zhou SH[85] 2018 | China | East Asian | HB | 145/88 | 3 | 18 | 8 | 4 | 13 | 11 | 15 | 90 | 40 | 11 | 48 | 29 | 0.194 | 4 |
| Cai[63] 2018 | China | East Asian | HB | 90/90 | – | – | – | 13 | 40 | 37 | 13 | 40 | 37 | 26 | 47 | 17 | 0.602 | 3 |
| Zuo YJ[80] 2018 | China | East Asian | HB | 154/294 | – | – | – | 33 | 59 | 62 | 33 | 59 | 62 | 95 | 138 | 61 | 0.406 | 4 |
| Ullah[32] 2019 | Pakistan | South Asian | HB | 232/114 | – | – | – | – | – | – | 169 | 53 | 10 | 99 | 12 | 3 | 0.003 | 3 |
| Xie C[88] 2019 | China | East Asian | HB | 167/78 | – | – | – | 23 | 82 | 62 | 23 | 82 | 62 | 33 | 39 | 6 | 0.229 | 4 |
| Suo F[89] 2019 | China | East Asian | HB | 715/572 | – | – | – | 126 | 326 | 264 | 126 | 326 | 264 | 134 | 272 | 166 | 0.272 | 6 |
| Shao LJ[90] 2019 | China | East Asian | HB | 167/65 | – | – | – | 52 | 71 | 44 | 52 | 71 | 44 | 30 | 28 | 7 | 0.903 | 4 |
| Song N[91] 2019 | China | East Asian | HB | 100/100 | – | – | – | – | – | – | 31 | 46 | 23 | 32 | 52 | 16 | 0.501 | 4 |
| Xu JJ[92] 2019 | China | East Asian | HB | 104/108 | – | – | – | – | – | – | 38 | 41 | 29 | 50 | 44 | 14 | 0.386 | 4 |
| MTHFR A1298C | ||||||||||||||||||
| Park[37] 2005 | Korea | East Asian | HB | 373/396 | – | – | – | – | – | – | 237 | 118 | 18 | 269 | 111 | 16 | 0.294 | 5 |
| Lee[38] 2006 | Korea | East Asian | HB | 360/325 | 109 | 57 | 8 | 113 | 63 | 10 | 222 | 120 | 18 | 213 | 98 | 14 | 0.526 | 5 |
| Dhillon[41] 2007 | India | South Asian | HB | 179/200 | – | – | – | 90 | 80 | 9 | 90 | 80 | 9 | 103 | 84 | 13 | 0.451 | 5 |
| Zhang[73] 2007 | China | East Asian | HB | 165/132 | – | – | – | – | – | – | 90 | 65 | 15 | 85 | 45 | 2 | 0.142 | 4 |
| Ravel[43] 2009 | French | Caucasian | HB | 250/113 | 34 | 28 | 7 | 97 | 66 | 18 | 131 | 94 | 25 | 54 | 46 | 13 | 0.501 | 4 |
| Farcas[65] 2009 | Romania | Caucasian | HB | 66/67 | – | – | – | – | – | – | 35 | 29 | 2 | 39 | 26 | 2 | 0.34 | 4 |
| Singh[44] 2010 | India | South Asian | HB | 151/141 | 66 | 76 | 9 | – | – | – | 66 | 76 | 9 | 64 | 74 | 2 | 0.0002 | 3 |
| Zhang[82] 2010 | China | East Asian | HB | 491/430 | – | – | – | – | – | – | 224 | 220 | 47 | 270 | 150 | 10 | 0.039 | 4 |
| Gava[45] 2011 | Brazil | Mixed | HB | 156/233 | 26 | 14 | 9 | 45 | 48 | 14 | 71 | 62 | 23 | 130 | 89 | 14 | 0.811 | 4 |
| Safarinejad[46] 2011 | Iran | West Asian | HB | 164/328 | – | – | – | 75 | 70 | 19 | 75 | 70 | 19 | 149 | 141 | 38 | 0.599 | 7 |
| Murphy[64] 2011 | Swede | Caucasian | HB | 153/184 | – | – | – | – | – | – | 58 | 77 | 11 | 87 | 62 | 27 | 0.007 | 5 |
| Eloualid[50] 2012 | Morocco | African | HB | 344/690 | 67 | 39 | 4 | 138 | 83 | 13 | 205 | 122 | 17 | 370 | 303 | 17 | < 0.001 | 5 |
| Gupta[30] 2013 | India | South Asian | HB | 611/136 | – | – | – | – | – | – | 165 | 320 | 126 | 27 | 74 | 35 | 0.283 | 7 |
| Stangler[66] 2013 | Slovene | Caucasian | PB | 100/111 | – | – | – | – | – | – | 44 | 35 | 21 | 48 | 50 | 13 | 0.997 | 6 |
| Weiner[29] 2014 | Russia | Caucasian | PB | 275/349 | 37 | 54 | 8 | 42 | 32 | 9 | 126 | 125 | 23 | 142 | 142 | 30 | 0.52 | 7 |
| Mfady[51] 2014 | Jordan | West Asian | HB | 150/150 | – | – | – | – | – | – | 71 | 61 | 18 | 59 | 75 | 16 | 0.273 | 5 |
| Vardarli[62] 2014 | Turkey | West Asian | HB | 100/50 | 21 | 23 | 6 | 24 | 18 | 8 | 45 | 41 | 14 | 19 | 22 | 9 | 0.556 | 4 |
| Balkan[81] 2014 | Turkey | West Asian | NR | 108/125 | 47 | 42 | 19 | – | – | – | 47 | 42 | 19 | 45 | 56 | 24 | 0.383 | 4 |
| Li SS[54] 2014 | China | East Asian | HB | 82/133 | – | – | – | – | – | – | 49 | 29 | 4 | 88 | 36 | 9 | 0.059 | 4 |
| Ni W[56] 2015 | China | East Asian | PB | 296/204 | – | – | – | – | – | – | 181 | 106 | 9 | 137 | 62 | 5 | 0.514 | 7 |
| Gurkan[57] 2015 | Turkey | West Asian | HB | 137/134 | 34 | 34 | 7 | 29 | 25 | 8 | 63 | 59 | 15 | 49 | 66 | 19 | 0.668 | 5 |
| Li XY[58] 2015 | China | East Asian | HB | 162/120 | 66 | 31 | 3 | 35 | 23 | 4 | 101 | 54 | 7 | 80 | 38 | 2 | 0.29 | 5 |
| Kurzawski[59] 2015 | Poland | Caucasian | HB | 284/352 | – | – | – | – | – | – | 128 | 130 | 26 | 156 | 156 | 40 | 0.916 | 5 |
| Kim[60] 2015 | Korea | East Asian | HB | 85/246 | 52 | 28 | 5 | – | – | – | 52 | 28 | 5 | 184 | 56 | 6 | 0.486 | 4 |
| Karimian[53] 2016 | Iran | West Asian | HB | 118/132 | – | – | – | 59 | 44 | 15 | 59 | 44 | 15 | 70 | 48 | 14 | 0.194 | 4 |
| Najafipour[74] 2017 | Iran | West Asian | HB | 280/120 | 27 | 30 | 13 | 102 | 114 | 22 | 129 | 116 | 35 | 57 | 50 | 13 | 0.683 | 4 |
| Ullah[32] 2019 | Pakistan | South Asian | HB | 235/109 | – | – | – | – | – | – | 59 | 133 | 43 | 47 | 59 | 3 | 0.002 | 3 |
| Xu JJ[92] 2019 | China | East Asian | HB | 104/108 | – | – | – | – | – | – | 77 | 14 | 13 | 78 | 15 | 15 | < 0.001 | 4 |
3.2. Quantitative synthesis
3.2.1. MTHFR C677T polymorphism
Table 4 shows the results of the association between the MTHFR C677T polymorphism and male infertility risk. Overall, a significantly increased male infertility risk (CT vs CC: OR = 1.27, 95% CI: 1.15–1.40, Ph < .001, I2 = 54.1%; TT vs CC: OR = 1.74, 95% CI: 1.47–2.07, Ph < .001, I2 = 65.3%; CT + TT vs CC: OR = 1.38, 95% CI: 1.24–1.54, Ph < .001, I2 = 66.6%; TT vs CC + CT: OR = 1.52, 95% CI: 1.33–1.74, Ph < .001, I2 = 56.4%; T vs C: OR = 1.33, 95% CI: 1.22–1.45, Ph < .001, I2 = 73.1%) was observed in all eligible studies.
Table 4.
The results of the association of MTHFR C667T polymorphism with male infertility.
| CT vs. CC | TT vs. CC | (CT + TT) vs. CC | TT vs. (CC + CT) | T vs. C | ||||||||||||
| Variable | n (Cases/Controls) | OR (95% CI) | Ph/I2 (%) | BFDP | OR (95% CI) | Ph/I2 (%) | BFDP | OR (95% CI) | Ph/I2 (%) | BFDP | OR (95% CI) | Ph/I2 (%) | BFDP | OR (95% CI) | Ph/I2 (%) | BFDP |
| Overall | 59 (11767/10591) | 1.27 (1.15–1.40)∗ | <.001/54.1 | 0.106 | 1.74 (1.47–2.07)∗ | <.001/65.3 | <0.001 | 1.38 (1.24–1.54)∗ | <.001/66.6 | 0.001 | 1.52 (1.33–1.74)∗ | <.001/56.4 | <0.001 | 1.33 (1.22–1.45)∗ | <.001/73.1 | <0.001 |
| Ethnicity | ||||||||||||||||
| African | 2 (451/797) | 0.82 (0.64–1.04) | .340/0.0 | – | 0.65 (0.40–1.04) | .843/0.0 | – | 0.78 (0.62–0.99) | .507/0.0 | 0.998 | 0.70 (0.44–1.11) | .661/0.0 | – | 0.80 (0.67–0.97) | .818/0.0 | 0.998 |
| Caucasian | 13 (1836/1689) | 0.99 (0.86–1.15) | .235/20.7 | – | 1.06 (0.71–1.57)∗ | .002/62.1 | – | 1.01 (0.83–1.24)∗ | .032/46.8 | – | 1.04 (0.74–1.47)∗ | .006/56.6 | – | 1.03 (0.86–1.24)∗ | .001/64.4 | – |
| East Asian | 27 (5587/4803) | 1.37 (1.21–1.56)∗ | .038/35.2 | 0.111 | 2.07 (1.70–2.51)∗ | <.001/57.0 | <0.001 | 1.57 (1.37–1.80)∗ | .001/52.1 | <0.001 | 1.70 (1.44–1.96)∗ | .001/51.5 | <0.001 | 1.45 (1.31–1.60)∗ | <.001/63.2 | <0.001 |
| West Asian | 7 (1199/1244) | 1.36 (1.14–1.61) | .471/0.0 | 0.928 | 2.15 (1.60–2.90) | .879/0.0 | 0.031 | 1.47 (1.25–1.74) | .653/0.0 | 0.268 | 1.86 (1.40–2.48) | .823/0.0 | 0.479 | 1.42 (1.26–1.62) | .895/0.0 | 0.012 |
| South Asian | 8 (2,464/1,741) | – | <.001/77.3 | – | 2.70 (1.14–6.40)∗ | .002/71.6 | 0.997 | – | <.001/80.6 | – | 2.42 (1.14–5.13)∗ | .011/63.9 | 0.997 | – | <.001/82.8 | – |
| Source of controls | ||||||||||||||||
| HB | 53 (10435/9444) | 1.25 (1.13–1.38)∗ | <.001/50.2 | 0.408 | 1.77 (1.48–2.12)∗ | <.001/65.3 | <0.001 | 1.37 (1.23–1.53)∗ | <.001/64.5 | 0.002 | 1.54 (1.34–1.77)∗ | <.001/56.5 | <0.001 | 1.33 (1.22–1.45)∗ | <.001/71.4 | <0.001 |
| PB | 6 (1,332/1,147) | – | .001/75.5 | – | 1.50 (0.79–2.86)∗ | .15/64.4 | – | – | <.001/ 81.4 | – | 1.31 (0.76–2.24)∗ | .049/ 55.0 | – | – | <.001/85.0 | – |
| Infertility type | ||||||||||||||||
| Azoospermia | 24 (2,241/4,952) | 1.27 (1.13–1.42) | .101/28.1 | 0.619 | 1.45 (1.09–1.93)∗ | .001/55.7 | 0.995 | 1.30 (1.11–1.53)∗ | .003/50.1 | 0.981 | 1.29 (1.00–1.66)∗ | .002/51.6 | 0.999 | 1.23 (1.07–1.42)∗ | <.001/65.4 | 0.993 |
| OAT∗ | 37 (5670/7062) | 1.25 (1.09–1.44)∗ | <.001/58.0 | 0.986 | 1.75 (1.39–2.19)∗ | <.001/63.4 | 0.049 | 1.37 (1.18–1.59)∗ | <.001/67.9 | 0.619 | 1.59 (1.33–1.89)∗ | <.001/52.1 | 0.009 | 1.35 (1.20–1.52)∗ | <.001/73.7 | 0.047 |
In subgroup analyses by ethnicity and source of controls, a significantly increased male infertility risk was found in Africans (CT + TT vs CC: OR = 0.78, 95% CI: 0.62–0.99, Ph = .507, I2 = 0.0%; T vs C: OR = 0.80, 95% CI: 0.67–0.97, Ph = .818, I2 = 0.0%), East Asians (CT vs CC: OR = 1.37, 95% CI: 1.21–1.56, Ph = .038, I2 = 35.2%, Fig. 2; TT vs CC: OR = 2.07, 95% CI: 1.70–2.51, Ph < .001, I2 = 57.0%; CT + TT vs CC: OR = 1.57, 95% CI: 1.37–1.80, Ph = .001, I2 = 52.1%; TT vs CC + CT: OR = 1.70, 95% CI: 1.44–1.96, Ph = .001, I2 = 51.5%; T vs C: OR = 1.45, 95% CI: 1.31–1.60, Ph < .001, I2 = 63.2%), West Asians (CT vs CC: OR = 1.36, 95% CI: 1.14–1.61, Ph = .471, I2 = 0.0%; TT vs. CC: OR = 2.15, 95% CI: 1.60–2.90, Ph = .879, I2 = 0.0%, Fig. 3; CT + TT vs. CC: OR = 1.47, 95% CI: 1.25–1.74, Ph = .653, I2 = 0.0%; TT vs CC + CT: OR = 1.86, 95% CI: 1.40–2.48, Ph = .823, I2 = 0.0%; T vs C: OR = 1.42, 95% CI: 1.26–1.62, Ph = .895, I2 = 0.0%), South Asians (TT vs CC: OR = 2.70, 95% CI: 1.14–6.40, Ph = .002, I2 = 71.6%; TT vs CC + CT: OR = 2.42, 95% CI: 1.14–5.13, Ph = .011, I2 = 63.9%), and hospital-based studies (CT vs CC: OR = 1.25, 95% CI: 1.13–1.38, Ph < .001, I2 = 50.2%; TT vs CC: OR = 1.77, 95% CI: 1.48–2.12, Ph < .001, I2 = 65.3%; CT + TT vs CC: OR = 1.37, 95% CI: 1.23–1.53, Ph < .001, I2 = 64.5%; TT vs CC + CT: OR = 1.54, 95% CI: 1.34–1.77, Ph < .001, I2 = 56.5%; T vs C: OR = 1.33, 95% CI: 1.22–1.45, Ph < .001, I2 = 71.4%). In subgroup analysis by infertility type, the MTHFR C677T polymorphism was also associated with increased azoospermia (CT vs. CC: OR = 1.27, 95% CI: 1.13–1.42, Ph = .101, I2 = 28.1%; TT vs CC: OR = 1.45, 95% CI: 1.09–1.93, Ph = .001, I2 = 55.7%; (CT + TT) vs. CC: OR = 1.30, 95% CI: 1.11–1.53, Ph = .003, I2 = 50.1%; TT vs (CC + CT): OR = 1.29, 95% CI: 1.00–1.66, Ph = .002, I2 = 51.6%; T vs C: OR = 1.23, 95% CI: 1.07–1.42, Ph < .001, I2 = 65.4%) and OAT risk (CT vs CC: OR = 1.25, 95% CI: 1.09–1.44, Ph < .001, I2 = 58.0%; TT vs. CC: OR = 1.75, 95% CI: 1.39–2.19, Ph < .001, I2 = 63.4%; CT + TT vs CC: OR = 1.37, 95% CI: 1.18–1.59, Ph < .001, I2 = 67.9%; TT vs (CC + CT): OR = 1.59, 95% CI: 1.33–1.89, Ph < .001, I2 = 52.1%; T vs C: OR = 1.35, 95% CI: 1.20–1.52, Ph < .001, I2 = 73.7%).
Figure 2.
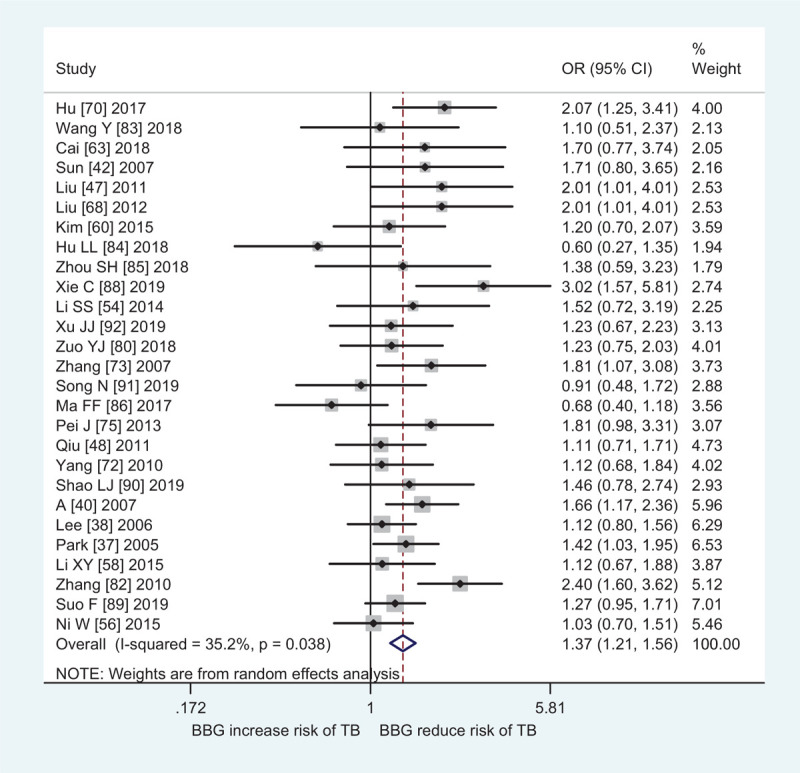
Forest plot of MTHFR C677T polymorphism and male infertile risk in East Asians (CT vs CC).
Figure 3.
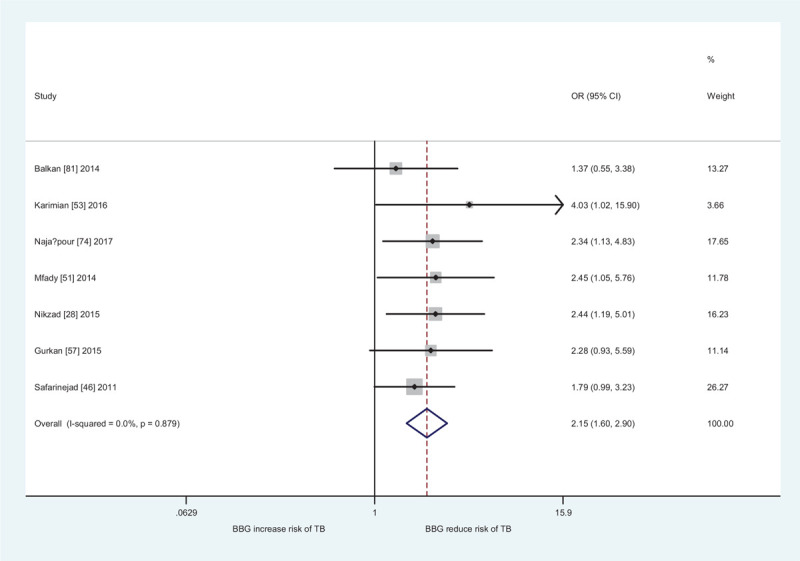
Forest plot of MTHFR C677T polymorphism and male infertile risk in West Asians (TT vs CC).
Obvious heterogeneity was observed in the current meta-analysis, as also shown in Table 2. I2 > 75% was found in South Asians (CT vs. CC: I2 = 77.3%, (CT + TT) vs. CC: I2 = 80.6%, T vs. C: I2 = 82.8%) and population-based studies (CT vs. CC: I2 = 75.5%, (CT + TT) vs CC: I2 = 81.4%, T vs C: I2 = 85.0%). Then, a meta-regression analysis method was applied to explore the sources of heterogeneity and the results indicate that ethnicity (TT vs CC: P = .014; TT vs (CC + CT): P = .008; T vs C: P = .021) and HWE (TT vs CC: P = .041; TT vs (CC + CT): P = .020) were sources of heterogeneity.
The results of sensitivity analysis were shown in Table 3. It is not clear whether the MTHFR C677T polymorphism is associated with increased male infertility risk in South Asians. The results did not pool because I2 > 75% was observed in any genetic model. Another results did not change, such as overall population, Africans, East Asians, West Asians, and so on.
No significant publication bias was found by Begg funnel plot shape (supplemental Figs. 1, –5) and Egger test (CT vs. CC: P = .418,TT vs CC: P = .203, CT + TT vs CC: P = .274, CT + TT vs CC: P = .179, T vs C: P = .402) in the overall analysis.
An BFDP test was used to further investigate significant associations in this study, as shown in Tables 4 and 5. Significantly increased male infertility risk was considered as “noteworthy” in the overall population (CT vs CC: BFDP = 0.106, TT vs CC: BFDP < 0.001, CT + TT vs. CC: BFDP = 0.001, TT vs. CT + CC: BFDP < 0.001, T vs. C: BFDP < 0.001), East Asians (CT vs. CC: BFDP = 0.111, TT vs. CC: BFDP < 0.001, CT + TT vs. CC: BFDP < 0.001, TT vs CT + CC: BFDP < 0.001, T vs C: BFDP < 0.001), West Asians (TT vs. CC: BFDP = 0.031, CT + TT vs CC: BFDP = 0.268, TT vs. CT + CC: BFDP = 0.479, T vs. C: BFDP = 0.012), hospital-based studies (CT vs CC: BFDP = 0.408, TT vs. CC: BFDP < 0.001, CT + TT vs. CC: BFDP = 0.002, TT vs. CT + CC: BFDP < 0.001, T vs C: BFDP < 0.001), azoospermia (CT vs. CC: BFDP = 0.619), and OAT (TT vs. CC: BFDP = 0.049, CT + TT vs CC: BFDP = 0.619, TT vs CT + CC: BFDP = 0.009, T vs. C: BFDP = 0.047) for MTHFR C677T polymorphism.
Table 5.
The results of sensitivity analysis between MTHFR C667T polymorphism with male infertility.
| CT vs. CC | TT vs. CC | (CT + TT) vs. CC | TT vs. (CC + CT) | T vs. C | ||||||||||||
| Variable | n (Cases/Controls) | OR (95% CI) | Ph/I2 | BFDP | OR (95% CI) | Ph/I2 | BFDP | OR (95% CI) | Ph/I2 | OR (95% CI) | Ph/I2 | OR (95% CI) | Ph/I2 | BFDP | ||
| Quality score ≥ 5 and HWE | ||||||||||||||||
| Overall | 23 (6827/6355) | 1.22 (1.06–1.41)∗ | <.001/65.1 | 0.995 | 1.60 (1.29–1.97)∗ | <.001/59.6 | 0.294 | 1.31 (1.13–1.52)∗ | <.001/72.1 | 0.937 | 1.41 (1.21–1.64)∗ | .035/37.9 | 0.300 | 1.27 (1.14–1.41)∗ | <.001/72.4 | 0.336 |
| African | 1 (344/690) | 0.77 (0.59–1.01) | – | – | 0.67 (0.39–1.15) | – | – | 0.75 (0.58–0.98) | – | 0.998 | 0.74 (0.44–1.26) | – | – | 0.80 (0.64–0.98) | – | 0.998 |
| Caucasian | 6 (1111/1202) | 1.03 (0.87–1.23) | .405/1.7 | – | 1.24 (0.95–1.64) | .188/33.0 | – | 1.07 (0.91–1.26) | .426/0.0 | – | 1.23 (0.95–1.59) | .167/36.0 | – | 1.09 (0.96–1.24) | .271/21.7 | – |
| East Asian | 7 (2753/2299) | 1.37 (1.13–1.68)∗ | .046/53.1 | 0.985 | 1.77 (1.39–2.24)∗ | .092/44.8 | 0.089 | 1.47 (1.19–1.81)∗ | .016/61.7 | 0.903 | 1.43 (1.25–1.64) | .864/ 0.0 | 0.020 | 1.33 (1.22–1.44) | .148/ 36.7 | <0.001 |
| West Asian | 4 (693/867) | 1.22 (0.99–1.52) | .397/0.0 | – | 2.16 (1.50–3.11) | .898/0.0 | 0.584 | 1.36 (1.11–1.66) | .584/0.0 | 0.985 | 1.93 (1.37–2.73) | .743/0.0 | 0.861 | 1.36 (1.17–1.59) | .876/0.0 | 0.829 |
| South Asian | 5 (1926/1297) | – | <.001/83.3 | – | – | <.001/78.5 | – | – | <.001/86.4 | – | 2.35 (0.97–5.68)∗ | .007/ 71.7 | – | – | <.001/88.5 | – |
| HB | 18 (5572/5321) | 1.15 (1.00–1.33)∗ | .001/59.8 | 0.999 | 1.59 (1.27–2.00)∗ | <.001/60.4 | 0.726 | 1.23 (1.06–1.44)∗ | <.001/69.1 | 0.996 | 1.43 (1.22–1.66)∗ | .083/ 33.5 | 0.126 | 1.21 (1.09–1.35)∗ | <.001/67.4 | 0.969 |
| PB | 5 (1255/1034) | 1.57 (1.07–2.30)∗ | .005/73.4 | 0.996 | 1.86 (0.92–3.76)∗ | .024/64.3 | – | – | .001/79.6 | – | 1.53 (0.83–2.81)∗ | .049/58.2 | – | – | <.001/84.6 | – |
| Azoospermia | 11 (1352/3370) | 1.31 (1.06–1.64)∗ | .021/52.3 | 0.997 | 1.79 (1.30–2.48)∗ | .074/41.3 | 0.927 | 1.40 (1.12–1.77)∗ | .004/ 61.3 | 0.991 | 1.48 (1.20–1.82) | .144/31.9 | 0.873 | 1.34 (1.12–1.61)∗ | .001/65.2 | 0.981 |
| OAT∗ | 14 (3191/4470) | 1.19 (0.98–1.44)∗ | <.001/66.0 | – | 1.46 (1.14–1.87)∗ | .045/ 42.7 | 0.984 | 1.25 (1.03–1.53)∗ | <.001/69.9 | 0.998 | 1.38 (1.19–1.60) | .226/ 21.0 | 0.494 | 1.24 (1.07–1.44)∗ | <.001/70.1 | 0.993 |
However, the positive results by sensitivity analysis (Table 5) were only considered as “noteworthy” in the overall population (TT vs. CC: BFDP = 0.294, CT + TT vs. CC: BFDP = 0.300, T vs. C: BFDP = 0.336), East Asians (TT vs. CC: BFDP = 0.089, TT vs. CT + CC: BFDP = 0.020, T vs. C: BFDP < 0.001), West Asians (TT vs. CC: BFDP = 0.584), hospital-based studies (TT vs. CC: BFDP = 0.726, TT vs. CT + CC: BFDP = 0.126), and OAT (TT vs. CT + CC: BFDP = 0.494) for MTHFR C677T polymorphism.
3.2.2. MTHFR A1298C polymorphism
Table 6 shows the results of meta-analysis on the association between the MTHFR A1298C polymorphism and male infertility risk. No significantly increased male infertility risk was found in all eligible studies. In subgroup analyses by ethnicity and source of controls, a significantly increased male infertility risk was found in East Asians (AC vs. AA: OR = 1.37, 95% CI: 1.20–1.56, Ph = 0.515, I2 = 0.0%; CC vs. AA: OR = 1.88, 95% CI: 1.10–3.20, Ph = 0.006, I2 = 62.7%; (AC + CC) vs. AA: OR = 1.42, 95% CI: 1.25–1.62, Ph = 0.106, I2 = 39.3%; CC vs. (AA + AC): OR = 1.69, 95% CI: 1.04–2.75, Ph = 0.020, I2 = 55.8%; C vs. A: OR = 1.35, 95% CI: 1.13–1.60, Ph = 0.016, I2 = 57.3%) and population-based studies (C vs. A: OR = 1.53, 95% CI: 1.28–1.83, Ph = 0.767, I2 = 0.0%). Moreover, no significant association was observed in subgroup analysis by infertility type.
Table 6.
Meta-analysis of the association of MTHFR A1298C polymorphism with male infertility.
| AC vs AA | CC vs AA | (AC + CC) vs. AA | CC vs. (AA + AC) | C vs. A | ||||||||||||
| Variable | n (Cases/Controls) | OR (95% CI) | Ph/I2 | BFDP | OR (95% CI) | Ph/I2 | BFDP | OR (95% CI) | Ph/I2 | BFDP | OR (95% CI) | Ph/I2 | BFDP | OR (95% CI) | Ph/I2 | BFDP |
| Overall | 28 (5,976/5,774 | 1.08 (0.96–1.22)∗ | .002/48.6 | – | 1.28 (0.99–1.67)∗ | <.001/63.5 | – | 1.11 (0.98–1.26)∗ | <.001/59.3 | – | 1.25 (0.99–1.58)∗ | <.001/58.4 | – | 1.11 (0.99–1.24)∗ | <.001/66.5 | – |
| Ethnicity | ||||||||||||||||
| South Asian | 4 (1,176/585) | 1.08 (0.75–1.55)∗ | .065/58.5 | – | – | <.001/87.2 | – | – | .004/77.3 | – | – | <.001/83.6 | – | – | <.001/87.9 | – |
| West Asian | 7 (1057/1039) | 0.86 (0.71–1.04) | .702/0.0 | – | 0.91 (0.69–1.21) | .816/ 0.0 | – | 0.87 (0.73–1.04) | .601/ 0.0 | – | 0.99 (0.76–1.29) | .940/ 0.0 | – | 0.93 (0.81–1.06) | .646/ 0.0 | – |
| East Asian | 9 (2123/2094) | 1.37 (1.20–1.56) | .515/0.0 | 0.111 | 1.88 (1.10–3.20)∗ | .006/ 62.7 | 0.996 | 1.42 (1.25–1.62) | .106/ 39.3 | 0.012 | 1.69 (1.04–2.75)∗ | .020/ 55.8 | 0.997 | 1.35 (1.13–1.60)∗ | .016/ 57.3 | 0.949 |
| Caucasian | 6 (1120/1133) | 1.06 (0.89–1.26) | .158/37.3 | – | 0.88 (0.65–1.17) | .551/0.0 | – | 1.02 (0.87–1.21) | .541/0.0 | – | 0.86 (0.65–1.13) | .159/37.2 | – | 0.98 (0.86–1.11) | .822/0.0 | – |
| Source of controls | ||||||||||||||||
| HB | 25 (5,306/5145) | 1.09 (0.96–1.24)∗ | .001/52.1 | – | 1.30 (0.97–1.74)∗ | <.001/66.7 | – | 1.11 (0.97–1.28)∗ | <.001/62.9 | – | 1.26 (0.97–1.63)∗ | <.001/61.2 | – | 1.11 (0.99–1.25)∗ | <.001/69.4 | – |
| PB | 3 (670/629) | 1.05 (0.83–1.33) | .309/14.7 | – | 1.15 (0.75–1.77) | .356/3.1 | – | 1.08 (0.86–1.35) | .471/ 0.0 | – | 1.19 (0.79–1.80) | .219/ 34.1 | – | 1.53 (1.28–1.83) | .767/ 0.0 | 0.139 |
| Infertility type | ||||||||||||||||
| Azoospermia | 12 (1,140/2,610) | 1.01 (0.86–1.18) | .316/13.1 | – | 1.21 (0.91–1.61) | .117/ 34.2 | – | 1.04 (0.90–1.21) | .212/ 23.5 | – | 1.21 (0.92–1.58) | .131/ 32.4 | – | 1.06 (0.95–1.19) | .109/35.1 | – |
| OAT∗ | 12 (1,664/2,759) | 0.98 (0.86–1.12) | .198/25.0 | – | 1.16 (0.91–1.47) | .248/19.9 | – | 1.01 (0.89–1.15) | .141/31.3 | – | 1.17 (0.93–1.47) | .375/7.2 | – | 1.04 (0.94–1.15) | .173/27.2 | – |
Obvious heterogeneity was observed in the current meta-analysis, as also shown in Table 6.
The results indicate that quality score of the eligible studies (AC vs. AA: P = .038, CC vs. AA: P = .013, (AC + CC) vs. AA: P = .009, CC vs. (AA +AC): P = .024, C vs. A: P = .003) was source of heterogeneity by a meta-regression analysis method.
The results of sensitivity analysis was shown in Table 7 indicating that the results are stable except in West Asians. Significant increased male infertility risk was observed in West Asians (AC vs AA: OR = 0.79, 95% CI: 0.62–1.00, Ph = .586, I2 = 0.0%).
Table 7.
The results of sensitivity analysis between MTHFR A1298C polymorphism with male infertility.
| AC vs AA | CC vs AA | (AC + CC) vs AA | CC vs (AA + AC) | C vs A | ||||||||||||
| Variable | n (Cases/Controls) | OR (95% CI) | Ph/I2 (%) | BFDP | OR (95% CI) | Ph/I2 (%) | BFDP | OR (95% CI) | Ph/I2 (%) | BFDP | OR (95% CI) | Ph/I2 (%) | BFDP | OR (95% CI) | Ph/I2 (%) | BFDP |
| Quality score ≥ 5 and HWE | ||||||||||||||||
| Overall | 13 (3198/2895) | 1.00 (0.89–1.11) | .371/7.5 | – | 0.92 (0.76–1.12) | .562/0.0 | – | 1.00 (0.89–1.11) | .296/14.7 | – | 0.96 (0.80–1.16) | .689/0.0 | – | 0.99 (0.91–1.08) | .292/15.1 | – |
| South Asian | 2 (790/336) | 0.90 (0.66–1.23) | .182/43.8 | – | 0.64 (0.40–1.02) | .581/0.0 | – | 0.85 (0.55–1.32)∗ | .148/52.2 | – | 0.75 (0.51–1.11) | .975/ 0.0 | – | 0.86 (0.70–1.06) | .257/ 22.1 | – |
| West Asian | 4 (559/737) | 0.79 (0.62–1.00) | .586/0.0 | 0.999 | 0.83 (0.58–1.18) | .786/0.0 | – | 0.80 (0.64–1.00) | .579/ 0.0 | 0.999 | 0.94 (0.68–1.31) | .862/ 0.0 | – | 0.87 (0.74–1.03) | .681/ 0.0 | – |
| East Asian | 4 (1191/1045) | 1.20 (1.00–1.45) | .973/0.0 | 0.999 | 1.36 (0.88–2.11) | .834/0.0 | – | 1.22 (1.03–1.46) | .985/ 0.0 | 0.998 | 1.28 (0.83–1.98) | .822/ 0.0 | – | 1.19 (1.03–1.38) | .981/ 0.0 | 0.998 |
| Caucasian | 3 (658/777) | 0.97 (0.78–1.21) | .701/0.0 | – | 0.96 (0.67–1.37) | .248/28.3 | – | 0.97 (0.79–1.20) | 1.000/ 0.0 | – | 1.04 (0.62–1.75)∗ | .114/ 53.9 | – | 0.98 (0.84–1.15) | .546/ 0.0 | – |
| HB | 10 (2528/2266) | 0.98 (0.86–1.11) | .321/13.2 | – | 0.87 (0.70–1.08) | .608/0.0 | – | 0.97 (0.86–1.10) | .214/ 24.9 | – | 0.92 (0.75–1.12) | .844/0.0 | – | 0.97 (0.88–1.06) | .273/ 18.5 | – |
| Azoospermia | 5 (556/1018) | 1.03 (0.82–1.29) | .292/19.2 | – | 0.86 (0.57–1.30) | .700/0.0 | – | 1.01 (0.82–1.26) | .267/ 23.1 | – | 0.88 (0.60–1.30) | .848/0.0 | – | 0.98 (0.83–1.16) | .403/0.5 | – |
| OAT∗ | 6 (736/1421) | 1.01 (0.84–1.23) | .427/0.0 | – | 1.03 (0.73–1.46) | .531/0.0 | – | 1.03 (0.85–1.23) | .349/10.4 | – | 1.06 (0.76–1.49) | .659/0.0 | – | 1.03 (0.89–1.19) | .370/7.3 | – |
Significant publication was observed by the Begg funnel plot shape (Figures not shown) and Egger test (CC vs. AA: P = 0.032; CC vs. (AA + AC): P = .024) in the overall analysis. Supplemental Figs.6 –7, list the Begg's funnel plots by the trim and fill method. Notably, log OR and 95% CI did not change.
An BFDP test was also applied to further investigate significant associations between MTHFR A1298C and male infertility risk, as shown in Tables 4 and 5. Significantly increased male infertility risk was considered as “noteworthy” in the East Asians (AC vs AA: BFDP = 0.111, AC + CC vs AA: BFDP = 0.012) and population-based studies (C vs A: BFDP = 0.139). However, we did not find that the positive results of sensitivity analysis were considered as “noteworthy” in the overall and all subgroup analyses.
4. Discussion
In 2001, Bezold et al.[33] first investigated the association between the MTHFR C667T polymorphism and male infertility risk. In 2005, Park et al.[37] first explored the MTHFR A1298C polymorphism with male infertility risk. Since then a lot of case–control studies have investigated the associations but the results are still inconsistent. Here, an updated and high quality meta-analysis was carried out to explore the above two gene polymorphism with male infertility risk.
Overall, the MTHFR C677T polymorphism was associated with increased male infertility risk in overall populations, Africans, East Asians, West Asians, South Asians, hospital-based studies, azoospermia and OAT. In addition, a significantly increased male infertility risk was also found in East Asians and population-based studies for the MTHFR A1298C polymorphism. The pooled data was analyzed using five different genetic models and several subgroup analyses in this study. Under the circumstances, the P-value must be adjusted to explain the multiple comparisons.[93] In addition, random error and bias were common in the studies with small sample sizes so that the results were unreliable, especially in molecular epidemiological studies. Wakefield et al.[101] in 2007 proposed a more precise Bayesian measure of false discovery in genetic epidemiology studies, for determining the “noteworthiness” of the positive association. Hence, we used BFDP test to assess the false discovery in the current meta-analysis. Finally, the positive results by sensitivity analysis were only considered as “noteworthy” in the overall population and OAT for MTHFR C677T polymorphism. We did not find that the positive results of sensitivity analysis were considered as “noteworthy” in the overall and all subgroup analyses for MTHFR A1298C.
Based on biochemical properties described for MTHFR C677T and A1298C polymorphisms, we expected that the two genes were associated with risk of male infertility risk risk in all races. However, we only observed that MTHFR C677T is associated with increased male infertility risk in East Asians and West Asians, but not other races (such as Caucasians and Africans). Moreover, no significant association was observed on MTHFR A1298C polymorphism with male infertility risk in any race. Hence, an ethnic variant in the frequency of MTHFR C677T polymorphism was demonstrated in different populations. The frequency of the 677T allele ranges from 30.5 to 42% among Asian population, from 32.2 to 44% in Caucasians. African population shows a lower frequency of T allele, ranging from 6 to 10.3%.[102,103] These results indicated that the same genes may play different roles in different races and countries, because infertility is a complicated multigenetic disease, and different genetic backgrounds and environmental factor (smoking or life style) may contribute to the discrepancy. Another possible explanation for the difference suggested the influence of the genetic variant might be masked by the presence of other as-yet unidentified causal genes involved in male infertility. The current studies demonstrated a clear north-to-south gradient in the effect of the MTHFR C677T variant in the determination of hyperhomocysteinemia, suggesting that diet is a relevant environmental agent, being the presence of folates in the food higher in the South of Europe than in the North. In addition, there was also the presence of folates in the food higher in the Caucasians than in the Asians. Obvious heterogeneity was observed in the current meta-analysis, as also shown in Tables 2 and 4. Ethnicity and HWE were sources of heterogeneity for MTHFR C677T polymorphism and quality score of the eligible studies was source of heterogeneity by a meta-regression analysis method. HWD may be genotyping errors and selection bias in molecular epidemiological studies. Small sample studies were easier to accept if there were positive reports as they tend to yield false-positive results because they may be not rigorous and are often of low-quality. Supplemental Fig. 3, indicated that the asymmetry of the funnel plot was caused by studies of low-quality small samples. Therefore, we performed a sensitivity analysis restricted to studies that only included high-quality articles and controls in HWE.
15 previous meta-analyses[15,17,19–29,31,32] have been reported on the MTHFR C677T polymorphism with male infertility risk (as shown in Table 6). Yang et al.[15] and Wei et al.[24] showed that the MTHFR C677T polymorphism was associated with a significantly increased male infertility risk in the overall and Asian populations. Zhu et al.[17] suggested the MTHFR C677T polymorphism is capable of causing male infertility susceptibility, especially in Asians, azoospermia and OAT. Hong et al[19] demonstrated that the MTHFR C677T polymorphism is associated with male infertility in East-asian populations, Middle-eastern populations, and mixed-race. Tüttelmann et al[20] and Nikzad et al[28] indicated that the MTHFR C677T polymorphism is associated with male infertility in overall populations. Wu et al.[21] supported that the MTHFR C677T polymorphism was capable of causing male infertility susceptibility in Asians and azoospermia. Gong et al[23] and Liu et al.[26]indicated that the MTHFR polymorphism was associated with an increased risk of male infertility in overall populations, especially in Asians and Caucasians and subgroups of azoospermia and OAT. Weiner et al.[29] suggested that the MTHFR C677T polymorphism was associated with an increased risk of male infertility in overall populations and subgroup of azoospermia. Gupta et al[31] supported that the MTHFR C677T polymorphism was associated with an increased risk of male infertility in overall populations and subgroups of azoospermia and OAT. Ullah et al[32] indicated that the MTHFR C677T polymorphism was associated with an increased risk of male infertility in Caucasians for middle income countries. Rai et al[22] and Shi et al[27] supported an association between C677T polymorphism and male infertility in Asians. Ren et al[25] suggested that the MTHFR C667T polymorphism may contribute to the genetic susceptibility to male infertility in the Chinese population. In addition, ten previous meta-analyses[15,16,18,20,24–27,30,32] have also been published on the MTHFR A1298C polymorphism with male infertility risk (as shown in Table 7). Among these publications, one study[32] investigated this issue in Caucasians, one study[27] in Asians, one study[25] in Chinese population, and seven studies[15,16,18,20,24,26,30] in overall populations. Ullah et al[32] indicated that the MTHFR A1298C polymorphism was associated with an increased risk of male infertility in Caucasians for low income countries. Shi et al[27] supported that MTHFR A1298C polymorphism was the risk factor with susceptibility to male infertility in Asians, especially in East Asians. Ren et al[25] demonstrated that MTHFR A1298C polymorphism may be unrelated to male infertility risk in Chinese population. Yang et al[15] suggested that there was a significant association between the A1298C polymorphism and male infertility risk in the Asian, Caucasian, and overall groups. Zhang et al[16] indicated that the MTHFR A1298C polymorphism may be a potential risk factor for male infertility, especially in the Asian population. Shen et al[18] and supported that the MTHFR A1298C polymorphism was capable of causing male infertility susceptibility, especially azoospermia. Tüttelmann et al,[20] Wei et al,[24] Gupta et al,[30]and Liu et al[26] indicated that the MTHFR A1298C polymorphism was not associated with male infertility susceptibility. However, quality assessment of the eligible studies was not performed in 13 previous meta-analyses.[15,17,18,20–24,28–32] In addition, the false-positive report probabilities of statistically significant association and statistical power was not evaluated in all previous meta-analyses except the study of Liu et al.[26] Moreover, many new studies have been published, therefore, an updated meta-analysis should be carried out.
This study has several advantages over previous meta-analyses.[15–32] First, the sample size was much larger, 59 studies on MTHFR C677T (11,767 male infertility cases and 10,591 controls) and 28 studies on MTHFR A1298C (5,976 male infertility cases and 5,774 controls) were identified in overall population. Second, this is the first meta-analysis to explore a false-positive report probability by BFDP method. Third, an important sensitivity analysis was performed on studies that were high-quality and HWE. Although we have put considerable effort and resources into testing possible associations between MTHFR C677T and A1298C polymorphisms and male infertility risk, there are still some limitations inherited from the published studies. First, the controls were not uniformly defined. Second, no data were extracted on exploring interaction between gene and environment.
In summary, this study indicates that the MTHFR C677T polymorphism is associated with increased male infertility risk in East Asians, West Asians, and OAT. Other significant association should be interpreted with caution and may most likely result from false-positive results, rather than from true associations or biological factors.
Author contributions
Conceptualization: Xiao-Feng He and Xiang-Hua Ye.
Data curation: Li-Juan Han and Xiao-Feng He.
Formal analysis: Xiao-Feng He.
Investigation: Li-Juan Han and Xiang-Hua Ye.
Methodology: Li-Juan Han and Xiao-Feng He.
Resources: Xiao-Feng He and Xiang-Hua Ye.
Software: Xiao-Feng He
Supervision: Xiao-Feng He and Xiang-Hua Ye.
Validation: Xiao-Feng He and Xiang-Hua Ye.
Visualization: Xiao-Feng He
Writing – original draft: Li-Juan Han
Writing – review & editing: Xiao-Feng He and Xiang-Hua Ye.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: BFDP = Bayesian false discovery probability, CIs = confidence intervals, HWD = Hardy-Weinberg dis-equilibrium, HWE = Hardy-Weinberg equilibrium, MTHFR = methylenetetrahydrofolate reductase, OAT = oligoasthenoteratozoospermia, ORs = odds ratios.
How to cite this article: Han LJ, He XF, Ye XH. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and male infertility risk: an updated meta-analysis. Medicine. 2020;99:51(e23662).
This is a meta-analysis, hence, ethical approval was waived or not necessary
This study was designed by Xiao-Feng He and Xiang-Hua Ye. Li-Juan Han and Xiao-Feng He did the literature search, study quality assessment, and data extraction. Xiao-Feng He performed the statistical analysis and drafted the tables and figures. Li-Juan Han wrote the first draft of this analysis, and Xiao-FH and XHY helped to finish the final version. All authors approved the conclusions of our study.
The authors have no funding and conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
Azoospermia.
Including oligoasthenoteratozoospermia (OAT), severe OAT, oligozoospermia, and teratozoospermia. NA = not available.
Azoospermia, 2 Including oligoasthenoteratozoospermia (OAT), severe OAT, oligozoospermia, and teratozoospermia.
1Including Oligoasthenoteratozoospermia (OAT), severe OAT, oligozoospermia, and teratozoospermia. HB = hospital-based studies, PB = population-based studies.
Including Oligoasthenoteratozoospermia (OAT), severe OAT, oligozoospermia, and teratozoospermia.
Including Oligoasthenoteratozoospermia (OAT), severe OAT, oligozoospermia, and teratozoospermia.
Including Oligoasthenoteratozoospermia (OAT), severe OAT, oligozoospermia, and teratozoospermia.
Including Oligoasthenoteratozoospermia (OAT), severe OAT, oligozoospermia, and teratozoospermia.
References
- [1].Oliva A, Spira A, Multigner L. Contribution of environmental factors to the risk of male infertility. Hum Reprod 2001;16:1768–76. [DOI] [PubMed] [Google Scholar]
- [2].Gava MM, Chagas Ede O, Bianco B, et al. Methylenetetrahydrofolate reductase polymorphisms are related to male infertility in Brazilian men. Genet Test Mol Biomarkers 2011;15:153–7. [DOI] [PubMed] [Google Scholar]
- [3].Lee HD, Lee HS, Park SH, et al. Causes and classification of male infertility in Korea. Clin Exp Reprod Med 2012;39:172–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hirsh A. Male subfertility. BMJ 2003;327:669–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brugh VM, Lipshultz LI. Male factor infertility: Evaluation and management. Med Clin North Am 2004;88:367–85. [DOI] [PubMed] [Google Scholar]
- [6].Kupis L, Dobronski PA, Radziszewski P. Varicocele as a source of male infertility-current treatment techniques. Cent European J Urol 2015;68:365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Miyamoto T, Tsujimura A, Miyagawa Y, et al. Male infertility and its causes in human. Adv Urol 2012;2012:384520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Anawalt BD. Approach to male infertility and induction of spermatogenesis. J Clin Endocrinol Metab 2013;98:3532–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fowler B. Homocysteine: overview of biochemistry, molecular biology, and role in disease processes. Semin Vasc Med 2005;5:77–86. [DOI] [PubMed] [Google Scholar]
- [10].Jacques PF, Bostom AG, Williams RR, et al. Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation 1996;93:7–9. [DOI] [PubMed] [Google Scholar]
- [11].Friso S, Choi SW. Gene-nutrient interactions in one-carbon metabolism. Curr Drug Metab 2005;6:37–46. [DOI] [PubMed] [Google Scholar]
- [12].van der Put NM, Gabreëls F, Stevens EM, et al. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet 1998;62:1044–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Weisberg I, Tran P, Christensen B, et al. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab 1998;64:169–72. [DOI] [PubMed] [Google Scholar]
- [14].Castro R, Rivera I, Ravasco P, et al. 5,10-Methylenetetrahydrofolate reductase 677C>T and 1298A>C mutations are genetic determinants of elevated homocysteine. QJM 2003;96:297–303. [DOI] [PubMed] [Google Scholar]
- [15].Yang Y, Luo YY, Wu S, et al. Association between C677T and A1298C polymorphisms of the MTHFR gene and risk of male infertility: a meta-analysis. Genet Mol Res 2016;15. [DOI] [PubMed] [Google Scholar]
- [16].Zhang Q, Yin GY, Liu J, et al. Association between MTHFR A1298C polymorphism and male infertility: a meta-analysis. J Huazhong Univ Sci Technolog Med Sci 2017;37:153–60. [DOI] [PubMed] [Google Scholar]
- [17].Zhu X, Liu Z, Zhang M, et al. Association of the methylenetetrahydrofolate reductase gene C677T polymorphism with the risk of male infertility: a meta-analysis. Ren Fail 2016;38:185–93. [DOI] [PubMed] [Google Scholar]
- [18].Shen O, Liu R, Wu W, et al. Association of the methylenetetrahydrofolate reductase gene A1298C polymorphism with male infertility: a meta-analysis. Ann Hum Genet 2012;76:25–32. [DOI] [PubMed] [Google Scholar]
- [19].Hong HH, Hu Y, Yu XQ, et al. Associations of C677T polymorphism in methylenetetrahydrofolate reductase (MTHFR) gene with male infertility risk: A meta-analysis. Eur J Obstet Gynecol Reprod Biol 2017;212:101–9. [DOI] [PubMed] [Google Scholar]
- [20].Tüttelmann F, Rajpert-De Meyts E, Nieschlag E, et al. Gene polymorphisms and male infertility-a meta-analysis and literature review. Reprod Biomed Online 2007;15:643–58. [DOI] [PubMed] [Google Scholar]
- [21].Wu W, Shen O, Qin Y, et al. Methylenetetrahydrofolate reductase C677T polymorphism and the risk of male infertility: a meta-analysis. Int J Androl 2012;35:18–24. [DOI] [PubMed] [Google Scholar]
- [22].Rai V, Kumar P. Methylenetetrahydrofolate reductase C677T polymorphism and risk for male infertility in Asian population. Indian J Clin Biochem 2017;32:253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gong M, Dong W, He T, et al. MTHFR 677C>T polymorphism increases the male infertility risk: a meta-analysis involving 26 studies. PLoS One 2015;10:e0121147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wei B, Xu Z, Ruan J, et al. MTHFR 677C>T and 1298A>C polymorphisms and male infertility risk: a meta-analysis. Mol Biol Rep 2012;39:1997–2002. [DOI] [PubMed] [Google Scholar]
- [25].Ren Z, Ren P, Yang B, et al. MTHFR C677T, A1298C and MS A2756G gene polymorphisms and male infertility risk in a chinese population: a meta-analysis. PLoS One 2017;12:e0169789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu K, Zhao R, Shen M, et al. Role of genetic mutations in folate-related enzyme genes on Male Infertility. Sci Rep 2015;5:15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shi TL, Wu Y, Li Y, et al. The relevance of MTHFR C677T, A1298C, and MTRR A66G polymorphisms with response to male infertility in Asians: a meta-analysis. Medicine (Baltimore) 2019;98:e14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nikzad H, Karimian M, Sareban K, et al. MTHFR-Ala222Val and male infertility: a study in Iranian men, an updated meta-analysis and an in silico-analysis. Reprod Biomed Online 2015;31:668–80. [DOI] [PubMed] [Google Scholar]
- [29].Weiner AS, Boyarskikh UA, Voronina EN, et al. Polymorphisms in folate-metabolizing genes and risk of idiopathic male infertility: a study on a Russian population and a meta-analysis. Fertil Steril 2014;101:87–94.e3. [DOI] [PubMed] [Google Scholar]
- [30].Gupta N, Sarkar S, David A, et al. Significant impact of the MTHFR polymorphisms and haplotypes on male infertility risk. PLoS One 2013;8:e69180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gupta N, Gupta S, Dama M, et al. Strong association of 677 C>T substitution in the MTHFR gene with male infertility--a study on an indian population and a meta-analysis. PLoS One 2011;6:e22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ullah N, Mansoor A, Micheal S, et al. MTHFR polymorphisms as risk for male infertility in Pakistan and its comparison with socioeconomic status in the world. Per Med 2019;16:35–49. [DOI] [PubMed] [Google Scholar]
- [33].Bezold G, Lange M, Peter RU. Homozygous methylenetetrahydrofolate reductase C677T mutation and male infertility. N Engl J Med 2001;344:1172–3. [DOI] [PubMed] [Google Scholar]
- [34].Stuppia L, Gatta V, Scarciolla O, et al. The methylenetethrahydrofolate reductase (MTHFR) C677T polymorphism and male infertility in Italy. J Endocrinol Invest 2003;26:620–2. [DOI] [PubMed] [Google Scholar]
- [35].Ebisch IM, van Heerde WL, Thomas CM, et al. C677T methylenetetrahydrofolate reductase polymorphism interferes with the effects of folic acid and zinc sulfate on sperm concentration. Fertil Steril 2003;80:1190–4. [DOI] [PubMed] [Google Scholar]
- [36].Singh K, Singh SK, Sah R, et al. Mutation C677T in the methylenetetrahydrofolate reductase gene is associated with male infertility in an Indian population. Int J Androl 2005;28:115–9. [DOI] [PubMed] [Google Scholar]
- [37].Park JH, Lee HC, Jeong YM, et al. MTHFR C677T polymorphism associates with unexplained infertile male factors. J Assist Reprod Genet 2005;22:361–8. [DOI] [PubMed] [Google Scholar]
- [38].Lee HC, Jeong YM, Lee SH, et al. Association study of four polymorphisms in three folate-related enzyme genes with non-obstructive male infertility. Hum Reprod 2006;21:3162–70. [DOI] [PubMed] [Google Scholar]
- [39].Paracchini V, Garte S, Taioli E. MTHFR C677T polymorphism, GSTM1 deletion and male infertility: a possible suggestion of a gene-gene interaction? Biomarkers 2006;11:53–60. [DOI] [PubMed] [Google Scholar]
- [40].ZC A, Yang Y, Zhang SZ, et al. Single nucleotide polymorphism C677T in the methylenetetrahydrofolate reductase gene might be a genetic risk factor for infertility for Chinese men with azoospermia or severe oligozoospermia. Asian J Androl 2007;9:57–62. [DOI] [PubMed] [Google Scholar]
- [41].Dhillon VS, Shahid M, Husain SA. Associations of MTHFR DNMT3b 4977 bp deletion in mtDNA and GSTM1 deletion, and aberrant CpG island hypermethylation of GSTM1 in non-obstructive infertility in Indian men. Mol Hum Reprod 2007;13:213–22. [DOI] [PubMed] [Google Scholar]
- [42].Sun HT, Zhang JY, Lu YJ. Association of the methylenetetrahydrofolate reductase gene C677T polymorphism with male infertility. Reprod Contracept 2007;27:443–6. [Google Scholar]
- [43].Ravel C, Chantot-Bastaraud S, Chalmey C, et al. Lack of association between genetic polymorphisms in enzymes associated with folate metabolism and unexplained reduced sperm counts. PLoS One 2009;4:e6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Singh K, Singh SK, Raman R. MTHFR A1298C polymorphism and idiopathic male infertility. J Postgrad Med 2010;56:267–9. [DOI] [PubMed] [Google Scholar]
- [45].Safarinejad MR, Shafiei N, Safarinejad S. Relationship between genetic polymorphisms of methylenetetrahydrofolate reductase (C677T, A1298C, and G1793A) as risk factors for idiopathic male infertility. Reprod Sci 2011;18:304–15. [DOI] [PubMed] [Google Scholar]
- [46].Liu L. The association between MTHFR C677T and MS A2756G polymorphisms and Hcy level and male infertility. Master's thesis Shantou University 2011;1–65. [Google Scholar]
- [47].Qiu XF, Hu XP, Li YJ, et al. Association of polymorphisms of MTHFR C677T with male infertility in Ningxia. J Ningxia Med Univ 2011;7:625–8. [Google Scholar]
- [48].Vani GT, Mukesh N, Rama Devi P, et al. Methylenetetrahydrofolate reductase C677T polymorphism is not associated with male infertility in a South Indian population. Andrologia 2012;44:252–9. [DOI] [PubMed] [Google Scholar]
- [49].Eloualid A, Abidi O, Charif M, et al. Association of the MTHFR A1298C variant with unexplained severe male infertility. PLoS One 2012;7:e34111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mfady DS, Sadiq MF, Khabour OF, et al. Associations of variants in MTHFR and MTRR genes with male infertility in the Jordanian population. Gene 2014;536:40–4. [DOI] [PubMed] [Google Scholar]
- [51].Naqvi H, Hussain SR, Ahmad MK, et al. Role of 677C→T polymorphism a single substitution in methylenetetrahydrofolate reductase (MTHFR) gene in North Indian infertile men. Mol Biol Rep 2014;41:573–9. [DOI] [PubMed] [Google Scholar]
- [52].Karimian M, Colagar AH. Association of C677T transition of the human methylenetetrahydrofolate reductase (MTHFR) gene with male infertility. Reprod Fertil Dev 2016;28:785–94. [DOI] [PubMed] [Google Scholar]
- [53].Li SS, Li J, Xiao Z, et al. Prospective study of MTHFR genetic polymorphisms as a possible etiology of male infertility. Genet Mol Res 2014;13:6367–74. [DOI] [PubMed] [Google Scholar]
- [54].Li XY, Ye JZ, Ding XP, et al. Association of polymorphisms of MTHFR A1298C and MS A2756G with male infertility in Sichuan males. Chin J Birth Healthy 2014;4:26–9. [Google Scholar]
- [55].Ni W, Li H, Wu A, et al. Lack of association between genetic polymorphisms in three folate-related enzyme genes and male infertility in the Chinese population. J Assist Reprod Genet 2015;32:369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gurkan H, Tozkir H, Göncü E, et al. The relationship between methylenetetrahydrofolate reductase c.677TT genotype and oligozoospermia in infertile male patients living in the Trakya region of Turkey. Andrologia 2015;47:1068–74. [DOI] [PubMed] [Google Scholar]
- [57].Li XY, Ye JZ, Ding XP, et al. Association between methionine synthase reductase A66G polymorphism and primary infertility in Chinese males. Genet Mol Res 2015;14:3491–500. [DOI] [PubMed] [Google Scholar]
- [58].Kurzawski M, Wajda A, Malinowski D, et al. Association study of folate-related enzymes (MTHFR, MTR, MTRR) genetic variants with non-obstructive male infertility in a Polish population. Genet Mol Biol 2015;38:42–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kim SY, Lim JW, Kim JW, et al. Association between genetic polymorphisms in folate-related enzyme genes and infertile men with non-obstructive azoospermia. Syst Biol Reprod Med 2015;61:286–92. [DOI] [PubMed] [Google Scholar]
- [60].Đorđević Valentina, Nikolić A, Ljujić M, et al. Combined effect of GSTM1 gene deletion, GSTT1 gene deletion and MTHFR C677T mutation in male infertility. Arch Biol Sci 2010;62:525–30. [Google Scholar]
- [61].Vardarli AT, Cetintas VB, Eroglu Z. Determination of the association between the C677T and A1298C polymorphisms of the MTHFR gene and the development risk of azoospermia and oligozoospermia in Turkish infertile men. Ege J Med 2014;53:124–8. [Google Scholar]
- [62].Cai LW, Sun WC. Relationship between distribution and frequency of methylenetetrahydrofolate reductase C677T gene polymorphism and male infertilit. The Chinese Journal of Human Sexuality 2018;27:18–21. [Google Scholar]
- [63].Murphy LE, Mills JL, Molloy AM, et al. Folate and vitamin B12 in idiopathic male infertility. Asian J Androl 2011;13:856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Farcas MF, Trifa AP, Militaru M. Methylenetetrahydrofolate reductase A1298C polymorphism and male infertility in a Romanian populatioin group. Maedica 2009;4:6. [Google Scholar]
- [65].Stangler Herodež S, Zagradišnik B, Erjavec Škerget A, et al. MTHFR C677T and A1298C genotypes and haplotypes in Slovenian couples with unexplained infertility problems and in embryonic tissues from spontaneous abortions. Balkan J Med Genet 2013;16:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kumar K, Venkatesh S, Sharma PR, et al. DAZL 260A4G and MTHFR 677C4T variants in sperm DNA of infertile Indian men. Indian J Biochem Biophys 2011;48:422–6. [PubMed] [Google Scholar]
- [67].Liu L, Cai ZM, Leng HM, et al. Association of MTHFR C677T and MS A2756G polymorphism with semen quality. J Cent South Univ 2012;37:1054–9. [DOI] [PubMed] [Google Scholar]
- [68].Chellat D, Rezgoune ML, Hamane D, et al. Influence of methylenetetrahydrofolate reductase C677T gene polymorphisms in Algerian infertile men with azoospermia or severe oligozoospermia. Genet Test Mol Biomarkers 2012;16:874–8. [DOI] [PubMed] [Google Scholar]
- [69].Hu F, Ai JH, Gu LJ. 186 oligoasthenospermia cases observation of MTHFR C677T gene polymorphism. J Reprod Med 2017;26:269–71. [Google Scholar]
- [70].Camprubi C, Pladevall M, Grossmann M, et al. Lack of association of MTHFR rs1801133 polymorphism and CTCFL mutations with sperm methylation errors in infertile patients. J Assist Reprod Genet 2013;30:1125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yang BH, Peng YF, Pi JP. Association of methylenetetrahydrofolatere-ductase gene 677C-4T polymorphism with asthenospermia in Han population of South Anhui. J Wannan Med Univ 2010;29:5–7. [Google Scholar]
- [72].Zhang XJ, Li C, Liu M. Association study of single Nucleotide polymorphisms in the genes of folate metabolism and idiopathic male infertility. Nanjing Normal University 2007;1–52. [Google Scholar]
- [73].Najafipour R, Moghbelinejad S, Aleyasin A, et al. Effect of B9 and B12 vitamin intake on semen parameters and fertility of men with MTHFR polymorphisms. Andrology 2017;5:1–7. [DOI] [PubMed] [Google Scholar]
- [74].Pei J. Association between MTHFR C677T polymorphism and male infertility in Han population of He Nan China. China Health Care Nutr 2013;7:629–30. [Google Scholar]
- [75].Tetik A, Aliyeva U, Cetintas VB, et al. Influence of methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C gene polymorphisms on male infertility in turkish infertile men with azoospermia and oligozoospermia. Eur Urol 2008;Suppl7: 92. [Google Scholar]
- [76].Hussein TM, Elneely DI. Y-chromosome microdeletions and the MTHFR C677 T polymorphism in Egyptian men with nonobstructive azoospermia. Hum Androl 2014;4:66–70. [Google Scholar]
- [77].Ng R, Louie K, Poon K. Association of single nucleotide polymorphisms (SNPS) in methylenetetrahydrofolate reductase (MTHFR) and male infertility. Fertil Steril 2014;102:e192. [Google Scholar]
- [78].Irfan M, Ismail M, Azhar Beg M, et al. Association of the MTHFR C677T (rs1801133) polymorphism with idiopathic male infertility in a local Pakistani population. Balk J Med Genet 2016;19:51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zuo YJ, Aireti Apizi Shao WM, Zhang JK, et al. Association between C677T polymorphism of MTHFR gene and severe oligozoospermia in Xinjiang. Chin J Androl 2018;32:34–7. [Google Scholar]
- [80].Balkan M, Atar M, Erdaf ME. The possible association of polymorphisms in MTHFR, MTRR, and MTHFD l genes with male infertility. Int Med J 2013;20:404–8. [Google Scholar]
- [81].Zhang WB. Association between seminal plasma folate and folic acid metabolism-related genepolymorphism and male infertility. Nanjing Normal Univ 2010;1–72. [Google Scholar]
- [82].Wang Y, Huang CY, Kang YL, et al. Association of the methylenetetrahydrofolatereductasegene C677T polymorphism with male infertility. J Cont Medl Educ 2018;32:142–3. [Google Scholar]
- [83].Hu LL, Niu XY, Bian JJ, et al. Association of MTHFR C677T polymorphism with idiopathic male infertile in Jining area of Han people. Chin J Birth Heal & Here 2018;26:114–6. [Google Scholar]
- [84].Zhou SH, Sun XQ, Niu XY, et al. Relationship between SNP of MTHFR gene and non-obstructive azoospermia and severe oligoasthenospermia in Jining Han population. Chin J Birth Heal & Here 2018;26:111–4. [Google Scholar]
- [85].Ma FF, Tan ZJ, Wang HZ. Association of MTHFR C677T polymorphism with male infertile in Xiamen area. Chin J Birth Heal & Here 2017;25:16–7. 24. [Google Scholar]
- [86].Gava MM, Kayaki EA, Bianco B, et al. Polymorphisms in Fo- late-related enzyme genes in idiopathic infertile Brazilian men. Reproductive Sci 2011;18:1267–72. [DOI] [PubMed] [Google Scholar]
- [87].Xie C, Ping P, Ma Y, et al. Correlation between methylenetetrahydrofolate reductase gene polymorphism and oligoasthenospermia and the effects of folic acid supplementation on semen quality. Transl Androl Urol 2019;8:678–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Suo F, Zhang Y, Wang Y, et al. Study on the association between MTHFR C677T gene polymorphisms and male infertility in Xuzhou. Chin J Birth Heal & Here 2019;27:1507–8. 26. [Google Scholar]
- [89].Shao LJ, Zhu DS, Chen W, et al. Relationships among MTHFR C677T genotype, serum homocysteine and idiopathic male infertility. Zhejiang Medical Journal 2019;41:1013–6. [Google Scholar]
- [90].Song N, Zhang D, Su TY, et al. Application of MTHFR gene C677T and A1298C double loci detection in male infertility. Xinjiang Medical Journal 2019;49:403–5. [Google Scholar]
- [91].Xu JJ, Chen WJ, Liang LZ, et al. Study on MTHFR gene polymorphism in primary male infertility. Journal of Reproductive Medicine 2019;28:777–80. [Google Scholar]
- [92].Shao LJ, Zhu DS, Chen LY, et al. Association between methyltetrahydrofolate reductase c677t gene polymorphism and idiopathic male infertility. Chinese Journal of Birth Health & Heredity 2019;27:397–400. 428. [Google Scholar]
- [93].Attia J, Thakkinstian A, D’Este C. Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol 2003;56:297–303. [DOI] [PubMed] [Google Scholar]
- [94].Davey SG, Egger M. Meta-analyses of randomized controlled trials. Lancet 1997;350:1182. [DOI] [PubMed] [Google Scholar]
- [95].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [97].DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [98].Thompson SG, Higgins JPT. How meta-regression analyses be undertaken and ihterpreted? Statist Med 2002;21:1559–73. [DOI] [PubMed] [Google Scholar]
- [99].Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088–101. [PubMed] [Google Scholar]
- [100].Egger M, Smith DG, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. Br Med J 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet 2007;81:208–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Rosenberg N, Murata M, Ikeda Y, et al. The frequent 5,10-methylenetetrahydrofolate reductase C677T polymorphism is associated with a common haplotype in whites, Japanese, and Africans. Am J Hum Genet 2002;70:758–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Sadewa AH Sunarti, Sutomo R, et al. The C677T mutation in the methylenetetrahydrofolate reductase gene among the Indonesian Javanese population. Kobe J Med Sci 2002;48:137–44. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


