Abstract
Gastroesophageal junction adenocarcinoma (GEJAC) is a malignant tumor with high mortality. Its incidence has increased sharply all over the world in recent years. The study aims to search for potential biomarkers for the diagnosis and prognosis of GEJAC based on the Gene Expression Omnibus database (GEO) database and The Cancer Genome Atlas (TCGA) database.
Microarray dataset (GSE96668 and GSE74553) of GEJAC was downloaded from the GEO. After screening overlapping differentially expressed genes (DEGs) by GEO2R and Wayne map, functional enrichment analysis of the DEGs was performed by the DAVID database. Then, a protein–protein interaction (PPI) network was constructed, and the hub gene was identified by using STRING and Cytoscape, as well as the diagnostic value of hub genes was evaluated by the receiver operating characteristic (ROC) curves. Finally, the gene transcriptome profiles of gastric cancer named TCGA-STAD were downloaded from TCGA database to screen the potential prognostic genes and construct the prognostic risk model using Cox proportional hazards regression. Meanwhile, the Kaplan–Meier curve and time-dependent ROC curve were adopted to test the prognostic value of the prognostic gene signature.
In this study, we identified 10 hub genes that might have high diagnostic value for GEJAC, and inferred that they might be involved in the occurrence and development of GEJAC. Moreover, we conducted a survival prediction model consisting of 6 genes and proved that they have value to some extent in predicting prognosis for GEJAC patients.
Keywords: diagnostic, GEJAC, GEO, prognostic, TCGA
1. Introduction
Gastroesophageal junction adenocarcinoma (GEJAC) is an adenocarcinoma occurred in the esophagogastric junction area. Siewert classification is most frequently used for GEJAC. GEJAC was divided into 3 types, each of which has specific biological behavior and poor prognosis.[1] In recent years, the incidence of GEJAC is increasing significantly in western countries.
In contrast, the incidence of GEJAC caused by eradication of Helicobacter pylori, obesity, and other factors have also continued to increase in Asian countries. GEJAC has the clinicopathological characteristics that differentiate itself from gastric adenocarcinoma and esophageal adenocarcinoma. It is special in classification, staging, surgical treatment, and prognosis.[2] Concealed onset and nonspecific clinical symptoms cause a late diagnosis of GEJAC and poor prognosis. And the tumor often has a larger size and more easily infiltrates adjacent tissues and occurs lymph node metastasis.[3] GEJAC has become an important clinical problem. Therefore, it is of great importance to explore new methods for its early diagnosis and treatment. Biomarkers contribute to an early, rapid, accurate, and sensitive determination of disease occurrence, development and prognosis, and play an important role in the early diagnosis of tumors. There have been previous studies on biomarkers of GEJAC. Toshihiko et al reported that STAT1, HLA-DRA, IFNG, IDO1, CXCL9, CXCL10 were associated with the occurrence of this tumor, but has limited practical significance.[4] HER2 and vascular endothelial growth factor can be used to conduct the clinical practice of trastuzumab and ramoximab for GEJAC patients, but they cannot be diagnosed early.[5,6] Pd-L1 is also a potential predictive biomarker of GEJAC.[7] However, the standard PD-L1 test cannot identify all patients who may benefit from this immunotherapy.[8] In summary, the discovery of new biomarkers is critical in helping us manage and understand this deadly disease.
Advances in gene chip and sequencing technology lead to a rapid increase in high-throughput data. We adopted the GEO and TCGA databases, and the GEO dataset was created by the National Center for Biotechnology Information (NCBI) containing high-throughput gene expression and gene chip dataset all over the world. The TCGA database established by the National Cancer Institute (NCI) and National Human Genome Research Institute (NHGRI) provides a variety of Omics data for cancers as well as clinical information. These 2 datasets were most frequently used in bioinformatic analysis.
In this study, we integrated analysis data based on the GEO and the TCGA database. After constructing the PPI network, we identified the top 10 hub genes. ROC curves evaluate the diagnostic value of top genes. Then, we constructed a prognostic gene signature for GEJAC patients using Cox regression analyses.
2. Materials and methods
2.1. Ethics and informed consent
Ethical approval and patient consent are not required because of no patient recruitment and personal information collection, and the data included in the study are derived from public database.
2.2. Date collection
Two Gene expression profile microarray datasets (GSE96668, GSE74553) were extracted from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) based on GPL10558 (Illumina Human HT 12 V4.0 expression bead chip) and GPL17692 (Affymetrix Human Gene 2.1ST Array). The datasets of GSE96668 includes 49 GEJAC tissues and 7 paired non-cancerous tissues. The database of GSE74553 contained 70 samples of GEJAC and 5 matched normal gastric mucosa tissues. There is no data on GEJAC in the TCGA database. Siewert I tumors are prone to esophageal cancer, while Siewert III tumors are inclined to gastric cancer. Siewert II tumors are true GEJAC, and they are more likely to gastric cancer in origin, surgical treatments, lymph node metastasis, recurrence model, and chemotherapy drugs.[9,10] Most of the data included in this study are type II and III, so we downloaded the RNA sequencing datasets (TCGA-STAD) of 375 gastric cancer and 32 adjacent normal tissues, including clinical information from TCGA to conduct survival analysis. (The data were by the year of December 2019.)
2.3. Identification of DEGs
GEO2R was adopted to identify DEGs between GEJACs samples and paired non-cancer samples. An adjust P < .05 and |log (foldchange)|>1.5 were considered to be statistically significant. Then, the online software Venny 2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to screen out overlapping DEGs between the 2 datasets. The Venn diagram was also drawn by Venny 2.1.0.
2.4. GO and KEGG functional enrichment analysis
To further analyze DEGs potential biological processes, cell components, physiological function, and enrichment signaling pathways, we conducted gene ontology analysis (GO) and Kyoto encyclopedia of genes and genomes analysis (KEGG) using the database for annotation, visualization, and integrated discovery (DAVID). The enrichment results of the top 10 of the count were taken.
2.5. PPI network analysis and hub gene screening
The identified DEGs were imported into the STRING database (https://stringdb.org/) to obtain the interaction among the product of overlapping DEGs, and the Cytoscape software was applied to construct and visualize a PPI network. Then, we utilized the plugin cytoHubba to calculate the node score of genes in the PPI network and pick up the top 10 hub genes of Degree scores. Finally, the ROC curves were used to evaluate the top 10 hub genes value in distinguishing GEJAC from healthy control groups as biomarkers.
2.6. Survival analysis
We download the TCGA-STAD dataset in the TCGA database. The “survival” package was applied for constructing univariate Cox proportional hazards regression to find candidate genes with P value <.05. And multivariate Cox proportional risk regression was used to analyze its value of survival prediction assessment further. Then, risk scores were calculated based on the expression level of screened genes and overall survival information. Risk score formula is as follows: Risk score = Σβi× ExpGenei (βi was the coefficient value and ExpGeneiwas the gene expression level). Finally, the survival ROC package of R software was utilized to draw the time-dependent receiver operating characteristic (ROC) curves and calculate the area under the curve (AUC) of the risk score for a 5-year survival prediction. Patients were classified into low-risk and high-risk groups based on the risk score, and Kaplan–Meier was implemented to compare the differences of survival outcomes between the 2 groups.
3. Results
3.1. Identification of DEGs
The dataset downloaded in GEO was processed by GEO2R with the thresholds of P < .05 and absolute logFC>1. A total of 1043 DEGs comprising 539 up-regulated and 504 down-regulated genes were obtained in GSE74553 (Fig. 1a). While 487 DEGs, including 293 up-regulated and 194 down-regulated genes, were screened out in GSE96668 (Fig. 1b). In the Volcano Plots, red dots denote up-regulated genes, and the green dots denote down-regulated genes. We verified the continuously up-regulated and down-regulated genes in both microarray datasets and drew a Venn diagram using Venny 2.1.0. Ultimately, there were 91 up-regulated and 84 down-regulated DEGs (Fig. 1c).
Figure 1.
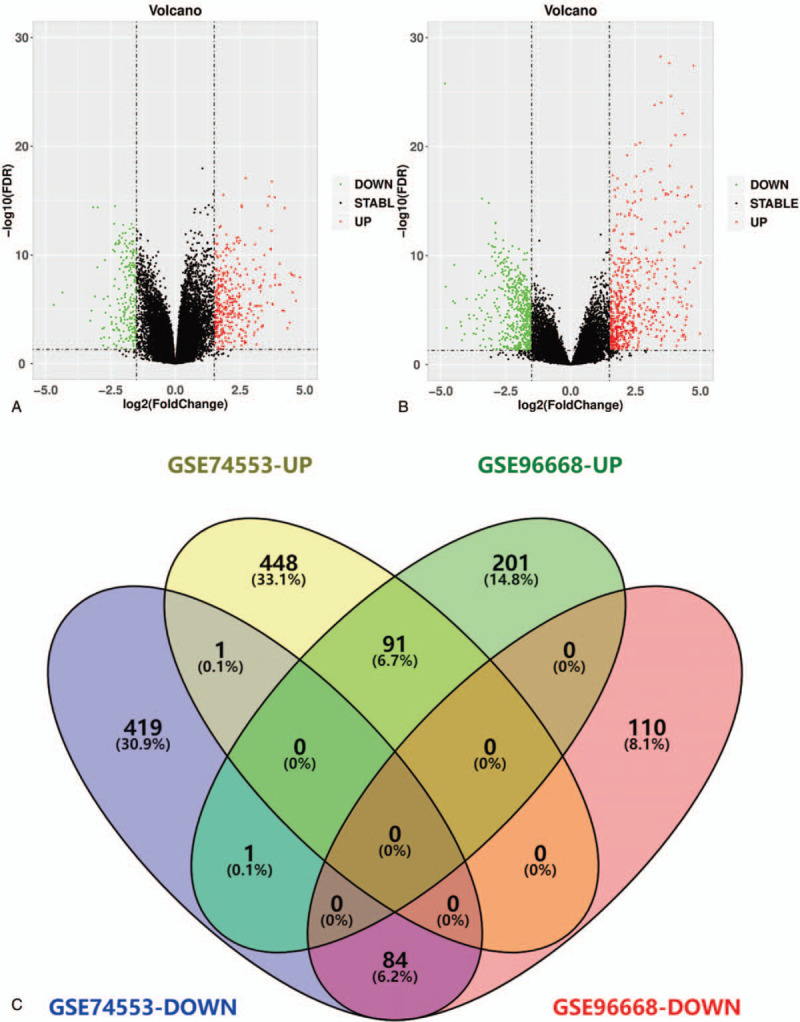
Identification of overlapping DEGs. (a) Volcano plots of GSE96668. (b) Volcano plots of GSE74553. (c) Venn plots of overlapping DEGs.
3.2. GO and KEGG pathway analysis
To further explore the biological functions of DEGs in GEJAC, we conducted GO function analysis and KEGG pathway analysis of 91 up-regulated and 84 down-regulated DEGs by using DAVID. DEGs were primarily concerned with cytoplasm, extracellular exosome, extracellular space in the aspect of cell components (CC) (Fig. 2a). DEGs were mainly concentrated in processes of cell division, mitotic nuclear division, the oxidation-reduction process in terms of biological process (BP) (Fig. 2b). And DEGs were mainly related to identical protein binding, microtubule-binding, and oxidoreductase activity in terms of molecular function (MF) (Fig. 2c). The results of KEGG analysis indicates that DEGs were mainly involved with the 4 processes: cell cycle, protein digestion, and absorption, metabolism of xenobiotics by cytochrome P450 and Chemical carcinogenesis (Fig. 2d).
Figure 2.
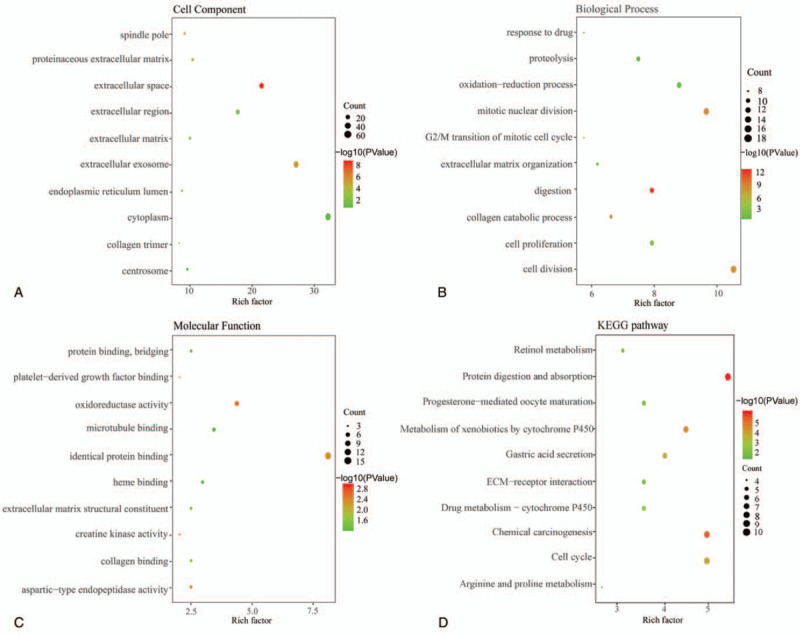
Functional analyses of overlapping DEGs for GEJAC. (a) Cell components. (b) Biological process. (c) Molecular function. (d) KEGG pathway analysis.
3.3. Establish PPI network and identify hub genes
The PPI network was established by the online website STRING and the Cytoscape software to further explored the interaction among DEGs. There were 133 nodes and 1137 edges in the network, among which incorporating 55 up-regulated and 77 down-regulated DEGs. Then the plugin cytoHubba was used to identify the top 10 hub genes, CCNA2, MAD2L1, UBE2C, CDK1, TOP2A, BIRC5, KIF11, CDC20, and CCNB2 from the PPI network (Figs. 3a and b). ROC curve is a comprehensive indicator to reflect the sensitivity and specificity of the continuous variables and reveal the correlation between sensitivity and specificity. The larger area the ROC curves encircles, the higher the accuracy of diagnostic is. The ROC curves were plotted according to the TCGA-STAD dataset obtained from the TCGA database to estimate the diagnostic value of the top 10 hub genes for GEJAC patients (Fig. 3c). The AUCs of CCNA2, MAD2L1, UBE2C, CDK1, BIRC5, KIF11, CDC20, and CCNB2 were 0.898, 0.936, 0.935, 0.935, 0.942, 0.906, 0.919, 0.896, 0.915, and 0.938, respectively which indicated that most of them had high diagnostic value for patients with GEJAC.
Figure 3.
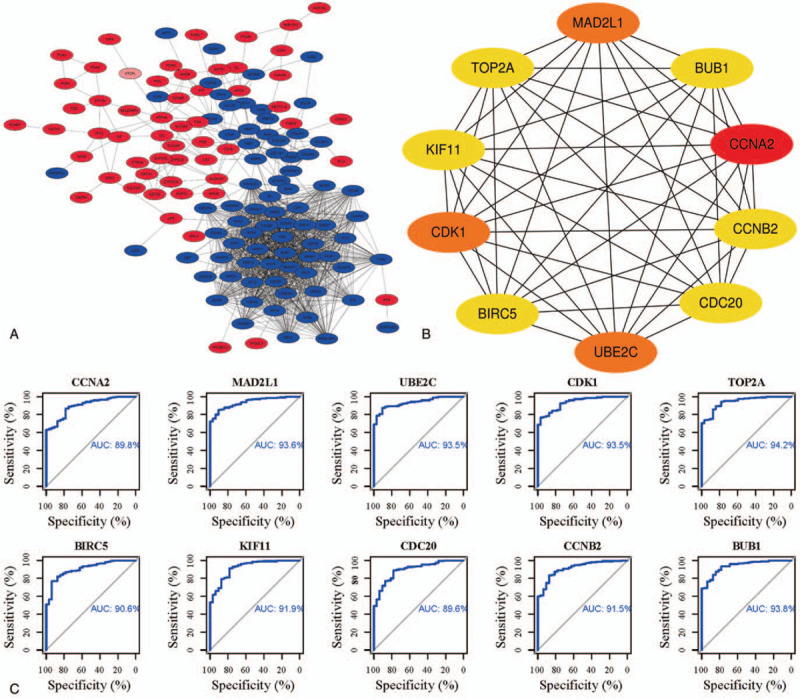
PPI network and top 10 hub genes. (a) PPI network, red denotes up-regulated, and the blue denotes down-regulated genes. (b) Top 10 hub genes. (c) The ROC curves of top 10 hub genes.
3.4. Survival model construction
Sixty nine survival-related genes were selected out (P < .05) to recognize the relationship between DEGs and clinical outcomes of GEJAC using univariate Cox proportional hazards regression (Table 1). Then the “step” method was used to identify the more valuable candidate genes further, and we picked 6 genes. The HR of these 6 genes were all greater than 1, so they were considered as risky prognostic genes (Table 2). As a result, a survival prediction model composing of AC008687.6, AC129507.1, RPS17P14, AC073323.1, LCN1, and MATN3 was structured. The risk score formula is: The riskscore = 2.361 × AC008687.6+2.752 × AC129507.1+2.167 × RPS17P14+2.189 × AC073323.1+2.274 × LCN1+2.438 × MATN3. It is found from results of the AUC of time-dependent ROC curve to predict 5-year survival was 0.674 (Fig. 4b), which demonstrated that this survival prediction model could to a certain extent predict the prognostic.
Table 1.
Univariate Cox regression analysis of the 69 genes.
| Gene | HR | lower 95% | upper 95% | P value |
| AC008677.3 | 1.118086 | 1.040617 | 1.201322 | .002314 |
| AC008687.6 | 1.100800 | 1.031423 | 1.174843 | .003834 |
| AC010528.1 | 1.119298 | 1.037765 | 1.207237 | .003494 |
| AC011352.1 | 1.097301 | 1.032588 | 1.166070 | .002754 |
| AC018653.2 | 1.141747 | 1.065213 | 1.223779 | .000181 |
| AC024581.1 | 1.087453 | 1.026091 | 1.152484 | .004668 |
| AC026369.2 | 1.441932 | 1.150072 | 1.807857 | .001515 |
| AC073323.1 | 1.133043 | 1.050230 | 1.222385 | .001257 |
| AC090283.1 | 1.140763 | 1.066838 | 1.219810 | .000117 |
| AC092625.1 | 1.098695 | 1.031154 | 1.170661 | .003641 |
| AC095350.1 | 1.124037 | 1.057665 | 1.194575 | .000166 |
| AC098799.3 | 1.201599 | 1.096782 | 1.316432 | .000080 |
| AC129507.1 | 1.396983 | 1.138973 | 1.713439 | .001332 |
| AC139491.3 | 1.107126 | 1.037110 | 1.181870 | .002265 |
| ADAMTS18 | 1.201032 | 1.076186 | 1.340361 | .001071 |
| AL022100.1 | 1.125997 | 1.051814 | 1.205412 | .000643 |
| AL049833.1 | 1.159414 | 1.058755 | 1.269643 | .001413 |
| AL354984.1 | 1.113946 | 1.048918 | 1.183006 | .000438 |
| AL645937.1 | 1.137149 | 1.060321 | 1.219543 | .000317 |
| AP000146.1 | 1.105275 | 1.039261 | 1.175482 | .001445 |
| AP000695.1 | 1.369570 | 1.120320 | 1.674274 | .002152 |
| AVP | 1.101834 | 1.038275 | 1.169284 | .001379 |
| C1QL2 | 1.114555 | 1.052207 | 1.180597 | .000222 |
| C8orf87 | 1.094530 | 1.031172 | 1.161782 | .002989 |
| CCDC144NL-AS1 | 1.201873 | 1.072529 | 1.346815 | .001549 |
| CGB2 | 1.093497 | 1.035351 | 1.154908 | .001345 |
| CGB3 | 1.099855 | 1.037445 | 1.166019 | .001406 |
| CGB5 | 1.108974 | 1.049316 | 1.172024 | .000246 |
| CLDN6 | 1.086492 | 1.027683 | 1.148667 | .003481 |
| CST2 | 1.198062 | 1.064152 | 1.348823 | .002807 |
| CTHRC1 | 1.293564 | 1.091584 | 1.532917 | .002962 |
| CYP19A1 | 1.264192 | 1.095606 | 1.458718 | .001326 |
| CYP4F30P | 1.094859 | 1.030247 | 1.163523 | .003499 |
| DCLK3 | 1.435878 | 1.135502 | 1.815712 | .002518 |
| DIRC1 | 1.121974 | 1.044720 | 1.204941 | .001567 |
| ELOVL2 | 1.220392 | 1.066407 | 1.396612 | .003800 |
| EVX2 | 1.113171 | 1.045354 | 1.185389 | .000829 |
| FRMD6-AS2 | 1.096199 | 1.030029 | 1.166620 | .003836 |
| GMCL1P2 | 1.098526 | 1.032825 | 1.168407 | .002823 |
| GPX3 | 1.375701 | 1.138930 | 1.661694 | .000933 |
| HSPD1P8 | 1.103310 | 1.031569 | 1.180040 | .004157 |
| LCN1 | 1.112318 | 1.035835 | 1.194449 | .003405 |
| LINC00307 | 1.127145 | 1.043203 | 1.217840 | .002436 |
| LINC02182 | 1.092182 | 1.031419 | 1.156526 | .002535 |
| LINC02621 | 1.090355 | 1.026688 | 1.157970 | .004833 |
| LNCOG | 1.487878 | 1.213429 | 1.824401 | .000134 |
| MATN3 | 1.355819 | 1.165906 | 1.576668 | .000077 |
| MED15P5 | 1.101678 | 1.034310 | 1.173434 | .002632 |
| MESTP4 | 1.107760 | 1.035836 | 1.184679 | .002809 |
| MFAP2 | 1.288003 | 1.085157 | 1.528766 | .003796 |
| MIR587 | 1.094695 | 1.031261 | 1.162030 | .002971 |
| MMP11 | 1.230360 | 1.066926 | 1.418829 | .004361 |
| NOX4 | 1.352024 | 1.104222 | 1.655436 | .003503 |
| OR1S2 | 1.123367 | 1.040159 | 1.213232 | .003049 |
| OR5H8 | 1.125691 | 1.050626 | 1.206119 | .000772 |
| OR5W1P | 1.129011 | 1.041917 | 1.223385 | .003052 |
| PGAM5P1 | 1.157638 | 1.071685 | 1.250485 | .000200 |
| PSAPL1 | 1.098183 | 1.030432 | 1.170388 | .003943 |
| RARRES2P9 | 1.114941 | 1.038982 | 1.196453 | .002509 |
| RNU6-173P | 1.161499 | 1.073690 | 1.256490 | .000189 |
| RPL12P40 | 1.116966 | 1.037860 | 1.202102 | .003162 |
| RPS12P10 | 1.102197 | 1.037577 | 1.170840 | .001596 |
| RPS17P14 | 1.114607 | 1.047712 | 1.185772 | .000591 |
| RPSAP52 | 1.357529 | 1.108672 | 1.662246 | .003092 |
| SSR1P2 | 1.129525 | 1.047199 | 1.218324 | .001608 |
| TMSB15A | 1.258675 | 1.090666 | 1.452564 | .001648 |
| TRARG1 | 1.105854 | 1.036711 | 1.179608 | .002255 |
| ZNF101P1 | 1.081605 | 1.025041 | 1.141291 | .004204 |
| ZNF734P | 1.089584 | 1.026974 | 1.156012 | .004490 |
Table 2.
Multivariate Cox regression analysis of 6 genes.
| Gene | HR | lower 95% | upper 95% | P value |
| AC008687.6 | 1.085743 | 1.014076 | 1.162476 | .018219 |
| AC129507.1 | 1.406373 | 1.103106 | 1.793014 | .005926 |
| RPS17P14 | 1.074518 | 1.006881 | 1.146698 | .030259 |
| AC073323.1 | 1.095614 | 1.009610 | 1.188943 | .028577 |
| LCN1 | 1.119104 | 1.032078 | 1.213469 | .006442 |
| MATN3 | 1.245422 | 1.043938 | 1.485792 | .014788 |
Figure 4.
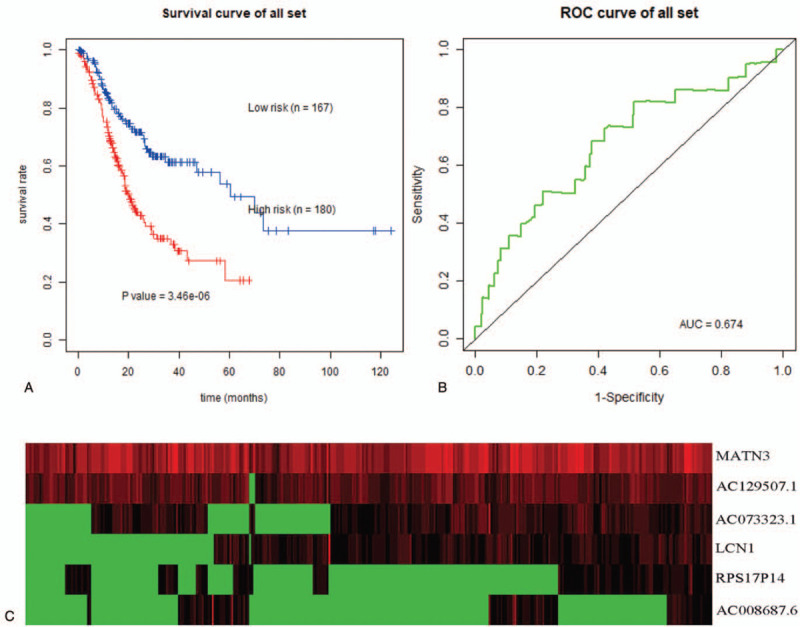
Survival prediction model of 6 genes for GEJAC. (a) Kaplan–Meier curve for high-risk and low-risk samples. (b) ROC curves for predicting 5-year OS in GEJAC. (c) Expression heat map of prognostic genes.
A total of 180 samples were categorized as a high-risk group, whereas the other 167 samples were classified as a low-risk group based on the median value of the risk score. Meanwhile, the Kaplan–Meier curve was drawn. The results showed that patients in the high-risk group had an increased risk of death compared to the low-risk group. And it was statistically significant (P < .05) (Fig. 4a). Finally, we plotted the expression heat map of the 6 prognostic genes in the low-risk and the high-risk group (Fig. 4 c).
4. Discussion
In this study, we conducted a bioinformatics analysis of DEGs effect on prognosis and diagnosis of GEJAC as biomarkers based on datasets from GEO and TCGA. A total of 175 DEGs consisting of 91 up-regulated and 84 down-regulated genes were identified. After conducting a GO function analysis of 175 DEGs, it is revealed that DEGs were concentrated in biological processes of cell division, oxidative stress, protein synthesis and decomposition, and cell adhesion, all of which were closely related to tumorigenesis and progression. We further conducted KEGG pathway analysis and it indicated that DEGs were mainly associated with the process of cell cycle, protein digestion and absorption, metabolism of xenobiotics by cytochrome P450 and chemical carcinogenesis. Cytochrome P450 is the metabolism pathway for numerous endogenous and exogenous substances. It is believed to play an important role in chemical carcinogenesis and anticarcinogen metabolic pathways. Previous studies have discovered that CYP1A and CYP3A, the subtypes of cytochrome P450, were overexpressed in gastric cancer.[11,12] This study indicated that identified DEGs might be involved in the occurrence and development of GEJAC. After constructing the PPI network, we identified 10 hub genes from DEGs, including CCNA2, CCNB2, MAD2L1, UBE2C, CDK1, BIRC5, KIF11, CDC20, TOP2A, and BUB1. These 10 hub genes were all related to cell cycle, indicating that this biological process may be the core of this PPI network.
Previous research has shown that CCNA2, CCNB2, MAD2L1, UBE2C, CDK1, BIRC5, KIF11, CDC20, and TOP2A might promote the occurrence and development of tumors. While this study indicates that the expressions of CCNA2, CCNB2, MAD2L1, UBE2C, CDK1, BIRC5, KIF11, CDC20, TOP2A, and BUB1 were all downregulated in GEJAC, CCNA2 is a regulatory molecule of the cell cycle. It can regulate the G1/S and G2/S transition of the cell cycle by binding and activating CDK2. There has been researched that CCNA2 is associated with the treatment and prognosis of tumors.[13] The increased expression of CCNA2 related to poor prognosis for gastric cancer patients (Zhang et al 2018).[14] CCNB2, which locates in dictyosome, facilitates G2/M transition by activating CDK1 and may also participate in the regulation of cell cycle mediated by TGF-β2. Besides, ISL1 may promote cell proliferation and tumor growth by activating CCNB2 in gastric cancer.[15]
As the crucial component of mitotic checkpoint complex and involved in the correct arrangement of chromosomes on the equatorial plate in mitotic metaphase, MAD2L1 has been found increased expression in gastric cancer (Wang et al, 2019).[16] UBE2C is associated with the complex. It benefits the degradation of mitotic cyclin in mitotic and participates in the separation of sister chromatids by inducing the degradation of securin. The increased expression of UBE2C in esophageal adenocarcinoma could affect the proliferating rate by regulating levels of CCNB1.[17] CDK1 is a critical regulatory molecule in G2/M transition, activating it is essential for entering mitosis. CDK1 phosphorylated mitotic substrates to stimulates nucleus reformation, chromosome condensation, and mitotic spindle formation.[18] Many studies have shown that the high-level expressions of CDK1 in gastric cancer were connected with cell proliferation and the mechanism of anticancer drugs.[19,20] It was a member of an inhibitor of apoptosis protein (IAP) family. BIRC5 was widely expressed in fetal and malignant tumor tissues, while seldom expressed in normal somatic cells. BIRC5 encodes the survivin protein, which has dual functions of inhibiting apoptosis and regulating the cell cycle. Researchers have found that the expression level of BIRC5 was positively related to the progression of esophagus cancer and it facilitated migration and invasion of esophageal carcinoma cells by regulating angiogenetic factors.[21]
Moreover, ZHU et al discovered that the low expression of BIRC5 might be associated with cisplatin resistance in gastric cancer cells.[22] KIF11 belongs to the kinesin superfamily (KIF). Its product functions include chromosome localization, centrosome separation, and formation of a bipolar spindle during mitosis. Overexpression of KIF11 led to an increase of aneuploid daughter cells, resulting in genetic instability that finally generated tumor progression.[23] In the research conducted by Imai et al, the expression of KIF11 increased in intestinal gastric cancer.[24] CDC20 is the regulatory protein that attended the cell cycle machinery and activated APC. The abnormal level or dysfunction of CDC20 might cause APCs inactivation, and leading to an early entrance to anaphase and aneuploidy in daughter cells.[25] Some studies have found that CDC20 is positively associated with tumor size, histological grade, lymph node involvement (LNI), and TNM stage of gastric cancer.[26] TOP2A encodes DNA topoisomerase. And it could affect cancer prognosis such as colon, ovarian, and breast cancer.[27] The expression level of TOP2A was positively correlated with hematogenous metastasis, lymph node metastasis, and peritoneal metastasis of advanced gastric cancer.[28] Meanwhile, in esophageal squamous carcinoma, the expression of TOP2A increased.[29]
BUB1 encodes serine/threonine kinases and plays a vital role in mitosis. It not only activates mitotic spindle checkpoint, ensures chromosome alignment and ensures the integrity of chromosome separation, but also involved in DNA damage. The expression of BUB1 is negatively related to tumor size and lymph node metastasis in gastric adenocarcinoma,[30] which is consistent with our research. Given that these 10 genes all affect the clinical characteristics of gastric and esophagus cancer, we inferred that they might also play an important role in GEJAC.
Certainly, the specific effects and mechanisms of these genes in GEJAC need further experiment research. Given the Top 10 genes significant impact on malignant tumors, we plotted ROC curves to verify their diagnostic value of GEJAC. The results showed that they all had a high diagnostic effect. Then, we use Cox proportional hazards regression to construct a survival prediction model composing of AC008687.6, AC129507.1, RPS17P14, AC073323.1, LCN1, and MATN3 genes. The Kaplan–Meier curve showed patients with GEJAC have significant differences in clinical outcomes after classifying with a risk score. The AUC of time-dependent ROC is 0.674, which indicated the specificity and sensitivity of the 6 prognostic markers in predicting prognosis for GEJAC patients.
In conclusion, we identify 10 hub genes about the diagnosis of GEJAC and conducted a survival prediction model composing of 6 genes by integrating gene expression datasets and clinical information of GEJAC. Combined with previous studies of these 10 hub genes, the results of our study also prove that GEJAC is a special tumor different from esophageal adenocarcinoma and gastric adenocarcinoma. Hopefully, the findings of this research would provide a theoretical reference for the exploration of potential biomarkers for the diagnosis and prognosis of GEJAC in the future.
Whereas the study has some limitations. Firstly, the number of samples we collect is limited due to the database. Meanwhile, we chose the gastric cancer dataset, which is more similar to GEJAC than esophagus cancer, from the TCGA database for survival analysis. It may also impact the results of this study. Secondly, the datasets we researched in our study were downloaded from public databases TCGA and GEO, which adds difficulty to assess the quality of data. Thirdly, we did not consider the patients characteristics including tumor grade and stage, gender, age, and race that might affect gene expression as well. Many studies have shown that race disparities commonly exist in incidence, progression, pathological classification, and prognosis of gastrointestinal tumors.[31,32] This phenomenon also occurs in gastric cancer and esophageal cancer.[33–36] Gu et al also found that gene expression levels in gastric cancer vary with race[37] in his studies. However, there is no relevant study in gastroesophageal junction adenocarcinoma. In our study, we found 9 genes expression were not consistent with previous studies about gastric cancer and esophageal cancer, involving CCNA2, CCNB2, MAD2L1, UBE2C, CDK1, BIRC5, KIF11, CDC20, TOP2A, and BUB1. It may cause by the different characteristics of gastroesophageal junction adenocarcinoma with these 2 cancers. On the other hand, racial disparity might also lead to the differences. But it still needs further investigation to clarify. At last, restrain by experimental foundations, our research is only based on bioinformatics analysis. Experimental verification is also needed in further research.
Author contributions
Conceptualization: Danlei Song, Yuping Wang, Yongning Zhou.
Data curation: Danlei Song, Yuping Hu, Yongning Zhou.
Formal analysis: Danlei Song.
Funding acquisition: Hong Lu, Quanlin Guan, Yongning Zhou.
Investigation: Jiming Tian.
Project administration: Quanlin Guan, Yongning Zhou.
Resources: Yuping Wang, Quanlin Guan, Yongning Zhou.
Software: Jiming Tian.
Supervision: Yuping Wang, Quanlin Guan.
Validation: Jiming Tian.
Visualization: Danlei Song, Jiming Tian.
Writing – original draft: Danlei Song, Yuping Hu, Yongjian Wei, Hong Lu.
Writing – review & editing: Danlei Song, Yongjian Wei, Yongning Zhou.
Footnotes
Abbreviations: AUC = the area under curve, BP = biological process, CC = cell components, DAVID = database for annotation, visualization, and integrated discovery, DEGs = differentially expressed genes, GEJAC = Gastroesophageal junction adenocarcinoma, GEO = Gene Expression Omnibus, GO = gene ontology, IAP = inhibitor of apoptosis protein family, KEGG = Kyoto encyclopedia of genes and genomes, KIF = kinesin superfamily proteins, LNI = lymph node involvement, MF = molecular function, NCBI = National Center for Biotechnology Information, NCI = National Cancer Institute, NHGRI = National Human Genome Research Institute, PPI = protein-protein interaction, ROC = receiver operating characteristic, TCGA = The Cancer Genome Atlas.
How to cite this article: Song D, Tian J, Hu Y, Wei Y, Lu H, Wang Y, Guan Q, Zhou Y. Identification of biomarkers associated with diagnosis and prognosis of gastroesophageal junction adenocarcinoma–a study based on integrated bioinformatics analysis in GEO and TCGA database. Medicine. 2020;99:51(e23605).
This work was supported by the National Natural Science Foundation of China (71964021), National Key R&D Program of China (2016YFC1302201 and 2017YFC0908302), the Natural Science Foundation of Gansu Province, China (18JR3RA351), the Fundamental Research Funds for the Central Universities (lzujbky-2020-kb16).
The authors have no conflicts of interests to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
- [1].Press MF, Sauter G, Buyse M, et al. Alteration of topoisomerase II-alpha gene in human breast cancer: association with responsiveness to anthracycline-based chemotherapy. J Clin Oncol 2011;29:859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lu B, Lu C, Sun Z, et al. Combination of apatinib mesylate and second-line chemotherapy for treating gastroesophageal junction adenocarcinoma. J Int Med Res 2019;47:2207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chevallay M, Bollschweiler E, Chandramohan SM, et al. Cancer of the gastroesophageal junction: a diagnosis, classification, and management review. Ann N Y Acad Sci 2018;1434:132–8. [DOI] [PubMed] [Google Scholar]
- [4].Spatola C, Tocco A, Pagana A, et al. Combined taxane-based chemotherapy and intensity-modulated radiotherapy with simultaneous integrated boost for gastroesophageal junction adenocarcinoma. Fut Oncol 2018;14(6s):47–51. [DOI] [PubMed] [Google Scholar]
- [5].Doi T, Piha-Paul SA, Jalal SI, et al. Safety and antitumor activity of the anti-programmed death-1 antibody pembrolizumab in patients with advanced esophageal carcinoma. J Clin Oncol 2018;36:61–7. [DOI] [PubMed] [Google Scholar]
- [6].Shitara K, Honma Y, Omuro Y, et al. Efficacy of trastuzumab emtansine in Japanese patients with previously treated HER2-positive locally advanced or metastatic gastric or gastroesophageal junction adenocarcinoma: a subgroup analysis of the GATSBY study. Asia Pac J Clin Oncol 2020;16:5–13. [DOI] [PubMed] [Google Scholar]
- [7].Das S, Gibson MK. Evolving management strategies for metastatic esophageal and gastroesophageal junction adenocarcinoma. Oncol Hematol Rev 2018;14:82–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brar G, Shah MA. The role of pembrolizumab in the treatment of PD-L1 expressing gastric and gastroesophageal junction adenocarcinoma. Therap Adv Gastroenterol 2019;12:1756284819869767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Weinberg BA, Xiu J, Hwang JJ, et al. Immuno-oncology biomarkers for gastric and gastroesophageal junction adenocarcinoma: why PD-L1 testing may not be enough. Oncologist 2018;23:1171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Brown AM, Giugliano DN, Berger AC, et al. Surgical approaches to adenocarcinoma of the gastroesophageal junction: the Siewert II conundrum. Langenbeck's Arch Surg 2017;402:1153–8. [DOI] [PubMed] [Google Scholar]
- [11].Hasegawa S, Yoshikawa T. Adenocarcinoma of the esophagogastric junction: incidence, characteristics, and treatment strategies. Gastric Cancer 2010;13:63–73. [DOI] [PubMed] [Google Scholar]
- [12].Li H, Chen X-L, Li H-Q. Polymorphism of CYPIA1 and GSTM1 genes associated with susceptibility of gastric cancer in Shandong Province of China. World J Gastroenterol 2005;11:5757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Murray GI, Taylor MC, Burke MD, et al. Enhanced expression of cytochrome P450 in stomach cancer. Br J Cancer 1998;77:1040–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dachineni R, Ai G, Kumar DR, et al. Cyclin A2 and CDK2 as novel targets of aspirin and salicylic acid: a potential role in cancer prevention. Mol Cancer Res 2016;14:241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang H-P, Li S-Y, Wang J-P, et al. Clinical significance and biological roles of cyclins in gastric cancer. Onco Targets Ther 2018;11:6673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shi Q, Wang W, Jia Z, et al. ISL1, a novel regulator of CCNB1, CCNB2 and c-MYC genes, promotes gastric cancer cell proliferation and tumor growth. Oncotarget 2016;7:36489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang Y, Wang F, He J, et al. miR-30a-3p targets MAD2L1 and regulates proliferation of gastric cancer cells. Onco Targets Ther 2019;12:11313–24. doi:10.2147/OTT.S222854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Palumbo AJ, Da Costa NM, De Martino M, et al. UBE2C is overexpressed in ESCC tissues and its abrogation attenuates the malignant phenotype of ESCC cell lines. Oncotarget 2016;7:65876–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rhind N, Russell P. Signaling pathways that regulate cell division. Cold Spring Harb Perspect Biol 2012;4: doi: 10.1101/cshperspect.a005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang L, Kang W, Lu X, et al. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle 2018;17:1886–900. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [21].Lee MH, Cho Y, Kim DH, et al. Menadione induces G2/M arrest in gastric cancer cells by down-regulation of CDC25C and proteasome mediated degradation of CDK1 and cyclin B1. Am J Transl Res 2016;8:5246–55. [PMC free article] [PubMed] [Google Scholar]
- [22].Shang X, Liu G, Zhang Y, et al. Downregulation of BIRC5 inhibits the migration and invasion of esophageal cancer cells by interacting with the PI3K/Akt signaling pathway. Oncol Lett 2018;16:3373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhu P, Shan X, Liu J, et al. miR-3622b-5p regulates cisplatin resistance of human gastric cancer cell line by targeting BIRC5. J Biomed Res 2019;33:382–90. [Google Scholar]
- [24].Jungwirth G, Yu T, Moustafa M, et al. Identification of KIF11 as a novel target in meningioma. Cancers (Basel) 2019;11: doi: 10.3390/cancers11040545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Imai T, Oue N, Nishioka M, et al. Overexpression of KIF11 in gastric cancer with intestinal mucin phenotype. Pathobiology 2017;84:16–24. [DOI] [PubMed] [Google Scholar]
- [26].Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature 2004;432:338–41. [DOI] [PubMed] [Google Scholar]
- [27].Ding Z-Y, Wu H-R, Zhang J-M, et al. Expression characteristics of CDC20 in gastric cancer and its correlation with poor prognosis. Int J Clin Exp Pathol 2014;7:722–7. [PMC free article] [PubMed] [Google Scholar]
- [28].Terashima M, Ichikawa W, Ochiai A, et al. TOP2A, GGH, and PECAM1 are associated with hematogenous, lymph node, and peritoneal recurrence in stage II/III gastric cancer patients enrolled in the ACTS-GC study. Oncotarget 2017;8:57574–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hanagiri T, Ono K, Kuwata T, et al. Evaluation of topoisomerase I/topoisomerase IIalpha status in esophageal cancer. J UOEH 2011;33:205–16. [DOI] [PubMed] [Google Scholar]
- [30].Stahl D, Braun M, Gentles AJ, et al. Low BUB1 expression is an adverse prognostic marker in gastric adenocarcinoma. Oncotarget 2017;8:76329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gad MM, Găman M-A, Saad AM, et al. Temporal trends of incidence and mortality in Asian-Americans with pancreatic adenocarcinoma: an epidemiological study. Ann Gastroenterol 2020;33:210–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gad MM, Saad AM, Faisaluddin M, et al. Epidemiology of cholangiocarcinoma; united states incidence and mortality trends. Clin Res Hepatol Gastroenterol 2020. [DOI] [PubMed] [Google Scholar]
- [33].Tramontano AC, Nipp R, Mercaldo ND, et al. Survival disparities by race and ethnicity in early esophageal cancer. Dig Dis Sci 2018;63:2880–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Laszkowska M, Tramontano AC, Kim J, et al. Racial and ethnic disparities in mortality from gastric and esophageal adenocarcinoma. Cancer Med 2020;9:5678–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Melkonian SC, Jim MA, Haverkamp D, et al. Disparities in cancer incidence and trends among American Indians and Alaska Natives in the United States, 2010-2015. Cancer Epidemiol Biomarkers Prev 2019;28:1604–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Renelus BD, Jamorabo DS, Kancharla P, et al. Racial disparities with esophageal cancer mortality at a high-volume university affiliated center: an all ACCESS invitation. J Natl Med Assoc 2019;doi: 10.1016/j.jnma.2019.04.005. [DOI] [PubMed] [Google Scholar]
- [37].Gu X, Zhang W, Xu L, et al. Quantitative assessment of the influence of prostate stem cell antigen polymorphisms on gastric cancer risk. Tumour Biol 2014;35:2167–74. [DOI] [PubMed] [Google Scholar]


