The central nervous system (CNS) has historically been viewed as an immune-privileged site, but recent data demonstrate a diverse landscape of immune cells within the meninges, the membranes that surround the brain and spinal cord1. Studies to date have focused on macrophages and T cells, but a detailed analysis of meningeal humoral immunity is lacking. Here, we show that the murine and human meninges contain IgA-secreting plasma cells during homeostasis. These cells are positioned adjacent to dural venous sinuses, regions of slow blood flow with fenestrations that can potentially permit blood-borne pathogens to access the brain2. Peri-sinus IgA plasma cells increased with age and following intestinal barrier breach. Conversely, they were scarce in germ-free mice, but their presence was restored by gut re-colonization. B cell receptor sequencing confirmed that meningeal IgA+ cells originated in the intestine. Specific depletion of meningeal plasma cells or IgA-deficiency resulted in reduced fungal entrapment in the peri-sinus region and increased spread into the brain following intravenous challenge, revealing a critical role for meningeal IgA in defending the CNS at this vulnerable venous barrier surface.
The meninges consist of three membranes that provide a protective covering to shield the CNS from physical trauma and pathogen infiltration. Adjacent to the skull is the dura mater that houses meningeal blood and lymphatic vessels as well as the dural venous sinuses3,4. The arachnoid mater adheres to the dura and is separated from the inner pia mater by the cerebrospinal fluid (CSF)-containing subarachnoid space. Healthy meninges also contain both innate and adaptive immune cells that provide a defense against pathogens4. Most meningeal immune cells are in the dura mater, including T cells, dendritic cells, mast cells, and meningeal macrophages. Meningeal macrophages lie adjacent to meningeal vessels, and together with the perivascular macrophages that line pial vessels, provide surveillance and protection along these barrier structures5,6. To date, few studies have focused on the phenotype or function of the humoral immune system in the meninges.
B cells develop in the bone marrow and circulate between blood and secondary lymphoid organs until they encounter a cognate antigen. Following activation, they undergo iterative rounds of proliferation, during which somatic hypermutation and class switching from IgM to other isotypes may occur7. The IgG-producing plasma cells generated home to bone marrow niches, where they may survive for decades8. In the mucosal-associated lymphoid tissue of the gut, specific cues lead to the development of IgA-producing plasma cells that localize to the lamina propria, secreting IgA dimers joined by a J-chain9. Circulating antibodies are thought to be largely excluded from the CNS in health, but increased levels of IgG and IgA antibodies have been observed in the CSF during many disease states10–13. It has been assumed that these antibodies are produced systemically and enter the CSF from the blood due to pathology-associated blood-brain barrier breach. A recent study extended this paradigm, showing the presence of gut-derived IL10-producing IgA+ plasma cells within the brain and spinal cord during inflammation in a mouse model of multiple sclerosis14.
To profile steady state meningeal humoral immunity, we investigated mouse dura mater. We found that meningeal B cells were mostly naïve (IgD+IgM+) (Extended Data Fig. S1a). In addition, there were CD138+ Blimp1-expressing plasma cells predominantly located adjacent to the superior sagittal and transverse venous sinuses (Fig. 1a). Surprisingly, in the unchallenged meninges, most of the peri-sinus plasma cells were IgA positive (Fig. 1b–e, Extended Data Fig. S1b–c), and RNA sequencing of murine meninges confirmed the presence of IgA heavy chain transcripts and plasma cell-supporting cytokines (Fig. 1e, Extended Data Fig. S1d). These meningeal IgA+ plasma cells also expressed CXCR4 (Extended Data Fig. S1e), a chemokine receptor important in regulating plasma cell homing to niches15, as well as J-chain (Fig. 1f), suggesting that they secrete dimeric IgA like intestinal IgA+ plasma cells. Intravital two-photon imaging confirmed that these Blimp1+ cells were adjacent to, rather than within, venous sinuses (Fig. 1g, Video 1). Dural IgA+ plasma cells increased with age (Fig. 1h), as has been observed in the intestine16, and were almost absent in T cell-deficient mice (Fig. 1i). In human dura mater, we similarly identified CD138+IgA+ plasma cells adjacent to the dural sinuses (Fig. 1j, Extended Data Fig. S1f).
Fig. 1: IgA plasma cells are localized adjacent to the dural sinuses in murine and human meninges.
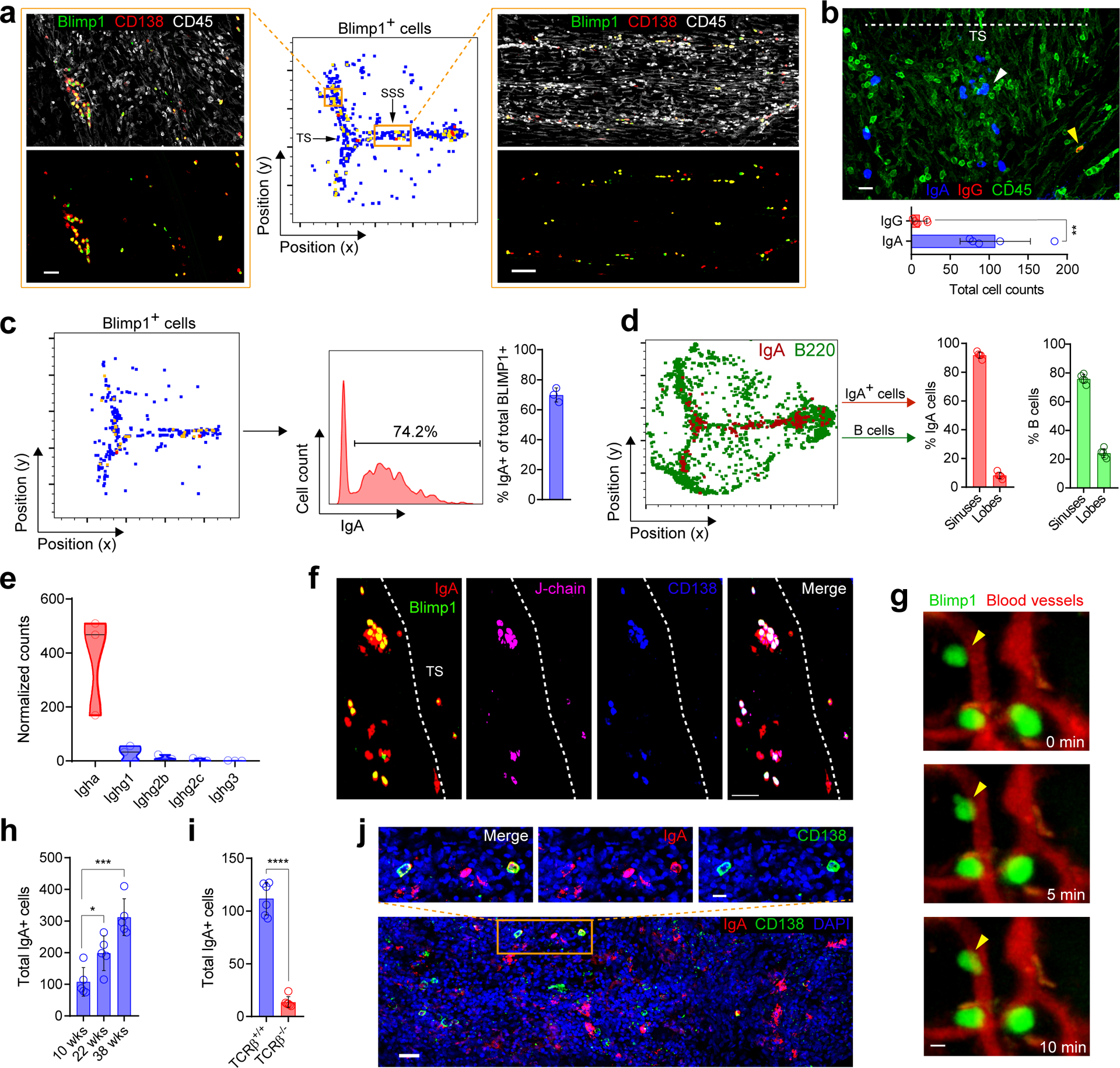
a, Confocal images of plasma cells along the transverse sinus (TS; scale bar, 70 μm) and superior sagittal sinus (SSS; scale bar, 100 μm) from murine whole-mount meninges stained with CD138 (red) and CD45 (white) with corresponding histo-cytometric plot of Blimp1-expressing cells (green) from a Prdm1EYFP/+ mouse. b, Representative image of IgA (blue), IgG (red) and CD45 (green) immunolabeling on whole-mount meninges (scale bar, 30 μm) and quantification of peri-sinus IgA+ and IgG+ cells below. White arrowhead depicts abluminal IgA+ cells and yellow arrowhead depicts an abluminal IgG+ cell; the dotted line shows the lumen of the TS (mean ± s.d., n = 5 mice; **P = 0.0016, unpaired two-tailed Student’s t-test; data are representative of two independent experiments). c, Representative histo-cytometric plot of Blimp1+ cells in a meningeal whole-mount immunolabeled for IgA from a Prdm1EYFP/+ mouse, with the proportion of Blimp1+ cells that co-express IgA quantified in the bar graph (mean ± s.d., n = 3 mice). d, Histo-cytometric plot from meningeal whole-mount stained with B220 (green) and IgA (red), with quantification of cells in the lobes (frontal, parietal and occipital lobes) and peri-sinus regions (mean ± s.d., n = 5 mice). e, Normalized counts of the denoted class-switched immunoglobulin heavy chain transcript expression from the B6 meninges analyzed by RNA-seq (n = 3 mice). f, Confocal image of the TS region from Prdm1EYFP/+ mouse whole-mount tissue immunolabeled for IgA (red), J-chain (pink) and CD138 (blue), with Blimp1-EYFP+ cells labeled in green (scale bar, 50 μm). g, Representative time lapse captured by two-photon microscopy through a thinned skull of a Prdm1EYFP/+ mouse, displaying Blimp1-EYFP+ cells (green) and peri-sinus vasculature (red) (scale bar, 5 μm); arrowheads denote relatively immobile Blimp1-EYFP+ cells adjacent to dural vasculature. h, Quantification of meningeal IgA+ cells in 10-week, 22-week and 38-week-old B6 mice (mean ± s.d., n = 5 mice per group; 10 weeks (wks) vs. 22 wks *P = 0.0215, 10 wks vs. 38 wks ***P = 0.0003, unpaired two-tailed Student’s t-test). i, Quantification of peri-sinus IgA-expressing cells by immunohistochemical analysis of meningeal whole-mounts of WT and Tcrb−/− mice (mean ± s.d., n = 6 mice per group; ****P = <0.0001, unpaired two-tailed Student’s t-test, data are representative of two independent experiments). j, Representative image of the luminal aspect of human whole-mount SSS tissue stained with IgA (red), CD138 (green) and DAPI (blue, 4′,6-diamidino-2-phenylindole) (scale bar, 50 μm). The inset above represents a higher magnification of the highlighted box (scale bar, 20 μm).
Since the presence of IgA+ cells in the gut is dependent on the microbiome17, we investigated whether treatment of specific pathogen free (SPF) mice with antibiotics to reduce their colonizing microbiome would impact dural plasma cells. Six weeks following oral antibiotic administration, there was a substantial reduction in IgA+ plasma cells in the meninges (Fig. 2a). IgA+ plasma cells were also almost completely absent in the meninges of germ free (GF) mice, but their presence was restored in GF mice reconstituted with either a murine microbiome or human gut microbiome (donor A (DA) and donor B (DB)) (Fig. 2b–d), which was also apparent by expression of IgA heavy chain transcripts (Fig. 2e). Remarkably, the phenotype of the plasma cells in the meninges of these two human microbiome-reconstituted mouse strains differed, with IgG+ plasma cells present in the GF-DA mice in addition to IgA+ cells (Fig. 2f). Metagenomic sequencing of the fecal content of GF-DA and GF-DB mice demonstrated a reduction in bacterial diversity in GF-DA mice (Extended Data Fig. S2a–b) and differing frequency of some bacterial species between GF-DA and GF-DB (Extended Data Fig. S2c). Together, these data show that the nature of, and variation in, the intestinal microbiome can significantly impact the magnitude and antibody isotype of the meningeal plasma cell pool.
Fig. 2: Meningeal IgA cells are clonally related to those in the gut and depend on intestinal microbiota.
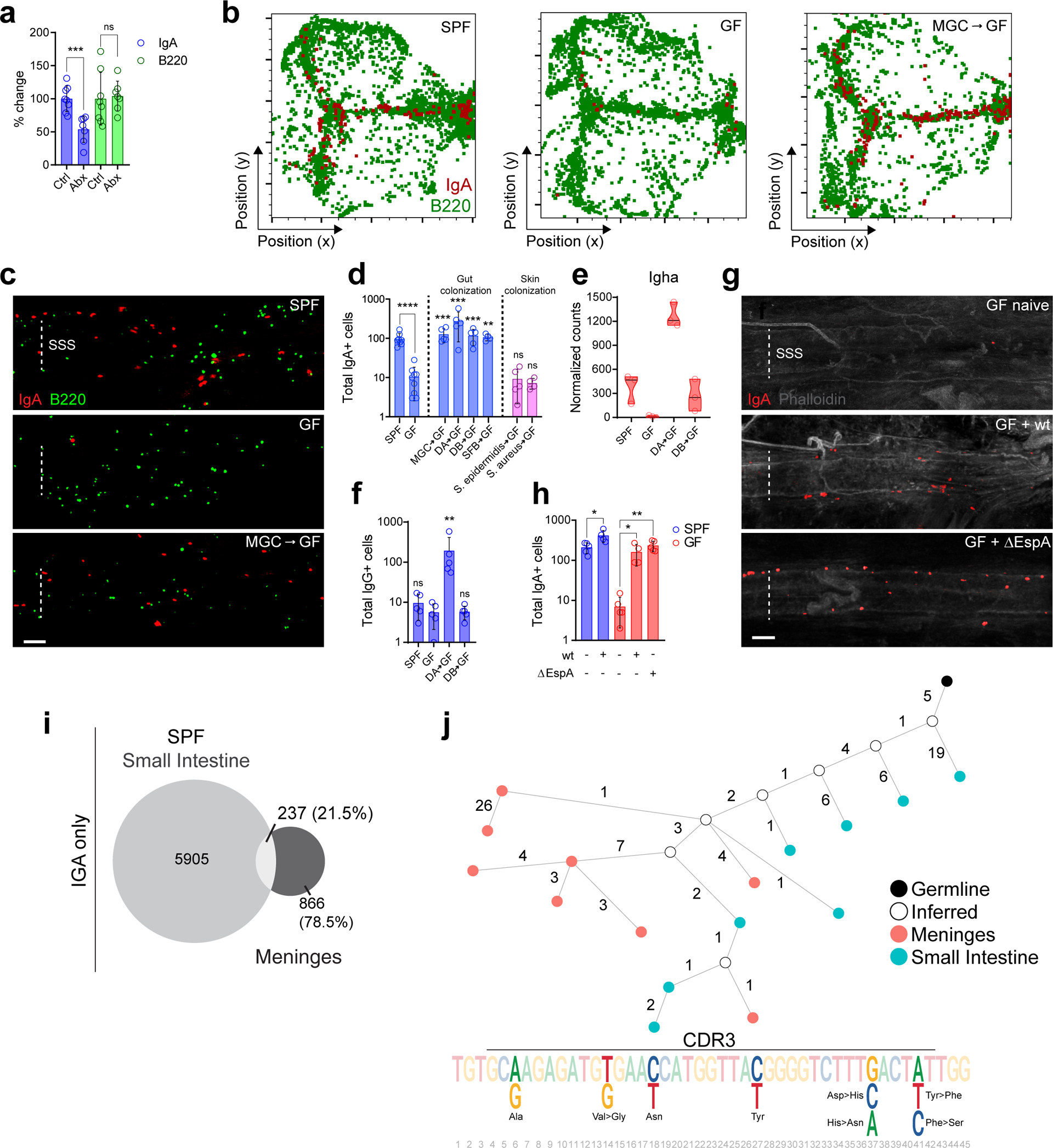
a, Immunohistochemistry based quantification of peri-sinus meningeal IgA+ and B220+ cells in whole-mount meninges from naive control (Ctrl) versus 6-week oral antibiotic (Abx)-treated B6 mice. Data are compiled from two independent experiments (mean ± s.d., n = 8 mice in Ctrl group and 7 in Abx group; IgA+ cells ***P = 0.0006, B220+ cells P = 0.8157, unpaired two-tailed Student’s t-test; ns, not significant). b, Representative histo-cytometry plots of meningeal whole-mounts stained for IgA (red) and B220 (green) from a specific-pathogen-free (SPF) mouse, a germ-free (GF) mouse and a GF mouse reconstituted with murine gut commensal bacteria by oral gavage (MGC-GF). c, Representative images of IgA (red) and B220 (green) immunolabeling of the SSS from whole-mount meninges from SPF, GF, and MGC-GF mice (scale bar, 100 μm). d, Quantification of peri-sinus IgA+ cells from SPF and GF (left panel); GF mice colonized by oral gavage with a murine gut commensal bacteria (MGC-GF), a human gut microbiome from two separate donors (DA-GF and DB-GF), or segmented filamentous bacteria (SFB-GF); or, GF mice with skin mono-colonized by S. epidermidis (S. epidermidis-GF) or S. aureus (S. aureus-GF) (mean ± s.d., n = 8 SPF mice; 9 GF mice; 5 MGC-GF, DA-GF, DB-GF and S. epidermidis-GF mice; and 4 SFB-GF and S. aureus-GF mice; SPF ****P = <0.0001, MGC-GF ***P = 0.0010, DA-GF ***P = 0.0010, DB-GF ***P = 0.0010, SFB-GF **P = 0.0028, S. epidermidis-GF P = 0.8731, S. aureus-GF P = 0.6350, all statistical comparisons were made with the indicated group against GF group, two-tailed Mann-Whitney test; ns, not significant). e, Normalized counts of meningeal Igha expression by RNA-seq in the denoted groups (n = 3 mice per group). f, Quantification of IgG+ cells along the dural sinuses in the denoted groups (mean ± s.d., n = 5 mice per group; SPF P = 0.2376, DA-GF **P = 0.0079, DB-GF P = 0.9163, all statistical comparisons were made with the indicated group against GF group, two-tailed Mann-Whitney; ns, not significant). g, Representative images of IgA (red) immunolabeling and phalloidin stain (gray) of the SSS region of meningeal whole-mounts from a naive GF mouse (GF naive), or a GF mouse infected with Citrobacter rodentium wild type (GF + wt) or a ΔEspA mutant (GF + ΔEspA) (scale bar, 100 μm). h, Quantification of peri-sinus IgA+ cells from the indicated groups (mean ± s.d., n = 5 mice in naive SPF, naive GF and GF ΔEspA groups and n = 4 in GF wt and SPF wt groups; SPF naive vs. SPF wt *P = 0.0206, GF naive vs. GF wt *P = 0.0159, GF naive vs. GF ΔEspA **P = 0.0079, two-tailed Mann-Whitney test). i, Frequency of IGA BCR clones in small intestines and meninges of SPF mice presented as proportional Euler diagrams. The intersect of the overlapping diagrams represents the frequency of BCR clones shared between the two tissues. The percentage values are calculated relative to total clones in meningeal tissues. j, Antibody lineage tree of representative IGA clone shared between meninges (red nodes) and small intestines (blue nodes) from a SPF mouse. The black node indicates the germline IGA sequences while white nodes indicate inferred sequences. The edge number indicates the distance (the number of the substitutions) between each connecting node. The CDR3 sequences of the shared clone is also shown. Positions of nucleotide bases that are different, resulting in synonymous and non-synonymous amino acid changes, are highlighted.
To test whether the presence of a single bacterial species within the intestine was sufficient to restore dural IgA+ plasma cells, we infected GF mice with wild type (WT) and an avirulent mutant (ΔEspA) of Citrobacter rodentium18, a model of human attaching-effacing enteropathogenic infection. In GF mice infected with ΔEspA C.rodentium, a mutant that is incapable of epithelial cell invasion19 and therefore of systemic spread18, we observed similar numbers of IgA+ plasma cells within the meninges as SPF mice or mice infected with WT C.rodentium (Fig. 2g–h). The meningeal IgA+ cell compartment was also replenished following reconstitution of GF mice with segmented filamentous bacteria (SFB) (Fig. 2d), another gut-restricted commensal20. By contrast, skin-limited monocolonization of GF mice with either S. epidermidis or S. aureus did not restore meningeal plasma cells (Fig. 2d). Collectively, these data suggest an intimate relationship between intestinal and meningeal plasma cells. We postulated the latter might be educated in the gut and arise from intestinal B cell clones that subsequently localize to the dura mater and differentiate into plasma cells in situ. Alternatively, plasmablasts or plasma cells might migrate to the meninges after differentiation in the intestines.
To better understand this relationship, we performed B cell receptor (BCR) sequencing on paired meningeal and small intestinal tissue (1cm of jejunum) samples. In SPF mice, 21.5% of meningeal clones were related to those in the small intestine, despite the limited proportion of intestine sampled relative to the total intestinal length (approximately 50cm)21, confirming a substantial overlap between intestinal and meningeal BCR specificities (Fig. 2i), significantly greater than would be expected by random chance (Extended Data Fig. S3a). Of the shared clones, a greater proportion of intestinal BCR sequences were found to lie closer to germline sequences compared to meningeal clones, (65% versus 35%), consistent with an intestinal origin of meningeal plasma cells, but raising the possibility of bi-directional movement between the meninges and intestine (Extended Data Fig. S3b). Overall, the BCR repertoire was less diverse in the meninges (Extended Data Fig. S3c), and there was evidence of clonal expansion, with dissimilarity in the most abundant meningeal and intestinal clones as well as some unique BCR sequences in the meninges (Fig. 2j, Extended Data Fig. S3d). Similarly, when comparing BCR sequences in intestine and meninges of GF-DA and GF-DB mice, there was overlap of BCR sequences detectable in intestine and meninges as well as evidence of clonal expansion in the meninges (Extended Data Fig. S3e–f). These data show that a considerable proportion of dural B cells and/or plasma cells present in homeostasis arise from gut-educated B cells or plasma cells and may undergo local expansion.
Given their intestinal origin, we next asked how intestinal inflammation and barrier breach would impact meningeal humoral immunity. Following oral administration of dextran sodium sulphate (DSS), a colitogen that causes osmotic damage to intestinal epithelial cells resulting in barrier breach and inflammation, there was a significant increase in meningeal B cells, particularly naïve and IgM+IgD- cells as well as IgA+ plasma cells (Extended Data Fig. S4a–d). Many B cells were found within clusters adjacent to the dural sinuses with IgA+ plasma cells located at the periphery of the clusters, and these cells were outside of Lyve-1+ dural lymphatics (Extended Data Fig. S4a). Some B cells within these clusters were Ki67+, indicating local proliferation (Extended Data Fig. S4e). RNA sequencing of meninges post-DSS colitis confirmed an increase in IgA heavy chain transcripts as well as a modest (but not significant) increase in the abundance of some IgG heavy chain transcripts (Extended Data Fig. S3g, S4d).
In the gut, IgA plays an important role in maintaining barrier integrity by trapping microbes within the mucus layer keeping them away from the epithelium22. In addition, protective IgA antibodies develop following vaccination and challenge with bacteria, viruses and fungi23,24. These antibodies may opsonize pathogens for uptake by phagocytes, induce antibody-dependent cellular cytotoxicity, and mediate proteasomal destruction of intracellular pathogen23,25. Given the localization of meningeal IgA+ plasma cells adjacent to dural sinuses, we hypothesized that the IgA secreted locally may form part of a peri-sinus barrier that prevents spread of intravenous pathogens from the slow-moving blood within the sinus into the CNS. To test this hypothesis in vivo, we challenged mice with intravenous Candida albicans, a cause of meningoencephalitis in neonates and the immunocompromised26. Candidal species also form part of the normal microbiome in mice and humans27. Following candida challenge, there was marked expansion of B and IgA+ plasma cells as well as fungal entrapment along the wall of the dural sinuses (Fig. 3a, Extended Data Fig. S5a, Video 2) within areas including CD45+ cells and diffuse extracellular IgA staining (Fig. 3b, Extended Data Fig. S5b). Some of the IgA+ cells also expressed Ki-67 (Extended Data Fig. S5c) and activation-induced cytidine deaminase (AID) (Extended Data Fig. S5d), suggesting local proliferation and somatic hypermutation.
Fig. 3. Meningeal IgA entraps fungi in the dural sinuses and protects the brain from infection.
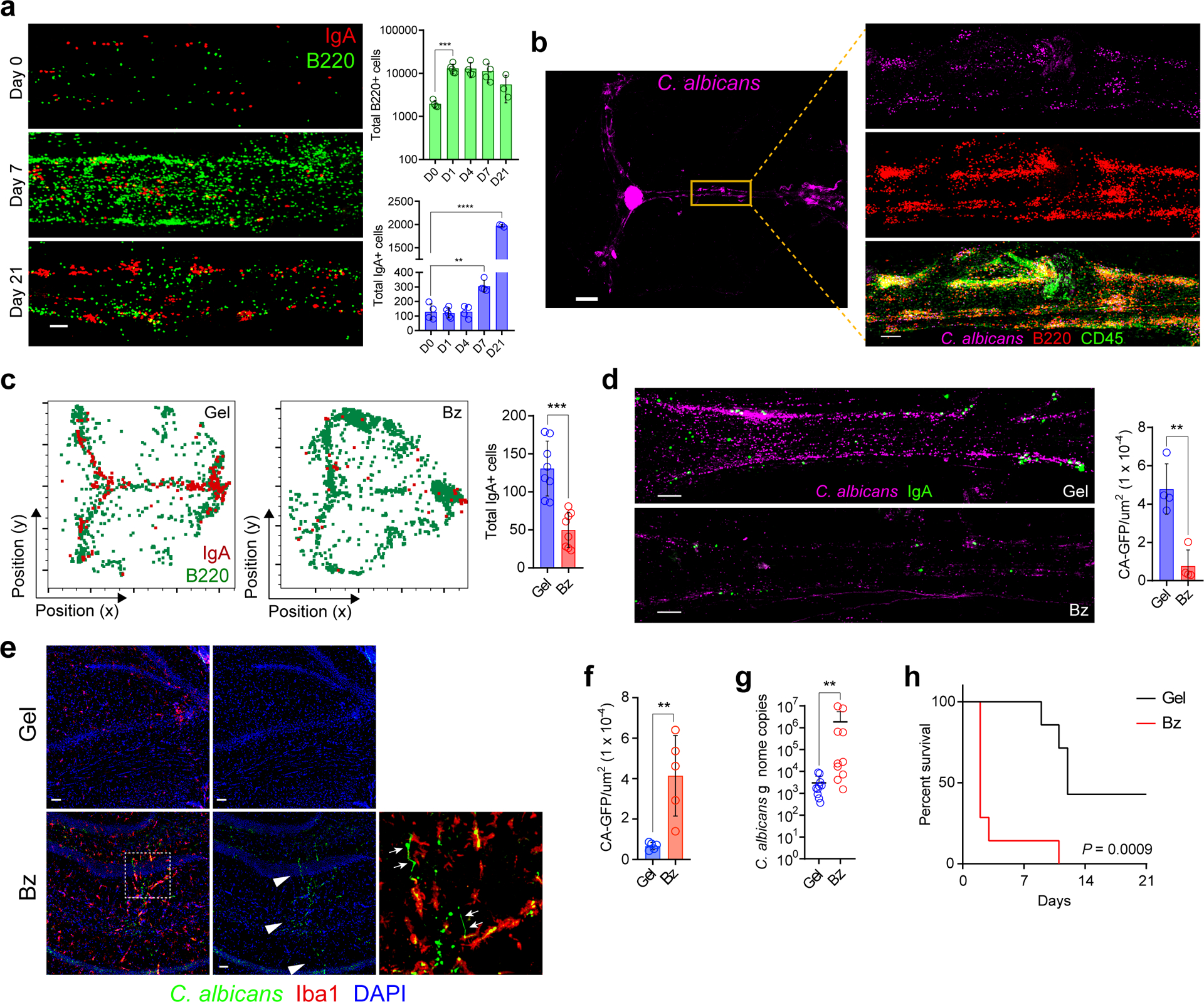
a, Representative images of the meningeal SSS from whole-mounts immunolabeled for IgA (red) and B220 (green). Mice were examined at day 0 (D0) after injection of sterile PBS or at day 7 (D7) and 14 (D14) after i.v. administration of 5 × 104 viable C. albicans cells (scale bar, 100 μm). Quantification of peri-sinus B220+ and IgA+ cells at the indicated time points post-infection are displayed in bar graphs (mean ± s.d., n = 4 mice in D0, D4 and D7 groups and 5 mice in D1 group and 3 mice in D21 group; B220+ cells D0 vs D1 *P = 0.0159, IgA+ cells D0 vs. D7 **P = 0.0021, IgA+ cells D0 vs. D21 ****P = <0.0001, unpaired two-tailed Student’s t-test). b, Representative image of a meningeal whole-mount from a mouse 2 days after i.v. administration of GFP-C. albicans displaying sequestration of the pathogen (purple) in the peri-sinus region (scale bar, 1000 μm); the inset is a higher magnification of the highlighted box showing B220+ and CD45+ cells (red) surrounding sequestered GFP-C. albicans (scale bar, 200 μm). c, Representative histo-cytometry plots of meningeal whole-mounts stained for B220+ (green) and IgA+ cells (red) from a mouse that received a sub-scalp injection of control hydrogel (Gel) or bortezomib hydrogel (Bz) to deplete meningeal plasma cells. Bar graph depicts quantification of peri-sinus IgA+ cells from each group (mean ± s.d., n = 8 mice per group; ***P = 0.0001, unpaired two-tailed Student’s t-test, data are representative of three independent experiments). d, Representative SSS images stained for IgA (green) from the indicated groups 2 days following i.v. administration of GFP-C. albicans shown in purple (scale bar, 150 μm). Bar graph shows quantification of GFP-C. albicans+ puncta along the SSS normalized by area in each group (mean ± s.d., n = 4 mice per group; **P = 0.0021, unpaired two-tailed Student’s t-test, data are representative of three independent experiments). e, Representative hippocampal images stained for Iba1 (red) from the indicated groups 2 days after GFP-C. albicans infection (shown in green; white arrowheads) (scale bar, 50 μm). f, Bar graph depicts quantification of hippocampal GFP-C. albicans+ puncta (mean ± s.d., n = 5 mice per group, **P =0.0046, unpaired two-tailed Student’s t-test). g, Quantification of C. albicans genome copies in the brain by qPCR 3 days after i.v. administration of C. albicans in the indicated groups (mean ± s.d., n = 10 mice per group **P = 0.0021, two-tailed Mann-Whitney test). h, Survival of mice in the indicated groups after i.v. injection of C. albicans (n = 7 mice per group; ***P = 0.0009, Mantel-Cox test, data shown as percent survival over time and representative of two independent experiments).
In IgA-deficient mice lacking meningeal IgA+ cells (Extended Data Fig. S6a), we observed a failure to confine candidal spread to the peri-sinus regions (Extended Data Fig. S6b) and a concomitant increase in fungal spread to the brain parenchyma (Extended Data Fig. S6c). To determine whether local rather than global IgA was required for pathogen defense in the meninges, we depleted meningeal plasma cells by applying transcranial bortezomib (Fig. 3c, Extended Data Fig. S7a), a proteasome inhibitor28. This treatment depleted meningeal plasma cells but had limited effects on other meningeal immune cells (Extended Data Fig. S7b–g) and distant plasma cells (Extended Data Fig. S8a–e). Following intravenous candida challenge, depletion of meningeal plasma cells decreased fungal entrapment adjacent to dural sinuses (Fig. 3d), increased fungal load in brain tissue (Fig. 3e–g) and increased mortality (Fig. 3h).
CNS barrier structures such as the meninges, choroid plexus, vasculature, and sinuses interface with the periphery and must be defended by a network of resident immune cells5,6,29. The dural venous sinuses are a vulnerable CNS barrier surface, because the slow flow of blood and limited shear forces within them provide an opportunity for blood-borne pathogens to adhere to the fenestrated sinus wall2. We demonstrate that IgA-secreting plasma cells contribute to an immunological barrier at this interface, providing protection against the spread of pathogens into the CNS (Extended Data Fig. S8f). Some of these IgA cells are educated in the gut, an organ containing trillions of microorganisms, including bacteria, fungi and viruses. While our study focuses on gut-education of meningeal IgA cells, our data do not rule out other sites of education (besides the skin). The intestinal immune system is orchestrated to permit the existence of the microbiome for its beneficial effects, while preventing mucosal barrier breach. IgA plays an important role in this process by enchaining microbes and facilitating their entrapment within the mucus layer22,30. Here we demonstrate that this paradigm also applies to a distinct internal barrier interface — the dural venous sinuses. We propose that evolutionary pressures have led the dural IgA antibody repertoire to mirror that of the gut because even a transient intestinal barrier breach can cause bacteremia with devastating consequences if there is spread into the CNS. Therefore, seeding the meninges with antibody-producing cells that are selected to recognize luminal commensals ensures defense against the most likely invaders. In conclusion, meningeal IgA cells represent a crucial immunological shield assembled during homeostasis and poised to prevent the spread of pathogens into the meninges and underlying CNS parenchyma.
Methods:
Human subjects.
Human dura mater tissue for Figure 1j was obtained from a 73-year-old man with combined small cell lung cancer/adenocarcinoma who was treated with the following regimens, in sequence: carboplatin/pemetrexed, carboplatin/etoposide, combined PD-1/CTLA4 blockade, and carboplatin/etoposide. Anti-PD-1/CTLA4 treatment was administered for 2 months and stopped 9 months before death. He underwent gamma-knife radiosurgery 19 months after diagnosis for a single cerebellar metastasis, followed by bilateral adrenalectomy 3 months later for symptomatic Cushing’s disease. He received 2 cycles of investigational agents at the National Cancer Institute but continued to have disease progression, including lymphangitic spread to both lungs and new bilateral multiple brain metastases. No metastatic involvement of the dura mater was noted on in vivo brain imaging. Tissue was harvested under an NIH IRB-approved protocol following consent from the next of kin.
The dura mater sample in Extended Data Figure 1f was obtained by a study approved by the Health Research Authority England (HRA) and Health and Care Research Wales (HCRW) (protocol (East of England LREC 17/EE/0485). A surgical tract meningeal biopsy was taken (with informed consent from the patient) from an otherwise healthy 72-year-old female patient undergoing resection of a benign intracranial tumor in the left frontal lobe at the Cambridge University NHS Foundation Trust Hospital. The patient did not receive any adjuvant chemo or radiotherapy prior to the operation. There was no tumor involvement of the resected dura mater on preoperative MRI imaging or post-hoc pathohistological analysis.
Mice.
C57BL/6J, B6.129P2-Tcrbtm1Mom/J (Tcrb−/−), B6.129P2-Cd19tm1(cre/ERT2)Rsky/J (CD19CreER/CreER) and B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (Stopfl/flTdTomato) mice were obtained from Jackson Laboratories. Blimp1EYFP/EYFP (Prdm1EYFP/EYFP) mice were obtained from M. Nussenzweig (Rockefeller University). CD19CreER/+ Stopfl/flTdTomato x Prdm1EYFP/+ reporter mice were generated by first crossing CD19CreER/CreER with Stopfl/flTdTomato mice, and offspring were then bred to Prdm1EYFP/EYFP mice. For induction of Cre recombinase, CD19CreER/+ Stopfl/flTdTomato x Prdm1EYFP/+ mice received tamoxifen formulated in chow (250 mg/kg; Envigo) for 3 to 4 weeks. IgA−/− mice were generously provided by Y. Belkaid (NIAID). All above mouse strains were bred and maintained under specific pathogen-free conditions at the National Institutes of Health (NIH). Germ-free C57BL/6 mice were bred and maintained in the NIAID Microbiome Program gnotobiotic animal facility or the Wellcome Sanger Institute germ free facility. Both male and female mice were used in experiments, and all were 10–18 weeks of age unless otherwise indicated. Sample size was chosen based on previously published animal experiments in the field of meningeal immunity. Experiments were not randomized or blinded.
Immunohistochemistry of murine meningeal whole-mounts and brain.
Immunostaining of mouse whole-mount meninges was performed as previously described31 with the following modifications. Immediately after death, animals were transcardially perfused with 20 mL of ice-cold PBS. Skull caps, with intact dura mater, were dissected as described above and placed in 2% paraformaldehyde for 16–18 hours at 4°C. The fixed tissue was then washed with PBS three times in PBS for 5 minutes per wash. Under a dissecting microscope, the dura mater was scored carefully with forceps from the skull cap, leaving the meninges intact, in a petri dish containing PBS. Meninges were then stored at 4°C in PBS within a 24-well plate until use for immunostaining.
For immunostaining of whole-mount meninges, the tissue was immersed in a block-permeabilization solution containing 2% normal rat serum, 1% bovine serum albumin, 0.1% Triton-X-100 and 0.05% Tween in PBS for 1 hour at room temperature. Subsequently, meninges were stained for antigens with anti-mouse antibodies in 1% bovine serum albumin and 0.5% Triton-X-100 in PBS at 4°C overnight. A combination of the following primary antibodies were used for immunostaining of whole-mount meninges: anti-CD138 (281–2, BD Biosciences), anti-CD45.2 (104, BioLegend), anti-IgA (catalog# 1040–09, SouthernBiotech), anti-IgG (catalog# 1030–02, SouthernBiotech), anti-B220 (RA3–6B2, Thermo Fisher), anti-IgA (RMA-1, BioLegend), anti-Lyve1 (223322, R&D), anti-Ki67 (SolA15, Thermo Fisher), anti-AICDA (catalog# ab93596, Abcam), anti-J-chain (SP105, Thermo Fisher), anti-GFP (catalog# A-21311, Thermo Fisher), and anti-rabbit IgG (catalog# A27040, Thermo Fisher). All primary antibodies were used at a concentration of 1:200 and secondary antibodies at 1:600. Following staining, tissue was washed 3 times in PBS for 5 minutes per wash at room temperature. When non-conjugated primary antibodies were used, anti-rabbit IgG (A27040, Thermo Fisher) was then applied in PBS for 1 hour at room temperature; the tissue was then washed three times in PBS for 5 minutes per wash. Using a paintbrush, the dura mater was flattened on a glass slide and mounted with FluorSave Reagent (Millipore Sigma). Slides were left in the dark at 4°C for 1 hour before imaging.
Upon perfusion and harvesting, brains were placed in 2% paraformaldehyde for 16–18 hours at 4°C, washed, and placed in 25% sucrose solution for 2 days for cryopreservation. Brains were then embedded in OCT medium, frozen over dry ice and 60 μm sections were made using a cryostat. Brain sections were blocked and stained on slides as above.
Tile scan images of whole-mount meninges or brain slices was carried out either on a Leica SP8 confocal microscope (Wetzlar, Germany) or Olympus FV1200 laser-scanning confocal microscope equipped with four detectors, six laser lines (405, 458, 488, 515, 559 and 635 nm) and five objectives (4×/0.16 NA, 10×/0.4 NA, 20×/0.75 NA and 40×/0.95 NA, and chromatic aberration-corrected 60×/1.4 NA). All images were analyzed using Imaris version 9.3 software (Bitplane).
Immunohistochemistry of human whole-mount dura mater.
Human dura mater was obtained post-surgically and fixed in 2% paraformaldehyde for 16–18 hours at 4°C. The tissue was then washed in PBS for 5 minutes at room temperature three times. The superior sagittal sinus and surrounding dura were dissected out using a scalpel and forceps, and any residual blood within the sinus was removed. The sinus was then segmented into 2 mm pieces and stored at 4°C in PBS within a 6-well plate. The immunostaining protocol is the same as above using the following anti-human antibodies at a concentration of 1:200: anti-IgA (catalog# ab97000, Abcam) and anti-CD138 (MI15, BioLegend). After staining, the sinus was cut open so that the luminal aspect of the vessel could be imaged and mounted with FluorSave Reagent (Millipore Sigma). Tile scan images were acquired with an Olympus FV1200 laser-scanning confocal microscope and analyzed using Imaris version 9.3 software (Bitplane).
Acute colitis model.
C57BL/6J mice were administered 2% weight/volume dextran sulfate sodium (MP Biomedicals) in drinking water for 7 days to induce acute colitis and intestinal barrier breach. Animals were then left on untainted drinking water for 14 days post DSS administration. Body weight and stool was monitored daily and used as indicators of severity of intestinal inflammation. On culling, animals were perfused transcardially with ice-cold PBS and appropriate tissues were collected for downstream processing.
Fungal challenge.
C. albicans-GFP (strain SC5314) was a generous gift from M. Lionakis (NIAID). A single colony grown 48 hours at 37°C on YPD (yeast extract peptone dextrose) agar plates was used to inoculate YPD broth containing penicillin and streptomycin (Mediatech). Yeast was grown at 30°C in a shaking incubator and serially passaged three times, with growth periods ranging from 18–24 hours at each passage. Upon the final passage, yeast cells were harvested by centrifugation at 1400 rpm for 7 minutes and washed twice with PBS. A hemocytometer was used to count yeast cells, and a suspension of 50,000 cells per 100 μL of sterile PBS was used for infections. To induce systemic candidiasis, 100 μL of C. albicans suspension was administered intravenously, and survival following infection was monitored daily.
Quantification of C. albicans by qPCR.
Quantitative PCR was used to assess fungal burden in brains after systemic infection with C. albicans as previously described32. Brains were harvested from mice transcardially perfused with 20 mL of ice-cold PBS and placed directly into lysis buffer from DNeasy Blood and Tissue Kit (Qiagen) and homogenized with tissue homogenizer (MP Biomedicals). Total DNA from 20 μg of brain homogenate was extracted using DNeasy Blood and Tissue Kit (Qiagen). Using a CFX96 Real-Time PCR machine (BioRad Laboratories), real-time PCR was performed with 40 ng of DNA per sample using SYBR Green (Applied Biosystems) or water (non-template negative control) with the following C. albicans-specific primers: forward primer - ACT TCT GTA AGA GTG CTG GTTC and reverse primer - GCA TGC CAG GAG AGT GTA AA (Integrated DNA Technologies). The following cycling conditions were used: an initial denaturation step of 95°C for 10 minutes followed by 40 cycles of 95°C for 5 seconds and 60°C for 30 seconds. Reactions were performed in duplicates and with generation of melting curves to confirm purity post DNA amplification. The number of C. albicans genome copies was quantified by normalizing against a series of standards that consisted of DNA isolated from brain homogenates from uninfected mice spiked with known CFU quantities of C. albicans.
Meningeal plasma cell depletion.
A stock solution of bortezomib (Santa Cruz Biotechnology) was made by dissolving 2.5 mg of bortezomib in 100 μL of sterile DMSO. Bortezomib was then diluted to a concentration of 40 μg/mL using sterile PBS and hydrogel was formulated by dissolving carboxymethycellulose (10 mg/mL, Sigma-Aldrich) in this solution inside of a beaker under agitation. Upon homogenization, bortezomib-containing hydrogel, as well as mock hydrogel made without bortezomib, were collected into a syringe. A subcutaneous injection of 300 μL of hydrogel under the scalp was administered to adult mice. Four days later, mice received a second dose of hydrogel, and the extent of plasma cell depletion was determined two days later. When indicated, animals were infected intravenously on the same day that the second dose of hydrogel was administered.
Flow cytometry.
For mononuclear cell isolation, mice were anesthetized and perfused transcardially with 20 ml of ice-cold PBS. Meningeal tissue was carefully removed from the underside of the scull using forceps under a dissecting microscope and subject to enzymatic digestion in 2.5 mg/mL collagenase D (Roche) and 0.1 mg/mL DNase (Roche) for 30 minutes at 37°C with gentle shaking every 10 minutes. Single-cell meningeal suspensions were then washed with PBS containing 2% fetal bovine serum before blocking with 5 μg/mL rat anti-mouse CD16/32 (BD Biosciences) and 0.5 mg/mL normal rat serum for 10 minutes on ice to reduce non-specific antibody binding. Splenocytes and femur bone marrow cells were subject to red blood cell lysis buffer, washed twice with PBS containing 2% fetal bovine serum, and resuspended in the same solution before blocking. Counting beads (Thermo Fisher) were added to each sample once a single cell suspension was made, when indicated. After washing, LiveDead fixable Blue Cell Staining Kit (Invitrogen) was used to exclude dead cells from the analysis. The cell suspensions were then washed and stained with the following cell surface anti-mouse antibodies: CD45.2 Brilliant Violet 785 (104, BioLegend), B220 FITC (RA3–6B2, Thermo Fisher), CD19 Pacific Blue (B4, BioLegend), CD79b APC (HM79–12, BioLegend), IgM Brilliant Violet 711 (RMM-1, BioLegend), IgD PE/Dazzle (11–26c.2a, BioLegend), IgA PE (catalog# 1040–09, SouthernBiotech), CD138 APC (281–2, BD Biosciences), CD3ε PE-Cy7 or FITC (145–2C11, BioLegend), CD4 APC/Cy7 (RM4–5, BioLegend), CD8 PECy7 (53–6.7, BioLegend), CD11b Pacific Blue or Brilliant Violet 605 (M1/70, BioLegend), Ly6C (HK1.4, BioLegend), I-A/I-E APC (M5/114.15.2, BioLegend) and CD11c (N418, BioLegend). Samples were washed and directly acquired on a BD FACSymphony A-5, and data were analyzed using FlowJo software version 9.7.2 (Tree Star).
Intravital two-photon microscopy.
Upon delivery of anesthesia, thinned skull windows were prepared as described previously33. To visualize meningeal vasculature, 50 μg of Evans blue in sterile PBS was administered intravenously before imaging, when indicated. A Leica SP8 two-photon microscope was used to capture three-dimensional time-lapse movies. This microscope is equipped with an 8,000 Hz resonant scanner, a x25 color-corrected water-dipping objective (1.0 NA), a quad HyD external detector array, a Mai Tai HP DeepSee Laser (Spectra-Physics) which was tuned to 930 nm (for Evans blue and EYFP) and an Insight DS laser (Spectra-Physics) tuned to 1,050 nm (for red fluorescent protein). Videos were captured in z-stacks of 15–35 planes (2 μm step size) at 1- to 2-minutes intervals. Signal contrast was optimized by averaging 8 video frames per plane in resonance scanning mode. Acquired videos were analyzed using Imaris version 9.3 software (Bitplane).
Confocal image analysis.
Confocal tile scan images were imported into Imaris version 9.3 (Bitplane). A surface of the peri-sinus region, including the superior sagittal and transverse sinuses, was made by manual drawing using the surface function, and the area in μm2 was determined by exporting surface statistics. A quantification of peri-sinusoidal cell numbers was generated by identifying and thresholding on positively stained cells within the three-dimensional surface of each respective channel using the spot-detection function. Quantification of GFP-C. albicans in hippocampal images was done by thresholding on and counting GFP+ puncta using the same function. Statistics were then exported into Excel (Microsoft) for further analysis.
Histocytometry plots were generated by either creating a surface of the peri-sinus region as above or the entire meningeal whole-mount, creating a value-based visual surface for all positively stained cells in each channel. This allows for quantification of fluorescent intensity and frequency of labeled and unlabeled cells, as well as their position in an x-y plane. Channel statistics were then exported into Excel (Microsoft), and position of each cell and mean voxel fluorescence, when indicated, was plotted in FlowJo software by utilizing the text to FCS conversion function (Tree Star).
Topical association.
Staphylococcus epidermidis strain NIHLM087 and Staphylococcus aureus strain 42F02 were cultured for 18 hours in tryptic soy broth at 37°C. As described previously34 topical association of bacteria was accomplished by placing 5 ml of the bacterial suspension (approximately 109 CFU/mL) across the entire skin surface (approximately 36 cm2) using a sterile cotton swab. For topical association of bacteria, each mouse was associated by placing 5 ml of the bacterial suspension (approximately 109 CFU/mL) across the entire skin surface (approximately 36 cm2) using a sterile cotton swab. Application of bacterial suspension was repeated every other day a total of four times. Bacteria were enumerated prior to topical application by assessing colony-forming units (CFU) using traditional bacteriology techniques and by measuring optical density (OD) at 600 nm using a spectrophotometer. In experiments involving topical application of various bacterial strains, 18-hour cultures were normalized using OD600 to achieve similar bacterial density (approximately 109 CFU/mL). Skin colonization was confirmed by counting CFUs from a skin swab after mono-association, and mice were analyzed 2 weeks after the final topical association.
Antibiotic treatment.
Intestinal commensal bacteria were depleted in C57BL/6J mice by provision of broad-spectrum antibiotics in drinking water for 6 weeks. The following regimen was used: ampicillin (1 g/L; Sigma-Aldrich), vancomycin (500 mg/L; Sigma-Aldrich), neomycin sulphate (1 g/L; Sigma-Aldrich) and metronidazole (1 g/L; Sigma-Aldrich).
Intestinal colonization.
The following bacteria were grown overnight in Fastidious Anaerobic Broth in anaerobic or aerobic conditions: Escherichia coli, Bacteroides finegoldii, Bacteroides fragilis, Bacteroides intestinalis, Bacteroides 1_1_30, Bacteroides caccae, Parabacteroides merdae, Lachnoclostridium clostridioforme, Hungatella hathewayi, Paraclostridium sp. AS15, and Bifidobacterium longum. Each culture was then back-diluted to an OD600 = 1, and 1 mL of each back-diluted culture was combined to make a master-mix, which was then used to gavage into germ-free mice at birth. Mice received a gavage of this bacterial community every other day for a total of three times. Mice were euthanized and studied 8 weeks post colonization.
For SFB-mono-association, frozen feces previously collected from SFB-mono-associated germ-free mice were homogenized in sterile PBS and the resulting suspension was filtered through a 40 mm cell strainer to obtain a solution cleared of solids. Germ-free C57Bl/6 mice were inoculated orally by gavage with 300 ml of this cleared solution inside gnotobiotic isolators. Gavage was repeated 48 hours later. The equivalent of the contents of 1–2 fecal pellets was inoculated in each mouse for each gavage. SFB colonization was confirmed by performing PCR on DNA extracted from feces collected from mice 4 days after the second gavage using primers specific for SFB 16S rRNA gene (SFB736F: 5’-GACGCTGAGGCATGAGAGCAT-3’; SFB844R: 5’-GACGGCACGGATTGTTATTCA-3’). Mice were studied 4 weeks after colonization.
Citrobacter infection.
Citrobacter rodentium ICC180 and C. rodentium ΔEspA bacterial inoculums (gifted from Gad Frankel, Imperial College London) were prepared by culturing bacteria overnight at 37°C in 100 mL of Luria Bertani broth (LB) supplemented with nalidixic acid (100 μg/mL), with shaking (220 rpm). Cultures were harvested by centrifugation and resuspended in a 1:10 volume of Dulbecco’s phosphate-buffered saline (D-PBS). Mice were orally inoculated under anesthesia by using a gavage needle with 200 μL of the bacterial suspension.
Human gut microbiome donor mouse lines.
Donor A and donor B mouse lines were generated as follows. Fresh donor feces were homogenized at 100 mg/mL in D-PBS.Fresh feces were processed within 1 hour of it reaching the lab and all preparation was done in an anaerobic cabinet (80% CO2, 10% H2, 10% N2). 5 male and 5 female germ-free mice were inoculated by oral gavage with 200 μL of the donor homogenates once a week for 3 weeks. 1 week after the final gavage mice were set up as breeding pairs in a decontaminated positive pressure isolator. All consumables that enter the isolator for maintenance of the colony were autoclaved 121°C for 15 mins. Feces of mice from the 2 lines were compared to the original donor feces to check engraftment of the human flora. Animals used for experiments were removed from the isolators in sealed ISOcages and maintained on a positive pressure ISOrack (Tecniplast).
RNA-seq.
Meningeal tissue was stabilized in RNAlater (Thermo Fisher) and then placed into lysis buffer and homogenized using a Precellys (Bertin instruments). RNA was subsequently extracted using the RNA pure link mini kit (Thermo Fisher) as per manufacturer’s instructions. RNA integrity was assessed using an RNA 6000 nano chip on a Bioanalyzer (Agilent). Libraries were prepared as per manufactures instructions using the TruSeq Stranded Total RNA kit with RiboZero. Libraries were checked for size with a high sensitivity DNA chip on a Bioanalyzer (Agilent), quantified using Kappa library quantification kit (Kappa biosystems) and pooled at an equimolar ratio. Library pools were sequenced on a Hiseq 4000 by Genewiz.
Data were demultiplexed using Casava (Illumina) to generate Fastq files. These were aligned to the mm10 genome with Hisat2 and a counts table produced using Featurecounts (RSubread). Normalization and differential expression analysis were carried out using DESeq2 and pathways analysis using the GSEA java application (Broad) against pathways found within the MSigDB database.
Sequencing data has been made available on GEO under accession number GSE135620 and GSE135733.
BCR-seq sample preparation.
RNA from paired whole meningeal and small intestinal tissue generated from 1 cm of jejunum sample was extracted as described above. Reverse transcription (RT) was performed in two steps: First, 1 μL barcoded reverse primer mix (10 μM per each primer), 1 μL dNTP (10 mM) and 14 μL RNA template (up to 500 ng) were mixed and heated to 65°C for 5 minutes followed by an immediate transfer to ice for at least one minute. Second, mixture 2 including 4 μL 5X First-Strand Buffer, 1uL DTT (0.1 M), 1 μL RNaseOUT™ (Thermo Scientific) and 1 μL SuperScript®III (Life Technologies) was added and incubated at 50°C for 60 minutes followed by 15 minutes inactivation at 70°C. cDNA was cleaned-up with Agencourt AMPure XP beads and PCR amplified with V-gene multiplex primer mix (10 μM each forward primer) and 3′ universal reverse primer (10 μM) using KAPA protocol (KAPA Biosystems) and the thermal cycling conditions: 1 cycle (95°C—5 minutes); 5 cycles (98°C—5 seconds, 72°C—2 minutes); 5 cycles (98°C—5 seconds, 65°C—10 seconds, 72°C—2 minutes); 30 cycles (98°C—20 seconds, 60°C—1 minute, 72°C—2 minutes) and 1 step (72°C—7 minutes). Primers are provided in Key Resource Table.
BCR-sequencing and analysis.
MiSeq libraries were prepared using Illumina protocols and sequenced using 300 bp paired-ended MiSeq (Illumina). Raw MiSeq reads were filtered for base quality (median Phred score > 32) using QUASR (http://sourceforge.net/projects/quasr/)35. MiSeq forward and reverse reads were merged together if they contained identical overlapping regions of >50 bp, or otherwise discarded. Universal barcoded regions were identified in reads and orientated to read from V-primer to constant region primer. The barcoded region within each primer was identified and checked for conserved bases (i.e. the T’s in NNNNTNNNNTNNNNT). Primers and constant regions were trimmed from each sequence, and sequences were retained only if there was >80% sequence certainty between all sequences obtained with the same barcode, otherwise discarded. The constant region allele with highest sequence similarity was identified by 10-mer matching to the reference constant region genes from the IMGT database36, and sequences were trimmed to give only the region of the sequence corresponding to the variable (V–D–J) regions. Isotype usage information for each IgH was retained throughout the analysis hereafter. Sequences without complete reading frames and non-immunoglobulin sequences were removed and only reads with significant similarity to reference IgH V and J genes from the IMGT database were retained using BLAST37. IGHV and IGHJ genes and mutational status were determined for each BCR using IMGT/V-QUEST38.
SHM levels (including silent and non-silent mutations) per unique IGHV-D-J region per isotype were calculated for each individual sample using observedMutation function within SHazaM package39. BCR clones were assigned using the changeo39 package after calculation of length normalized distance of each sequence in the data and its nearest-neighbor using the single nucleotide Hamming distance model. The network generation algorithm and network properties were calculated as previously described40. Briefly, each vertex represents a unique sequence. Edges are generated between vertices that differ by single-nucleotide, non-indel differences and clusters are collections of related, connected vertices. Lineage trees were generated using buildPhylipLineage function within Alakazam package39 after merging sequences from paired meningeal and small intestinal samples. The tissue assignment (meningeal or small intestinal) for the most recent common ancestor, defined as the node/BCR that is the most proximal to the germline node, of each reconstructed lineage tree was extracted. Statistical analysis on the difference in proportion of MRCA assigned to each tissue was performed in Prism using an unpaired two-tailed Student’s t test. VDJtools41 was used to analyze the BCR sequencing data for diversity estimation of CDR3 sequences (Chao1); the diversity estimates were adjusted for sequencing depth via subsampling with 1000 random iterations. Statistical tests were performed in R using Wilcoxon tests for significance (non-parametric test of differences between distributions).
Shotgun metagenomic sequencing and analysis.
Whole-genome metagenomic sequencing was performed on the Illumina HiSeq 2500 as described previously42. Taxonomic classification from filtered metagenomics reads was performed using Kraken v1.0 and a database comprising complete, high-quality reference bacterial genomes. Metagenomic samples were compared at the species levels by relative abundance. The R package microbiome and R scripts described previously42 were used for metagenomic data analysis and results were visualized using R package ggplot2.
Statistical analysis.
Results herein are represented as mean ± standard deviation (s.d.). Statistical significance was determined using GraphPad Prism 8.0 by two-tailed Student’s t-test or Mann-Whitney rank sum test. Statistical significance was determined by a P < 0.05.
Ethics statement.
For experiments performed in the UK, mice were maintained in specific pathogen-free conditions at a Home Office-approved facility in the UK. All procedures were carried out in accordance with the United Kingdom Animals (Scientific Procedures) Act of 1986. For experiments performed in the US, all work was conducted in strict accordance with recommendations within the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All protocols were approved by the NINDS Animal Care and Use Committee (Protocol Number: 1295–17).
Data Availability.
All data collected in this study are included in the primary figures and supplementary files. The sequencing data has been made available on GEO under accession number GSE135620 and GSE135733. The corresponding authors can be contacted for any additional requests.
Extended Data
Extended Data Fig. 1: Anatomical localization of dural B cells and IgA+ cells.
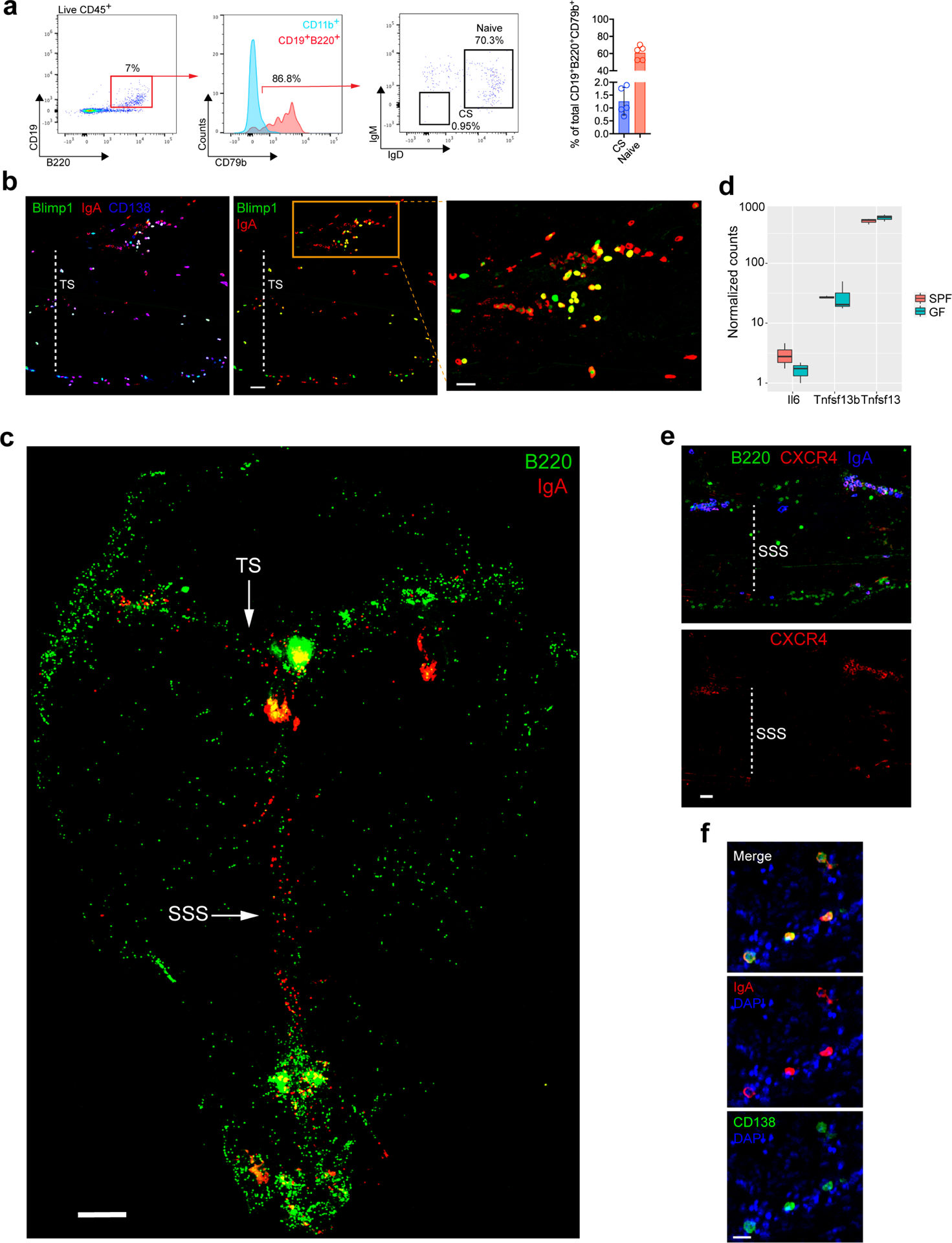
a, Representative flow cytometric plot of meningeal CD19+B220+ cells from a naïve B6 mouse with a histogram depicting CD79b expression on this population of cells relative to CD11b+ myeloid cells. The flow cytometric plot depicts the proportion of naïve (IgD+IgM+) and class-switched (CS, IgD−IgM−) B cells of total CD19+B220+ CD79b+ cells, which is quantified in the bar graph (mean ± s.d., n = 5 mice). b, Representative image of a meningeal whole-mount from a Prdm1EYFP/+ mouse stained with IgA (red) and CD138 (blue) showing co-localization with Blimp1-EYFP+ cells (green) (scale bar, 50 μm). Inset represents a higher magnification of the highlighted box (scale bar, 30 μm); the dotted white line depicts the lumen of the transverse sinus (TS). c, Image of a meningeal whole-mount from a naive B6 mouse immunolabeled for IgA (red) and B220 (green) displaying the localization of tissue-resident B cells and IgA+ cells in the dura mater (scale bar, 1000 μm). The arrows depict the TS and superior sagittal sinus (SSS). d, Normalized counts from RNASeq of meninges from SPF and GF mice for the indicated genes. Normalized counts calculated using DESeq (n=3 mice per group as a boxplot). e, Representative image of the peri-SSS region from a naïve B6 mouse stained for CXCR4 (red), B220 (green) and IgA (blue) (scale bar, 50 μm). f, Representative image of IgA (red) and CD138 (green) immunolabeling of whole-mount human dura mater in the lobe area with DAPI (blue, 4′,6-diamidino-2-phenylindole) in blue (scale bar, 30 μm).
Extended Data Fig. 2: Bacterial diversity in the gut of DA-GF and DB-GF mouse lines.
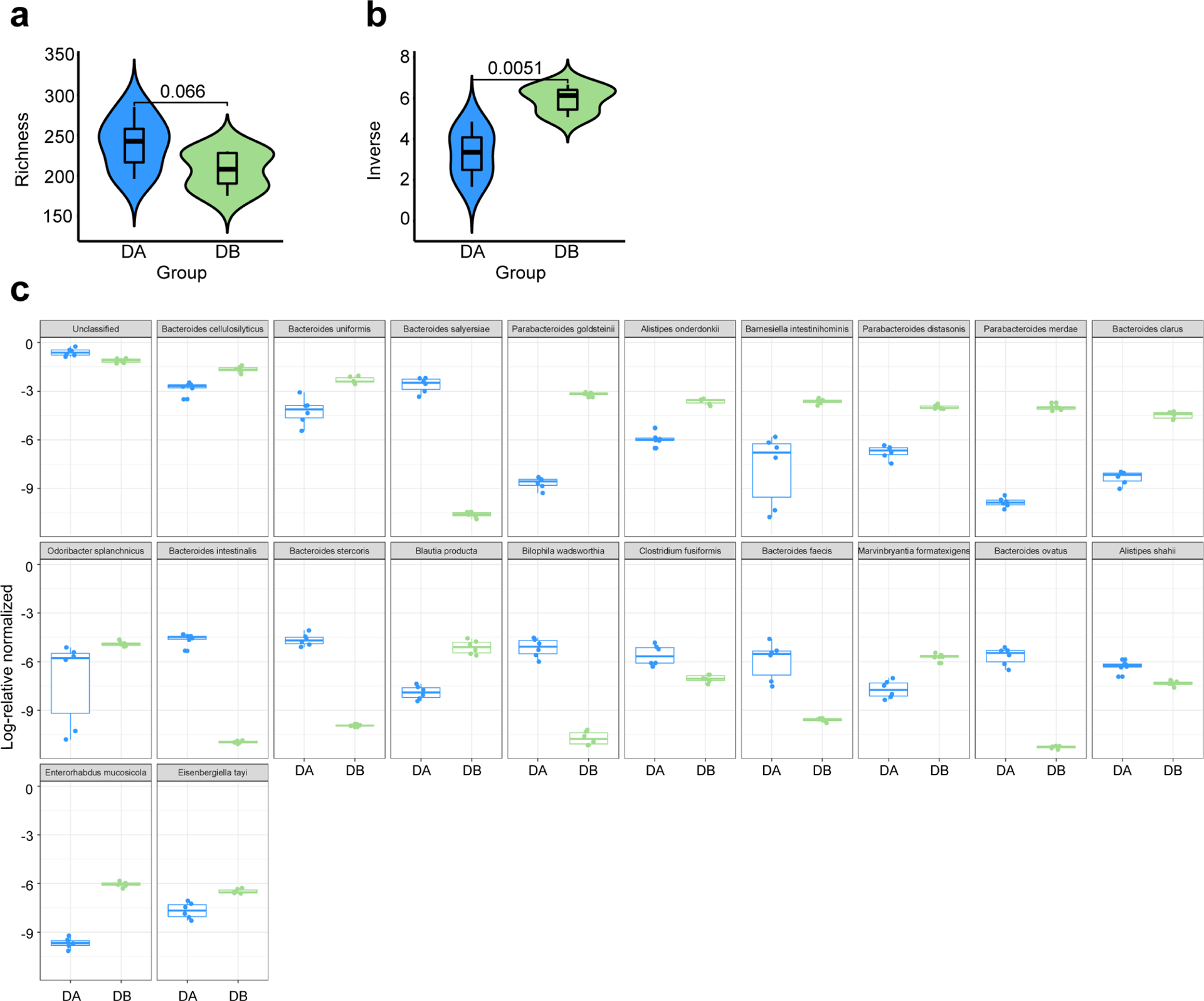
a-b, Comparison of microbial richness using Chao1 (a) and diversity using Inverse Simpson indexes (b) between two groups (DA-GF: blue, n=6; DB-GF: green, n=6) is shown. Violin plots are density plots where the width indicates frequency and the box plot shows median, 95% CI and IQR (two-sided Wilcoxon signed-rank test). c, Log 2-fold change in the relative abundance of taxa between two groups (DA-GF: blue, n=6; DB-GF: green, n=6). Only taxa that are significantly different between two groups are shown (Kruskal-Wallis test).
Extended Data Fig. 3: Clonal relatedness of IgA+ cells in the gut and meninges of SPF, DA GF and DB GF mice, and meningeal Ig heavy chain gene expression in DSS colitis mice.
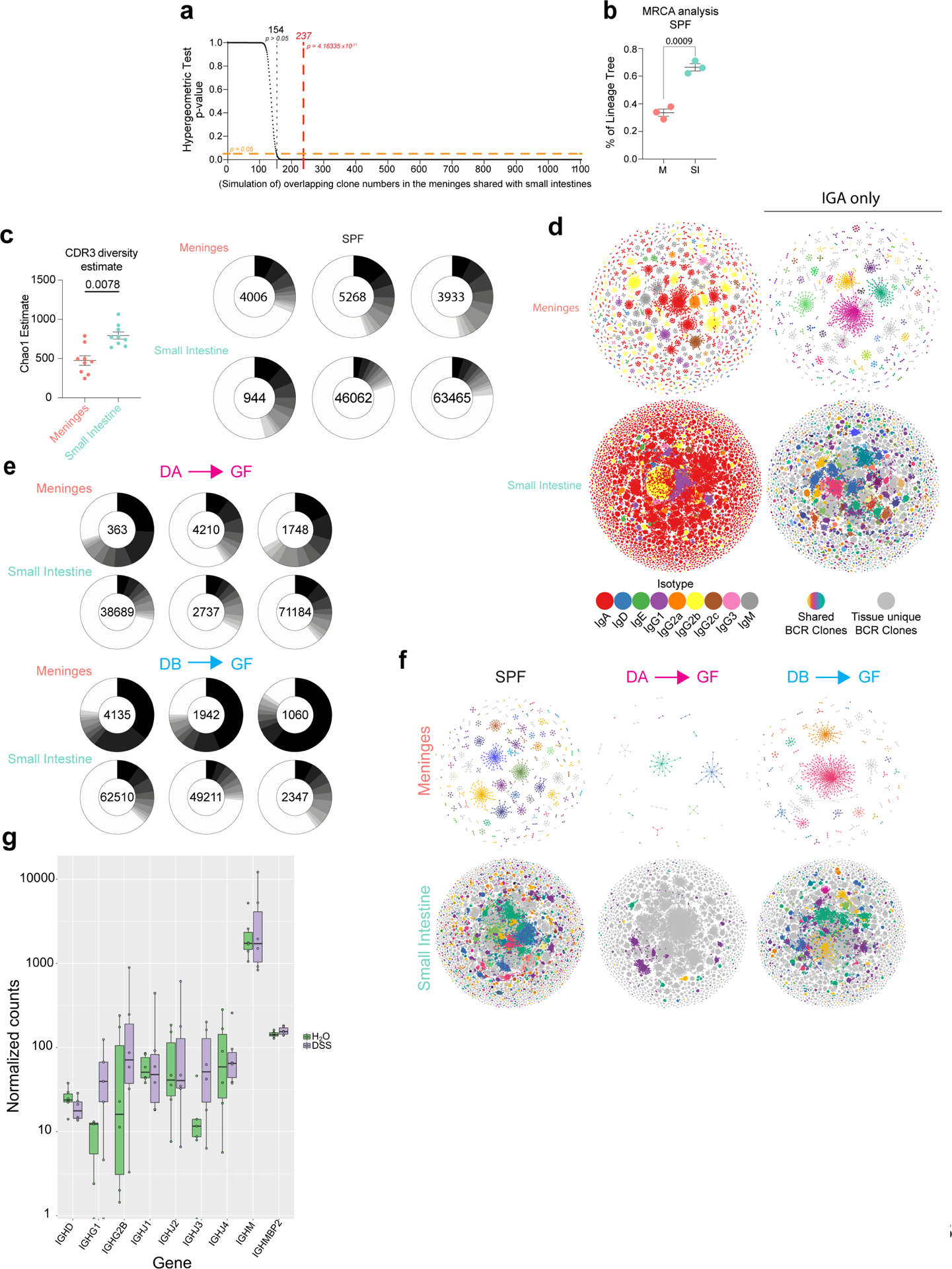
a, Hypergeometric test for enrichment (over-representation) of the shared clones between small intestines and meninges. An overlap occurring by random chance would be < 13% or < 154 clones (dotted black line). The observed overlap in clones between meninges and small intestine = 237/1103 clones (red dotted line) and is non-random (p= 4.2 × 10−21). b, Percentage of lineage trees with most ancestral sequences beginning from meninges (M) or small intestines (SI) in SPF mice inferred after BCR lineage reconstruction. Statistical analysis was performed using an unpaired two-tailed Student’s t test where p<0.05 was considered statistically significant. c, Diversity estimation of CDR3 repertoire represented as a calculated Chao1 estimate after subsampling with 1000 random iterations (left panel). Wilcoxon Rank Sum test was used to determine statistical significance (p < 0.05 considered statistically significant). (Right panel) Relative proportions (frequency of reads) of unique CDR3 amino acid sequences present in each SPF sample is shown, and the top ten most frequent sequences are colored in decreasing shades of grey. Clones after top 10 are not colored (white). d, (Left panels) BCR network of meninges and small intestines from a representative SPF mouse. Colors indicate isotype of BCR sequences. (Right panels) BCR network of IGA clones only from the same mouse. Shared clones between meninges and small intestines are highlighted with the same colors in both networks. Grey nodes and edges indicate BCR sequences/clones not shared between the two tissues. e, Relative proportions (frequency of reads) of unique CDR3aa sequences present in each DA-GF and DB-GF sample is shown, and the top ten most frequent CDR3aa sequences are colored in decreasing shades of grey. Clones after top 10 are not colored (white). f, BCR network of IGA clones from meninges and small intestines in SPF (left panel), DA-GF (middle panel), and DB-GF (right panel) mice. BCR clones shared between meninges and small intestines are highlighted with the same colors in both networks. Grey nodes and edges indicate BCR sequences/clones not shared between the two tissues. g, Normalized counts for the indicated Ig heavy chain genes from bulk RNA-seq of meninges from either DSS-colitic mice or control mice (n = 6 mice per group). Note that, with exception to IGHA (Extended Data Fig. S4d), heavy chain transcript expression did not significantly differ between DSS and control mice.
Extended Data Fig 4: Accumulation and proliferation of meningeal IgA+ cells after gut-epithelial barrier breach.
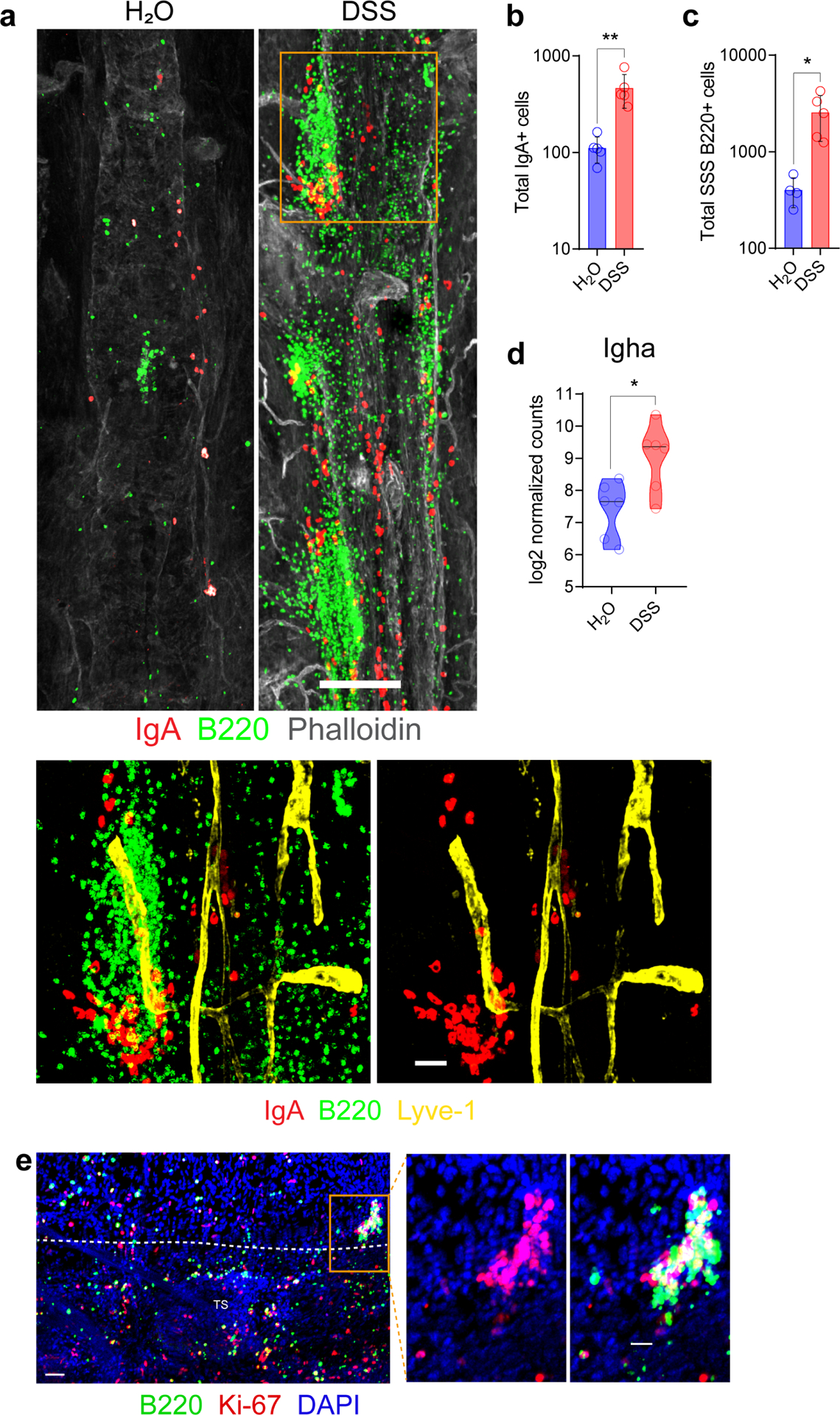
a, Representative confocal images of IgA (red) and B220 (green) immunolabeling and Phalloidin staining (grey) of the SSS from a mouse receiving normal drinking water and from a mouse receiving 2% DSS in drinking water for 7 days followed by normal water for 2 weeks to induce acute colitis. (scale bar, 200 μm); the inset is a higher magnification of the highlighted box depicting a cluster of B cells and IgA+ cells associating adjacent to Lyve-1+ lymphatic vessels (yellow) (scale bar, 40 μm). b, Quantification of peri-sinus IgA+ cells from control vs. 2% DSS mice (mean ± s.d., n = 5 mice per group; **P = 0.0079, two-tailed Mann-Whitney test). c, Quantification of B220+ cells along the SSS in control vs. 2% DSS mice (mean ± s.d., n = 4 mice in control group and 5 mice in DSS group; *P = 0.0159, two-tailed Mann-Whitney test). d, Normalized counts of meningeal Igha expression determined by RNA-seq in the denoted groups (n = 6 mice per group). e, A representative image of the TS region of whole-mount tissue from a mouse receiving 2% DSS in drinking water for 7 days followed by normal water for 2 weeks to induce acute colitis. Tissue was stained for B220 (green), Ki-67 (red) and DAPI (blue) (scale bar, 50 μm). The inset to the right depicts a cluster of proliferating B cells (scale bar, 20 μm).
Extended Data Fig 5: B cell expansion in meninges following systemic candidiasis.
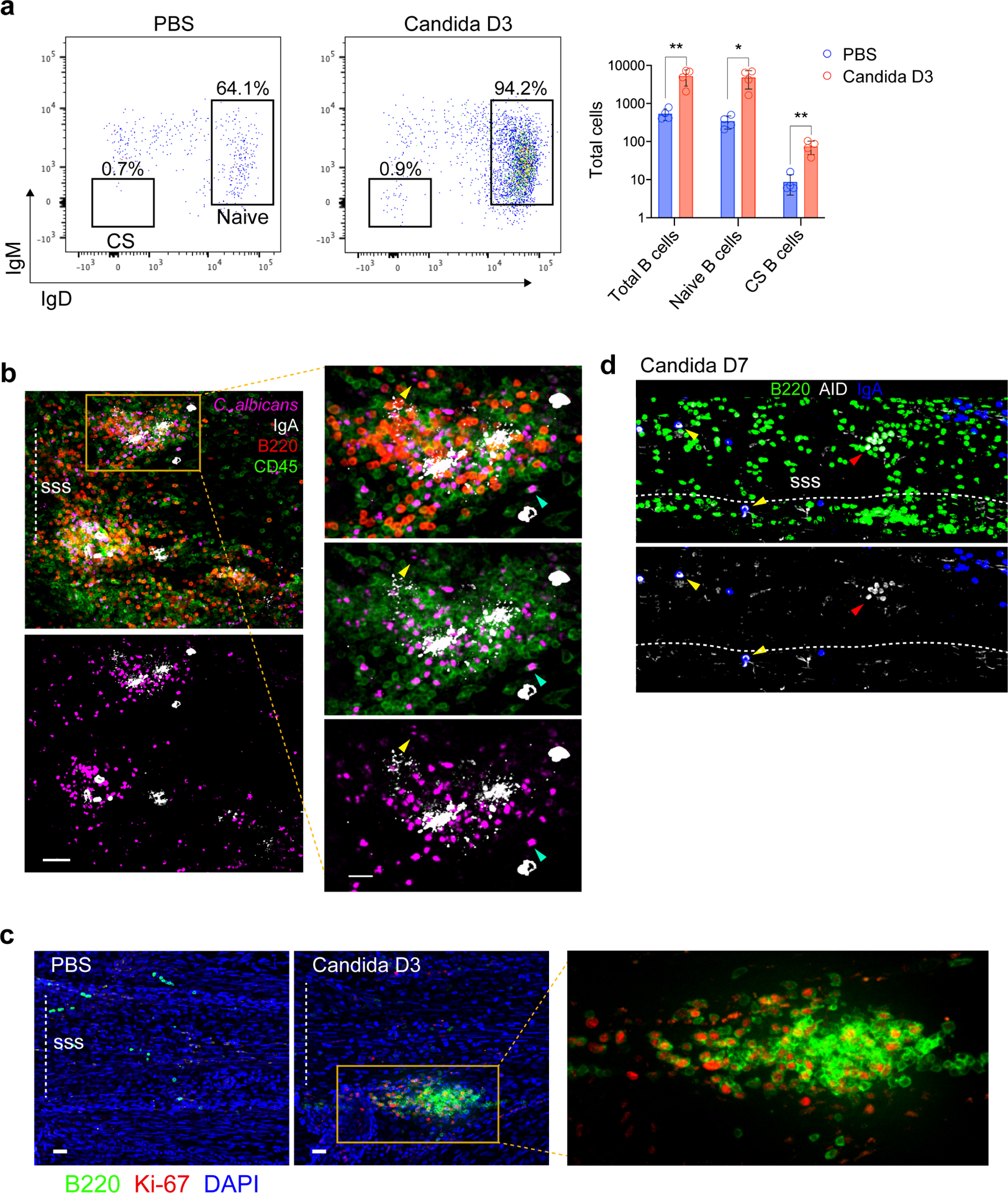
a, Representative flow cytometric plot of CD19+B220+CD79b+ meningeal B cells after intravenous administration of C. albicans, with a bar graph to the right depicting an increase in the total number of naïve (IgD+IgM+) and class-switched (CS, IgD−IgM−) B cells 3 days after infection (mean ± s.d., n = 4 mice per group Total CD19+B220+CD79b+ B cells **P = 0.0074, Total IgD+IgM+ B cells *P = 0.0103, Total IgD−IgM− B cells **P = 0.0040, unpaired two-tailed Student’s t-test). b, Representative image of peri-sinus immune cell clusters from whole-mount tissue stained with IgA (white), B220 (red) and CD45 (green) from a mouse 2 days post systemic candida infection showing GFP-C. albicans in purple (scale bar, 50 μm). The inset represents a higher magnification image of the highlighted region displaying IgA+ cells and diffuse IgA signal in regions with pathogen sequestration (scale bar, 20 μm); yellow arrowhead depicts an extracellular GFP-C. albicans cell, whereas the turquoise arrowhead shows GFP-C. albicans signal within a CD45+ leukocyte. c, Representative image of the superior sagittal sinus (SSS) region from the indicated groups, stained for B220 (green) and Ki-67 (red), showing a peri-SSS cluster of Ki-67+B220+ cells 3 days after infection (scale bar, 50 μm). The inset to the right represents a higher magnification of the highlighted box. d, Representative image of the SSS region in a B6 mouse one week after systemic candida infection immunolabelled for IgA (blue), B220 (green) and AID (white). The yellow arrowheads depict AID-expressing IgA+ cells and the red arrowhead depicts AID-expressing B220+ cells (scale bar, 40 μm).
Extended Data Fig. 6: Distribution of C. albicans in IgA-deficient mice.
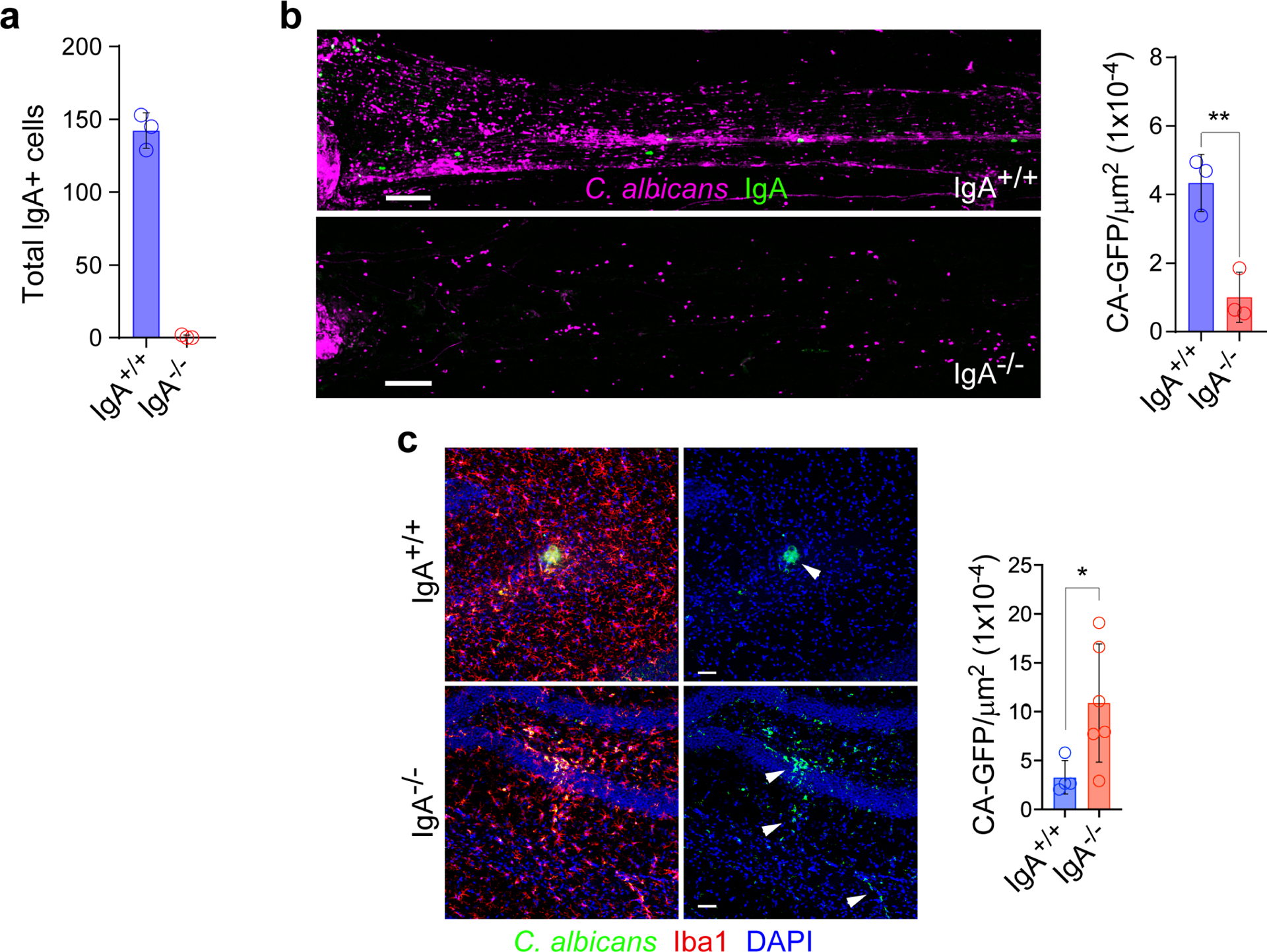
a, Quantification of peri-sinus IgA+ cells in wild type (WT) vs. IgA-deficient animals by confocal imaging of IgA-stained whole-mount tissue (n = 3 mice per group). b, Representative images of the SSS immunolabeled for IgA (green) from a wild type (WT) vs. IgA-deficient mouse 2 days after i.v. administration of GFP-C. albicans shown in purple (scale bar, 150 μm). Quantification of GFP-C. albicans+ puncta along the SSS normalized by area in each group is shown in the bar graph (mean ± s.d., n = 3 mice per group, **P = 0.0065, unpaired two-tailed Student’s t-test). c, Representative images captured in the hippocampus of a WT vs. IgA-deficient mouse stained for Iba1 (red) 2 days after i.v. administration of GFP-C. albicans (shown in green; white arrowheads) (scale bar, 50 μm). Quantification of hippocampal GFP-C. albicans+ puncta normalized by area is shown in the bar graph. Each data point represents the average of four serial hippocampal images from a single animal (mean ± s.d., n = 4 WT mice and 6 IgA−/− mice, *P =0.0428, unpaired two-tailed Student’s t-test).
Extended Data Fig. 7: Local effects of bortezomib-mediated meningeal plasma cell depletion.
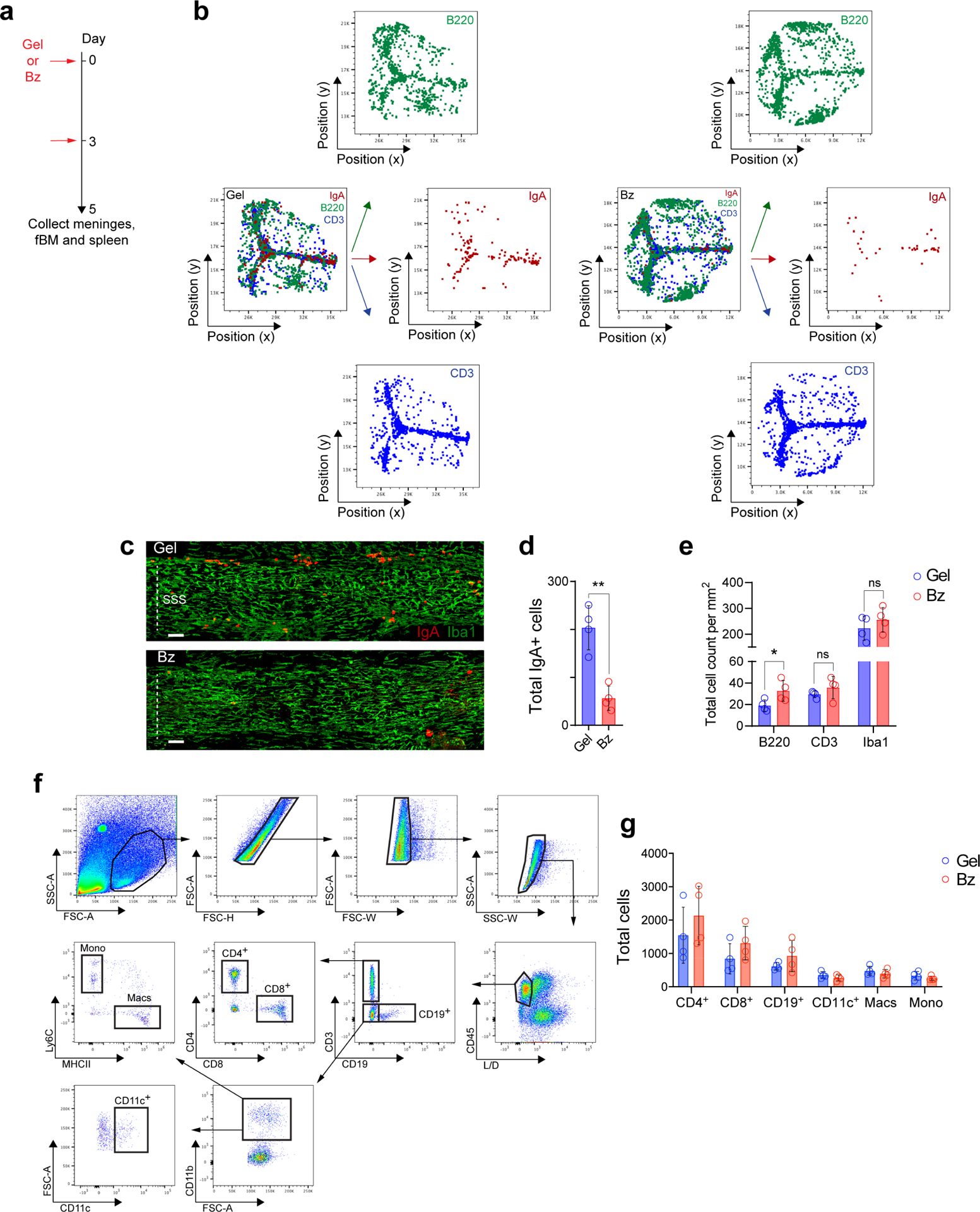
a, Schematic paradigm of sub-scalp administration of either vehicle control hydrogel (Gel) or bortezomib-containing hydrogel (Bz) for meningeal plasma cell depletion. b, Representative histo-cytometry dot plots from meningeal whole-mounts immunolabeled for IgA (red), B220 (green) and CD3 (blue) from either a mouse injected with Gel or Bz. c, Representative confocal images of IgA (red) and Iba1 (green) immunolabeling in whole-mount meninges of the SSS region, depicting the presence of peri-sinus IgA+ cells and macrophages in a mouse administered either Gel or Bz (scale bar, 100 μm). d, Quantification of peri-sinus IgA+ cells in mice injected with either Gel or Bz, as assessed by immunohistochemical analysis of whole-mount tissue (mean ± s.d., n = 4 mice per group; **P = 0.0014, unpaired two-tailed Student’s t-test). e, Quantification of peri-sinus B220+ cells (B cells), CD3+ cells (T cells) and Iba1+ cells (macrophages) from immunolabeling of meningeal whole-mounts in mice administered either Gel or Bz. Total cell counts were normalized by area (mean ± s.d., n = 4 mice per group; B220+ cells *P = 0.0473, CD3+ cells P = 0.2897, Iba1+ cells P = 0.3716, unpaired two-tailed Student’s t-test; ns, not significant). f, Gating strategy for flow cytometric analysis of meningeal immune cell subsets following Bz treatment. g, Bar graph depicting the total cell counts of the indicated immune cell populations in Gel- or Bz-treated animals (mean ± s.d., n = 4 mice per group; no statistically significant changes were noted in each group).
Extended Data Figure 8: Effects of bortezomib-mediated meningeal plasma cell depletion on bone marrow and spleen compartments.
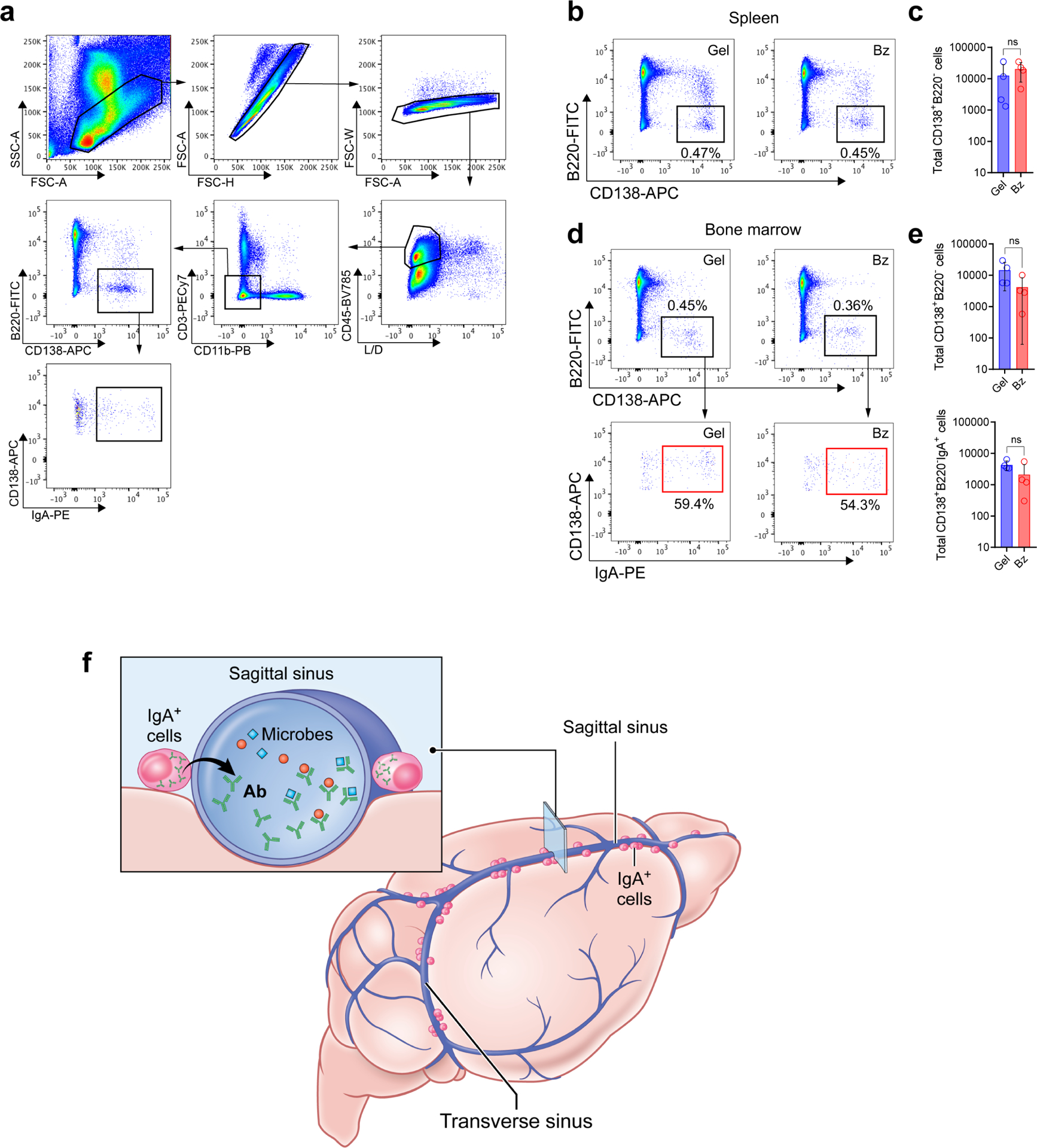
a, Gating strategy for flow cytometric analysis of spleen and bone marrow plasma cells in the protocol depicted in Fig. S5b. b, Representative flow cytometric plots showing CD138+ plasma cells in the spleen from either Gel- or Bz-treated mice. c, Quantification of absolute numbers of splenic plasma cells by flow cytometry in either Gel- or Bz-treated mice (mean ± s.d., n = 4 mice per group; P = 0.4693, unpaired two-tailed Student’s t-test; ns, not significant). d, Representative FACS plots of bone marrow plasma cells and IgA+ plasma cells in either Gel or Bz treated mice. e, Flow cytometric quantification of absolute numbers of total bone marrow plasma cells and IgA+ plasma cells in Gel- or Bz-treated mice (mean ± s.d., n = 4 mice per group; Total CD138+B220− cells P = 0.1369, Total CD138+B220−IgA+ cells P = 0.1556, unpaired two-tailed Student’s t-test; ns, not significant). f, Illustration highlighting the role of gut-educated IgA+ plasma cells along the dural sinuses in entrapping blood-borne microbes and protecting the brain from infection.
Supplementary Material
Acknowledgements
The Clatworthy Lab is based in the University of Cambridge Molecular Immunity Unit in the MRC Laboratory of Molecular Biology and is grateful for the use of the core facilities. Funding: J.R.F., R.J.M. and M.R.C. are supported by the National Institute of Health Research (NIHR) Cambridge Biomedical Research Centre and the NIHR Blood and Transplant Research Unit and M.R.C. by a Medical Research Council New Investigator Research Grant (MR/N024907/1), a Chan-Zuckerburg Initiative Human Cell Atlas Technology Development Grant, a Versus Arthritis Cure Challenge Research Grant (21777), and an NIHR Research Professorship (RP-2017-08-ST2-002). D.B.M., D.S.R., M.L.N., and Z.F. are supported by the Intramural Research Program of NINDS. Y.B. and N.B. are supported by the Intramural Research Program of NIAID. We would like to thank Prof Roger Barker for helpful discussions, and Anish Thomas (NCI) and Govind Nair (NINDS) for the provision of human dura mater in this study. We thank the NIAID Microbiome Program gnotobiotic animal facility staff (D. Trageser-Cesler and C. Acevedo). We would also like to acknowledge Siddharth Krishnamurthy for his work contributing to the colonization of germ-free mice with commensal bacteria.
Footnotes
Competing interests. The authors declare no competing interests.
References
- 1.Korin B et al. High-dimensional, single-cell characterization of the brain’s immune compartment. Nat Neurosci 20, 1300–1309, doi: 10.1038/nn.4610 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Schuchardt F et al. In vivo analysis of physiological 3D blood flow of cerebral veins. Eur Radiol 25, 2371–2380, doi: 10.1007/s00330-014-3587-x (2015). [DOI] [PubMed] [Google Scholar]
- 3.Louveau A et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341, doi: 10.1038/nature14432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rua R & McGavern DB Advances in Meningeal Immunity. Trends Mol Med 24, 542–559, doi: 10.1016/j.molmed.2018.04.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herz J, Filiano AJ, Smith A, Yogev N & Kipnis J Myeloid Cells in the Central Nervous System. Immunity 46, 943–956, doi: 10.1016/j.immuni.2017.06.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polfliet MM et al. Meningeal and perivascular macrophages of the central nervous system play a protective role during bacterial meningitis. J Immunol 167, 4644–4650, doi: 10.4049/jimmunol.167.8.4644 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Gitlin AD, Shulman Z & Nussenzweig MC Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature 509, 637–640, doi: 10.1038/nature13300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moser K, Tokoyoda K, Radbruch A, MacLennan I & Manz RA Stromal niches, plasma cell differentiation and survival. Curr Opin Immunol 18, 265–270 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Mora JR & von Andrian UH Differentiation and homing of IgA-secreting cells. Mucosal Immunol 1, 96–109, doi: 10.1038/mi.2007.14 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Henriksson A, Kam-Hansen S & Link H IgM, IgA and IgG producing cells in cerebrospinal fluid and peripheral blood in multiple sclerosis. Clin Exp Immunol 62, 176–184 (1985). [PMC free article] [PubMed] [Google Scholar]
- 11.Roberg M, Forsberg P, Tegnell A & Ekerfeldt K Intrathecal production of specific IgA antibodies in CNS infections. J Neurol 242, 390–397 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Doss S et al. High prevalence of NMDA receptor IgA/IgM antibodies in different dementia types. Ann Clin Transl Neurol 1, 822–832, doi: 10.1002/acn3.120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westman G et al. Clinical significance of IgM and IgA class anti-NMDAR antibodies in herpes simplex encephalitis. J Clin Virol 103, 75–80, doi: 10.1016/j.jcv.2018.04.007 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Rojas OL et al. Recirculating Intestinal IgA-Producing Cells Regulate Neuroinflammation via IL-10. Cell 176, 610–624 e618, doi: 10.1016/j.cell.2018.11.035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biajoux V et al. Efficient Plasma Cell Differentiation and Trafficking Require Cxcr4 Desensitization. Cell Rep 17, 193–205, doi: 10.1016/j.celrep.2016.08.068 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Senda S, Cheng E & Kawanishi H Aging-associated changes in murine intestinal immunoglobulin A and M secretions. Scand J Immunol 27, 157–164, doi: 10.1111/j.1365-3083.1988.tb02334.x (1988). [DOI] [PubMed] [Google Scholar]
- 17.Hapfelmeier S et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709, doi: 10.1126/science.1188454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamada N et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329, doi: 10.1126/science.1222195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knutton S et al. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J 17, 2166–2176, doi: 10.1093/emboj/17.8.2166 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnupf P et al. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature 520, 99–103, doi: 10.1038/nature14027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casteleyn C, Rekecki A, Van der Aa A, Simoens P & Van den Broeck W Surface area assessment of the murine intestinal tract as a prerequisite for oral dose translation from mouse to man. Lab Anim 44, 176–183, doi: 10.1258/la.2009.009112 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Moor K et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 544, 498–502, doi: 10.1038/nature22058 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Wills S et al. HIV-1-Specific IgA Monoclonal Antibodies from an HIV-1 Vaccinee Mediate Galactosylceramide Blocking and Phagocytosis. J Virol 92, doi: 10.1128/JVI.01552-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai XD, Liu XH & Tong QY Intestinal colonization with Candida albicans and mucosal immunity. World J Gastroenterol 10, 2124–2126, doi: 10.3748/wjg.v10.i14.2124 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bidgood SR, Tam JC, McEwan WA, Mallery DL & James LC Translocalized IgA mediates neutralization and stimulates innate immunity inside infected cells. Proc Natl Acad Sci U S A 111, 13463–13468, doi: 10.1073/pnas.1410980111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakrabarti A Epidemiology of central nervous system mycoses. Neurol India 55, 191–197 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Limon JJ, Skalski JH & Underhill DM Commensal Fungi in Health and Disease. Cell Host Microbe 22, 156–165, doi: 10.1016/j.chom.2017.07.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moran E et al. Proteasome inhibitors as immunosuppressants: biological rationale and clinical experience. Semin Hematol 49, 270–276, doi: 10.1053/j.seminhematol.2012.04.004 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Mastorakos P & McGavern D The anatomy and immunology of vasculature in the central nervous system. Sci Immunol 4, doi: 10.1126/sciimmunol.aav0492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fagarasan S Evolution, development, mechanism and function of IgA in the gut. Curr Opin Immunol 20, 170–177, doi: 10.1016/j.coi.2008.04.002 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Rua R et al. Infection drives meningeal engraftment by inflammatory monocytes that impairs CNS immunity. Nat Immunol 20, 407–419, doi: 10.1038/s41590-019-0344-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.St Leger AJ et al. An Ocular Commensal Protects against Corneal Infection by Driving an Interleukin-17 Response from Mucosal gammadelta T Cells. Immunity 47, 148–158 e145, doi: 10.1016/j.immuni.2017.06.014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manglani M & McGavern DB Intravital Imaging of Neuroimmune Interactions Through a Thinned Skull. Curr Protoc Immunol 120, 24 22 21–24 22 12, doi: 10.1002/cpim.46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naik S et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108, doi: 10.1038/nature14052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson SJ et al. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos Trans R Soc Lond B Biol Sci 368, 20120205, doi: 10.1098/rstb.2012.0205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefranc MP IMGT, the international ImMunoGeneTics information system, http://imgt.cines.fr. Novartis Found Symp 254, 126–136; discussion 136–142, 216–122, 250–122 (2003). [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J Mol Biol 215, 403–410, doi: 10.1016/S0022-2836(05)80360-2 (1990). [DOI] [PubMed] [Google Scholar]
- 38.Brochet X, Lefranc MP & Giudicelli V IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36, W503–508, doi: 10.1093/nar/gkn316 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta NT et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358, doi: 10.1093/bioinformatics/btv359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bashford GR, Burnfield JM & Perez LC Physical activity discrimination improvement using accelerometers and wireless sensor network localization - biomed 2013. Biomed Sci Instrum 49, 243–250 (2013). [PubMed] [Google Scholar]
- 41.Shugay M et al. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput Biol 11, e1004503, doi: 10.1371/journal.pcbi.1004503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forster SC et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol 37, 186–192, doi: 10.1038/s41587-018-0009-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data collected in this study are included in the primary figures and supplementary files. The sequencing data has been made available on GEO under accession number GSE135620 and GSE135733. The corresponding authors can be contacted for any additional requests.


