Abstract
Uncaria rhynchophylla is a vine plant belonging to the family Rubiaceae and has been used as medicine for a long time in China. In this study, the complete chloroplast (cp) genome sequence of U. rhynchophylla was first reported and characterized. The cp genome was 154,605 bp in length and contains a pair of inverted repeats (IRs, 34,165 bp each) separated by a large (84,327 bp) and small (12,966 bp) single-copy regions. A total of 113 unique genes were predicted, including 79 protein-coding genes, 30 tRNA genes and 4 rRNA genes. The phylogenetic analysis suggested that U. rhynchophylla was closer to Neolamarckia cadamba.
Keywords: Chloroplast genome, phylogenetic analysis, Rubiaceae, Uncaria rhynchophylla
Uncaria rhynchophylla (Miq.) Jacks is a vine plant of the Rubiaceae family. The dry branches bearing hooks of this species known as Gouteng are officially listed in the Chinese Pharmacopeia and are used as an antiaggregation, antidepressant, antipyretic, and anticonvulsant for the treatment of headache, vertigo, and epilepsy (Fujiwara et al. 2006; Geng et al. 2019; Yang et al. 2019). The major active components of U. rhynchophylla are alkaloids, terpenoids, and flavonoids (Yang et al. 2019). To provide genomic resources for investigating the evolution of U. rhynchophylla, the complete chloroplast (cp) genome of this species was analyzed from high-throughput Illumina sequencing reads.
The fresh leaves of U. rhynchophylla were collected from Guiyang (Guizhou, China, N26°37′53″, E106°43′23″, 1,280 m) and the specimen (lpssy0287) was deposited in the herbarium of the Liupanshui Normal University (LPSNU). The genomic DNA was extracted and sequenced as previously described (Zhang et al. 2019). Approximately 2 Gb raw data were generated and used for de novo cp genome assembly with SPAdes (Bankevich et al. 2012) and all predicted genes were annotated using PGA (Qu et al. 2019). The complete cp genome sequence of U. rhynchophylla was deposited in GenBank database (accession number MN723865).
The complete U. rhynchophylla cp genome is 154,605 bp in length, including a large single copy (LSC) region of 84,327 bp, a small single copy (SSC) region of 12,966 bp, and a pair of inverted repeats (IRs) of 34,165 bp each. The cp genome shows the GC content of 37.7% and contains 113 unique genes, including 79 protein-coding genes, 30 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA) genes. Most of these genes are in a single copy; however, four rRNA genes (4.5S, 5S, 16S and 23S rRNA), six protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7, and ycf2), and seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) occur in double copies. Fifteen distinct genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contain one intron and three genes (clpP, rps12, and ycf3) have two introns.
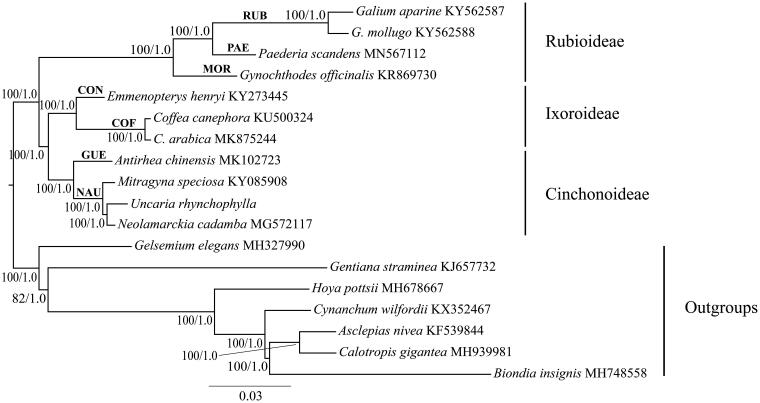
Rubiaceae is one of the five largest families of flowering plants with over 13,000 species and is divided into three subfamilies (subfam. Cinchonoideae, subfam. Ixoroideae and subfam. Rubioideae) and over 63 tribes (Stevens 2001; Bremer 2009). To understand the phylogenetic position of Uncaria within the family Rubiaceae, U. rhynchophylla and other eight genera (Galium, Paederia, Gynochthodes, Emmenopterys, Coffea, Antirhea, Mitragyna, and Neolamarckia) representing seven tribes of Rubiaceae were used for phylogenetic analysis based on their complete cp genomes. Seven species from the other families of Gentianales were used for outgroups. The phylogenetic tree was constructed by maximum likelihood (ML) and Bayesian inference (BI) methods using RAxML (Stamatakis 2014) and MrBayes (Ronquist et al. 2012). As can be seen from Figure 1, a framework of the phylogeny with support for three subfamilies was obtained. Uncaria rhynchophylla was closer to Neolamarckia cadamba, which was formed a sister group with another Naucleeae tribe species, Mitragyna speciose.
Figure 1.
The maximum likelihood (ML) tree of 11 species from the Rubiaceae family inferred from the complete chloroplast genome sequences. Numbers at nodes correspond to ML bootstrap percentages (1,000 replicates) and Bayesian inference (BI) posterior probabilities. COF: Coffeeae; CON: Condamineeae; GUE: Guettardeae; MOR: Morindeae; NAU: Naucleeae; PAE: Paederieae; RUB: Rubieae.
Funding Statement
This study was supported by Scientific Elitists Project of Ordinary Colleges and Universities of Guizhou Province [QJH KY [2019] 061], Science and Technology Platform and Talent Team Project of Science and Technology Department of Guizhou Province [QKH Platform & Talent [2017] 5721] and Natural Science Foundation of Guizhou Education Department [KY [2013] 175].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer B. 2009. A review of molecular phylogenetic studies of Rubiaceae. Ann Mo Bot Gard. 96(1):4–26. [Google Scholar]
- Fujiwara H, Iwasaki K, Furukawa K, Seki T, He M, Maruyama M, Tomita N, Kudo Y, Higuchi M, Saido TC, et al. 2006. Uncaria rhynchophylla, a Chinese medicinal herb, has potent antiaggregation effects on Alzheimer's β-amyloid proteins. J Neurosci Res. 84(2):427–433. [DOI] [PubMed] [Google Scholar]
- Geng CA, Yang TH, Huang XY, Ma YB, Zhang XM, Chen JJ.. 2019. Antidepressant potential of Uncaria rhynchophylla and its active flavanol, catechin, targeting melatonin receptors. J Ethnopharmacol. 232(25):39–46. [DOI] [PubMed] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS.. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens P F. 2001. Angiosperm Phylogeny Website. Version 14, July 2017. [Google Scholar]
- Yang W, Ip SP, Liu L, Xian YF, Lin ZX.. 2019. Uncaria rhynchophylla and its major constituents on central nervous system: a review on their pharmacological actions. Curr Vasc Pharmacol. [Epub ahead of print]. doi: 10.2174/1570161117666190704092841. [DOI] [PubMed] [Google Scholar]
- Zhang SD, Zhang C, Ling LZ.. 2019. The complete chloroplast genome of Rosa berberifolia. Mitochondrial DNA B. 4(1):1741–1742. [Google Scholar]