Abstract
The long-armed scarab Cheirotonus gestroi is an endangered large insect in southwestern China and neighboring countries. Herein, we present the first complete mitochondrial genome of C. gestroi. The 16,899 bp long circular genome consists of 2 ribosomal RNA genes, 22 transfer RNA genes, 13 protein-coding genes, and a non-coding control region. Phylogenetic analysis showed that C. gestroi shared the closest evolutionary relationship with C. jansoni and that Scarabaeidae and Lucanidae were correctly identified within superfamily Scarabaeoidea. The complete mitogenome sequence will provide a basis for further phylogenetic studies and conservation genetics of the genus Cheirotonus.
Keywords: Cheirotonus gestroi, mitochondrial genome, phylogenetic analysis, Endangered insect
The long-armed scarab, Cheirotonus gestroi (Coleoptera: Euchiridae), is an endangered large insect, mainly living in the mountainous areas of southwestern China and neighboring countries (Yi et al. 2015). The adult individuals of C. gestroi possess fascinating metallic color, large body size and disproportionate long arms (mainly males), which make them the top list of insect fans’ collections. In last few decades, the population size of C. gestroi has substantially decreased due to illegal collections and habitats fragmentation. The species has been listed as the State Second-Class Protected Animal in China since 1989 (http://www.forestry.gov.cn/main/3951/20180104/1063898.html). Mitogenomics play important roles in conservation biology (Shamblin et al. 2012; Themudo et al. 2015). Herein, we report and characterize the complete mitogenome of C. gestroi, which will provide useful genetic evidence for conservation genetics.
In this study, the sample was collected from Huanglian Mountain Nature Reserve (22°53′35″N, 102°11′49″E), Yunnan, China in June 2018 and deposited in the herbarium of Southwest Forestry University (specimen code: BJG_HL2018_032). Genomic DNA was extracted from the muscles of a single adult’s chest using the QIAamp DNA Micro Kit (Qiagen, Hilden, Germany) and then sequenced using Illumina HiSeq X10 (Illumina, San Diego, CA). Genes were assembled by the Assembly by Reduced Complexity (ARC) pipeline (http://ibest.github.io/ARC/) (Hunter et al. 2015), using C. jansoni. (NC_023246) as the reference genome (Shao et al. 2014). Protein-coding genes (PCGs), transfer RNA genes (tRNAs) and ribosomal RNA genes (rRNAs) were predicted using the MITOS web server (Bernt et al. 2013).
The complete mitogenome of C. gestroi is circular and 16,899 bp in length (Genbank: MN893347). The base composition is 38.8% for A, 31.9% for T, 18.5% for C and 10.7% for G. The genome contains 37 genes, including 13 PCGs, 22 tRNAs, two rRNAs, and a non-coding control region (D-loop). J-strand (majority strand) encodes nine PCGs (NAD2-3, NAD6, COX1-3, CYTB, ATP6, and ATP8), while N-strand (minority strand) encodes four PCGs (NAD1, NAD4, NAD4L, and NAD5). The gene arrangement of C. gestroi mitogenome is identical to the most insect mitogenomes (Timmermans and Vogler 2012).
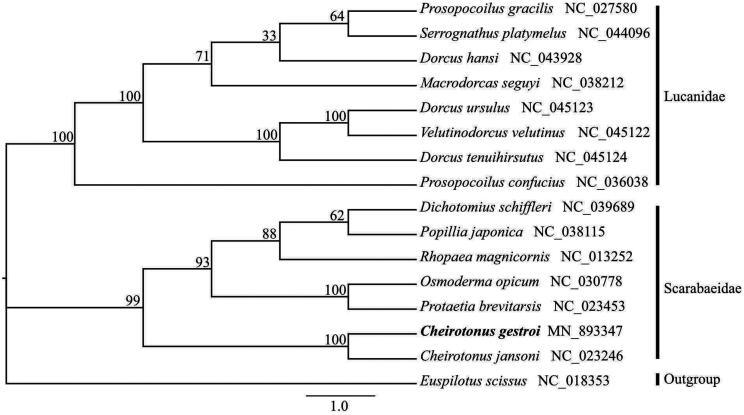
In order to validate the phylogenetic position of C. gestroi, the mitogenomes of 15 species in superfamily Scarabaeoidea and the outgroup Euspilotus scissus (Coleoptera: Histeridae) were used to construct the maximum likelihood (ML) tree. The ML inference was performed using IQ-TREE (Nguyen et al. 2015) in PhyloSuite (Zhang et al. 2020) with 10,000 ultrafast bootstrap replicates. The result indicates that C. gestroi has the closest evolutionary relationship with the same genus species C. jansoni. Furthermore, two clades of Scarabaeidae and Lucanidae were correctly identified with high bootstrap values (Figure 1). The mitochondrial genome data of C. gestroi present in this study will be beneficial to the further phylogenetic studies of Scarabaeidae and the conservation genetics of the genus Cheirotonus species.
Figure 1.
Phylogenetic tree for Cheirotonus gestroi and the related species based on concatenated 13 protein-coding genes (PCGs) by Maximum Likelihood (ML) method. Numbers above branches represent Bootstrap support values. The position of C. gestroi is marked with bold font.
Acknowledgments
We thank the Huanglian Mountain National Reserve Administration for the assistance on the collection of specimens.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31660631]; and the biodiversity investigation, observation and assessment program (2019–2023) of Ministry of Ecology and Environment of China.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Hunter SS, Lyon RT, Sarver BA, Hardwick K, Forney LJ, Settles ML. 2015. Assembly by reduced complexity (ARC): a hybrid approach for targeted assembly of homologous sequences. Biorxiv. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamblin BM, Bjorndal KA, Bolten AB, Hillis-Starr ZM, Lundgren IAN, Naro-Maciel E, Nairn CJ. 2012. Mitogenomic sequences better resolve stock structure of southern Greater Caribbean green turtle rookeries. Mol Ecol. 21(10):2330–2340. [DOI] [PubMed] [Google Scholar]
- Shao LL, Huang DY, Sun XY, Hao JS, Cheng CH, Zhang W, Yang Q. 2014. Complete mitochondrial genome sequence of Cheirotonus jansoni (Coleoptera: Scarabaeidae). Genet Mol Res. 13(1):1047–1058. [DOI] [PubMed] [Google Scholar]
- Themudo GE, Rufino AC, Campos PF. 2015. Complete mitochondrial DNA sequence of the Endangered giant sable antelope (Hippotragus niger variani): Insights into conservation and taxonomy. Mol Phylogenet Evol. 83:242–249. [DOI] [PubMed] [Google Scholar]
- Timmermans MJ, Vogler AP. 2012. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea). Mol Phylogenet Evol. 63(2):299–304. [DOI] [PubMed] [Google Scholar]
- Yi CH, Chen Y, He QJ, Zhou Y, Wang L, Chen XM. 2015. Morphometric characteristics Study on Cherirotonus gestroi Pouillaud. J Northwest For Univ. 30:154–157. [Google Scholar]
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT.. 2020. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. [DOI] [PubMed] [Google Scholar]