Abstract
Amomum villosum Lour. (Zingiberaceae) is an important edible and medicinal crop. The complete chloroplast (cp) genome of A. villosum was determined using Illumina sequencing platform. The size of whole cp genome was 164,069 bp, containing a small single copy (SSC) region of 15,353 bp and a large single copy (LSC) region of 88,798 bp, which were separated by a pair of inverted repeat (IRs) regions (29,959 bp). The A. villosum cp genome contained 133 genes, including eight ribosomal RNA genes (4 rRNA species), 38 transfer RNA genes (30 tRNA species) and 87 protein-coding genes (79 PCG species). The overall GC content of A. villosum cp genome is 36.05%. To investigate the evolution status of A. villosum, as well as Zingiberales, a phylogenetic tree with A. villosum and other 21 species was constructed based on their complete chloroplast genomes. Phylogenetic analysis revealed that A. villosum was closely related to Amomum krervanh.
Keywords: Amomum villosum, complete chloroplast genome, phylogenetic analysis
Amomum villosum Lour. belongs to the ginger family of Zingiberaceae, the dry ripe fruit is called “Sharen” in China, is a commonly used traditional Chinese medicine herb but also used as cooking condiments (Wu and Larsen 2000; Doh et al. 2019). In clinical, “Sharen” is widely used to treat digestive diseases such as abdominal pain, vomiting, and dysentery (Chinese Pharmacopoeia Commission 2015). At present, the research on A. villosum mainly focuses on the chemical composition and pharmacological effects, while there are few studies on the molecular aspects (Suo et al. 2018; Ao et al. 2019). The complete chloroplast genome will be useful to shed light on the phylogenetic relationships and will be beneficial for DNA molecular studies in genus Amomum.
Fresh leaves of A. villosum were collected from Jinping County (22°73′99.86″N, 103°21′43.19″E), Yunnan Province, China. The voucher specimen (LBY20190425) was deposited in Herbarium of Honghe University, China. Approximately 5 g of fresh leaves was harvested for chloroplast DNA isolation (McPherson et al. 2013). After DNA isolation, purified cp DNA was used for short-insert libraries construction (Borgstrom et al. 2011), the whole cp genome sequencing was conducted by BIOZERON Co., Ltd. (Shanghai, China) on the Illumina Hiseq 4000 platform. Then we used the software SOAPdenovo2.04 to assemble the complete cp genome of A. villosum (Luo et al. 2012) and the genes were annotated using an online DOGMA tool (Wyman et al. 2004). Finally, The assembled and annotated chloroplast genome was submitted to GenBank database (accession no. MN931250).
The complete cp genome of the A. villosum was 164,069 bp, containing a small single copy (SSC) region of 15,353 bp and a large single copy (LSC) region of 88,798bp, which were separated by a pair of inverted repeat (IRs) regions (29,959 bp). The A. villosum circular cp genome contained 133 genes, including eight ribosomal RNA genes (4 rRNA species), 38 transfer RNA genes (30 tRNA species) and 87 protein-coding genes (79 PCG species). The most of gene species occurred in a single copy, while 20 gene species occurred in double copies, including four rRNA species (23S, 16S, 5S and 4.5S rRNA), eight tRNA species (trnA-UGC, trnI-CAU, trnI-GAU, trnH-GUG, trnL-CAA, and trnN-GUU, trnR-ACG, trnV-GAC) and eight PCG species (rps7, rps12, rps19, rpl2, rpl23, ycf1, ycf2 and ndhB). Eighteen genes contain intron (12 protein-coding genes and 6 tRNA genes). The overall GC content of the circular genome was 36.05%.
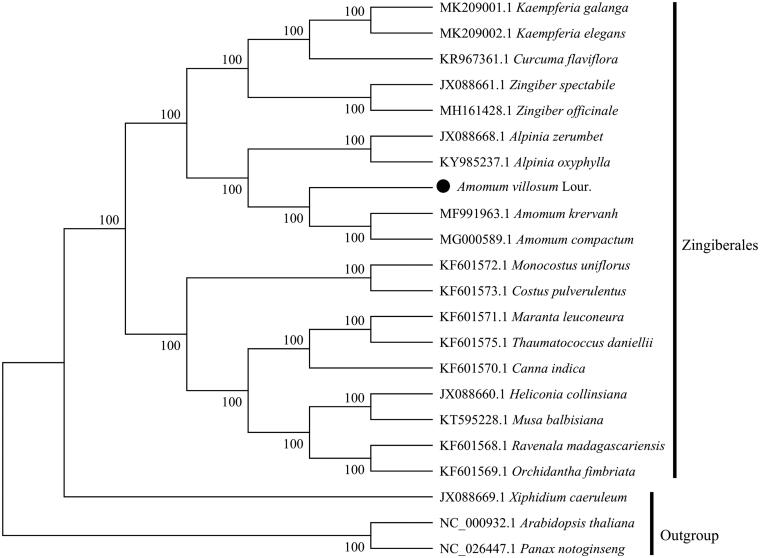
To obtain its evolution status of A. villosum within the order Zingiberales, the phylogenetic relationships were constructed by complete chloroplast genomes of 22 species (Xiphidium caeruleum, Arabidopsis thaliana and Panax notoginseng as the outgroup). The alignment was performed using software MAFFT (Katoh and Standley 2013). A maximum likelihood (ML) tree was generated by MEGA6.0 (Tamura et al. 2013) using 1000 bootstrap replicates. ML bootstrap values of all nodes were 100%, 10 Zingiberaceae species form one branch, A. villosum is a sister species to A. krervanh and A. compactum (Figure 1).
Figure 1.
Phylogenetic tree based on the complete chloroplast genome sequences of A. villosum and 21 other species (contain 3 outgroup Xiphidium caeruleum, Arabidopsis thaliana and Panax notoginseng). Numbers on the nodes indicate bootstrap values.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ao H, Wang J, Chen L, Li SM, Dai CM. 2019. Comparison of volatile oil between the fruits of Amomum villosum Lour. and Amomum villosum Lour. var. xanthioides TL Wu et Senjen based on GC-MS and chemometric techniques. Molecules. 24(9):1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgstrom E, Lundin S, Lundeberg J. 2011. Large scale library generation for high throughput sequencing. PLoS One. 6:e19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese Pharmacopoeia Commission. 2015. Pharmacopoeia of the People’s Republic of China. Vol. 1. Beijing: China Medical Science and Technology Press; p. 253. [Google Scholar]
- Doh EJ, Kim JH, Lee G. 2019. Identification and monitoring of Amomi fructus and its adulterants based on DNA barcoding analysis and designed DNA markers. Molecules. 24(22):4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson H, van der Merwe M, Delaney S K, Edwards M A, Henry R J, McIntosh E, Rymer P D, Milner M L, Siow J, Rossetto M. 2013. Capturing chloroplast variation for molecular ecology studies: a simple next generation sequencing approach applied to a rainforest tree. BMC Ecol. 13(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo S, Lai Y, Li M, Song Q, Cai J, Zhao J, Yang Q, Ung C O L, Hu H. 2018. Phytochemicals, pharmacology, clinical application, patents, and products of Amomi fructus. Food Chem Toxicol. 119:31–36. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Larsen K. 2000. Zingiberaceae Vol 24. Flora of China. Beijing: Science Press; p. 322–377. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]