Abstract
Rhamnus taquetii (family Rhamnaceae) is a shrub , endemic to Korea. Here, the R. taquetii plastid genome was found to be 161,205 bp long and consists of quadripartite structures; a large single-copy region of 89,373 bp, a small single-copy region of 18,936 bp, and a pair of inverted repeat regions of 26,448 bp each. The GC content of the sequence was found to be 37.1%. The plastid genome was found to harbor 129 genes, including 84 protein-coding genes, 37 transfer RNA genes, and 8 ribosomal RNA genes. On the phylogenetic tree of Rosales (based on 63 protein-coding genes), Rhamnaceaewas found to be monophyletic.
Keywords: Endemic species, Rhamnaceae, Rhamnus taquetii, plastid genome
The genus Rhamnus L. s.l. (Rhamnaceae) is composed of about 150 species, that range from shrubs to trees (Chen and Schirarend 2007), and occurs mainly in the tropical to temperate zones of the Northern Hemisphere (Chen and Schirarend 2007). Rhamnus has a complex taxonomic history, and several species within the genus have recently been considered as distinct genera (Hauenschild et al. 2016). Among seven Rhamnus species in Korea (Chang et al. 2011), Rhamnus taquetii (H. Lev.) H. Lev. is an endemic deciduous shrub, growing only on Jeju Island. This species is distinguished from other Korean Rhamnus species by the presence of spines at the terminal of branches, alternating leaves, and small stature (Chang et al. 2011). However, these features occasionally overlap with those of Rhamnus parvifolia Bunge and Rhamnus rugulosa Hemsl. (Chang et al. 2011), taxonomic position of R. taquetii has been controversial. Here, we report the complete plastid genome (plastome) of R. taquetii, which could help in the conclusive taxonomic classification of R. taquetii.
Leaves of R. taquetii were collected from Jeju Island, Korea (Voucher specimen: 33°21′N, 126°29′E, D. P. Jin & J. W. Park 1905248, KH: Korea National Arboretum). Genomic DNA was extracted using a Qiagen DNeasy Kit (QIAGEN, Seoul, Korea) and then sequenced on the Illumina MiSeq platform (Macrogen, Seoul, Korea). A total of 19,539,837 paired reads were produced and mapped onto the plastome of Berchemia berchemiifolia (Makino) Koidz. (GenBank: MG739656). Regions of this draft genome with <1000× coverage and four junctions between the large single-copy (LSC) region, small single-copy (SSC) region, and two inverted repeats (IRs), were verified using Sanger sequencing. Referencing the plastome of Berchemia berchemiifolia, genes of the draft genome were annotated using Geneious 7.1.8 (Biomatters Ltd., Auckland, NZ), but some genes were manually confirmed. Transfer RNAs (tRNAs) were confirmed using tRNAscan-SE (Lowe and Chan 2016). The plastome of R. taquetii (GenBank: MN901522) is 161,205 bp long and consists of an LSC (89,373 bp) region, an SSC (18,936 bp) region, and a pair of IRs (26,448 bp), with an overall GC content of 37.1% and a protein-coding region covering 76.0% of the plastome. The total number of genes in the plastome was 129 (83 protein-coding genes, 38 tRNA genes, and 8 ribosomal RNA genes). Rhamnus taquetii plastid gene content and arrangement closely coincided with those of B. berchemiifolia (Cheon et al. 2018), but the psbL gene of R. taquetii was determined as an intact gene.
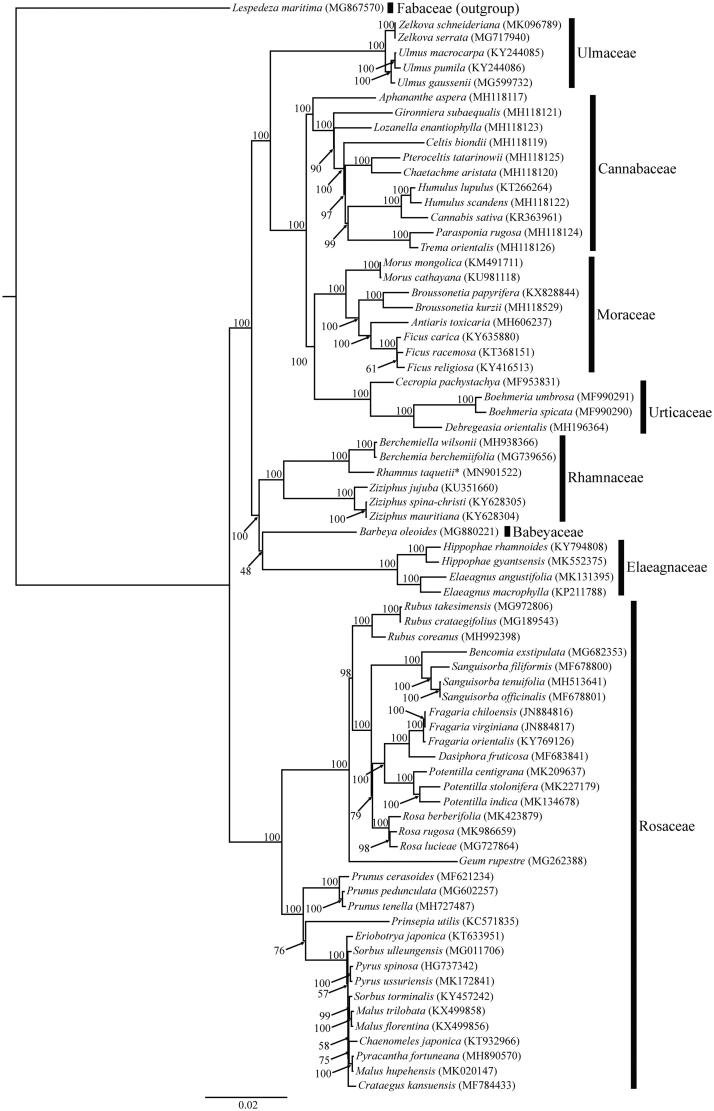
The maximum-likelihood tree of Rosales was constructed based on 63 protein-coding genes with RAxML 8.2.11 (Stamatakis 2014) (Figure 1). GTR + G + I was selected as the nucleotide substitution model according to the Akaike Information criterion throughout jModelTest 2.1.6 (Darriba et al. 2012). Lespedeza maritima Nakai (Fabaceae) was used as the outgroup. On the phylogenetic tree (Figure 1), Rhamnaceae was supported as monophyletic, and R. taquetii was closely clustered with Berchemia berchemiifolia and Berchemiella wilsonii (C.K. Schneid.) Nakai (GenBank: MH938366). Rhamnaceae was a sister to Elaeagnaceae and Barbeyaceae, supporting the result of Zhang et al. (2011).
Figure 1.
A maximum-likelihood tree of Rosales based on 63 coding-genes on plastid genomes. The NCBI accession number for each species is given after its scientific name. The number at each node indicates a bootstrap value. Taxon sequenced here is marked with an asterisk.
Funding Statement
This work was supported by a scientific research fund from an Inha University Research Grant (2019).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Chang CS, Kim H, Chang KS. 2011. Illustrated encyclopedia of fauna and flora of Korea, Vol. 43—woody plants. Paju, South; Designpost Press Co.; p. 330–334. Korean. [Google Scholar]
- Chen Y, Schirarend C. 2007. Rhamnus L. In: Flora China Vol. 12. Beijing (China): Science Press; St. Louis (MO): Missouri Botanical Garden Press; p. 139–162. [Google Scholar]
- Cheon KS, Kim KA, Yoo KO. 2018. The complete chloroplast genome sequence of Berchemia berchemiifolia (Rhamnaceae). Mitochondrial DNA B. 3(1):133–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Meth. 9(8):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenschild F, Favre A, Salazar GA, Muellner-Riehl AN. 2016. Analysis of the cosmopolitan buckthorn genera Frangula and Rhamnus s.l. supports the description of a new genus. Ventia Taxon. 65(1):65–78. [Google Scholar]
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SD, Soltis DE, Yang Y, Li DZ, Yi TS. 2011. Multi-gene analysis provides a well-supported phylogeny of Rosales. Mol Phylogenet Evol. 60(1):21–28. [DOI] [PubMed] [Google Scholar]