Abstract
The complete chloroplast genome of medicinally important plant Adenium obseum (Forssk.) Roem. & Schult. (Apocynaceae) was sequenced. A total of 5,028,192,488 paired-end reads were obtained. One-hundred-and-twenty-seven unique coding genes including 96 protein-coding, 28 tRNA, and 3 rRNA genes were annotated. The length and GC content of the plastome were 154,437 bp (GenBank accession number MN765097) and 38.1%, respectively.
Keywords: Chloroplast genome, medicinal plant, Adenium obesum, Apocynaceae
The drought-deciduous succulent poisonous shrub Adenium obesum (Forssk.) Roem. & Schult. (commonly known as Desert Rose) belongs to the family Apocynaceae, subfam. Apocynoideae, tribe Nerieae, subtribe Neriinae. In the wild, it is mainly distributed in the tropical and subtropical regions of Africa to Arabia and used in traditional medicines and contains toxic cardiac glycosides (Versiani et al. 2014). The recent advances in next-generation sequencing (NGS) such as Illumina, PacBio, and Nanopore sequencing platforms, and bioinformatics tools and techniques for the sequence data analysis in the current decade (Shendure et al. 2017) have greatly helped to understand species relationships in the tree of life to gene function and genomes evolution (Daniell et al. 2016; Leebens-Mack et al. 2019), and have transformed the discipline from phylogenetics into phylogenomics (McKain et al. 2018). The genus Adenium is very closely related to Nerium (subfam. Apocynoideae, tribe Nerieae, subtribe Alafiinae), the morphological characters such as inflorescence, corolla, carpels, fruits and the usually slightly exserted appendices of the stamens of Adenium and Nerium resembles each other (Endress et al. 2014). Hence, the complete plastome of A. obesum was sequenced for the first time in order to assess the systematic relationships.
The whole chloroplast genome was sequenced in the present study on a Illumina sequencing platform from the DNA extracted from the young and green leaves of A. obesum collected from the botanical garden of the King Saud University [Voucher: MAA 15 (KSUH), (24°43′10′′N, 46°36′55′′E), Riyadh, Saudi Arabia using DNeasy DNA extraction kit (QIAGEN, Hilden, Germany). The total number of bases, reads, GC (%), Q20 (%), and Q30 (%) were calculated. The filtered data statistics revealed 3,785,889,890 total read bases with 25,235,946 total reads with Q30 (the percentage of bases that have a quality score of 30 or above) being 95.96%. De novo assembly was performed using NOVOPlasty (Dierckxsens et al. 2017). A total of 127 unique coding genes including 96 protein-coding, 28 tRNA, and 3 rRNA genes were annotated using AGORA (Jung et al. 2018). The length and GC content of the plastome were 154,437 bp (GenBank accession number MN765097) and 38.1%, respectively.
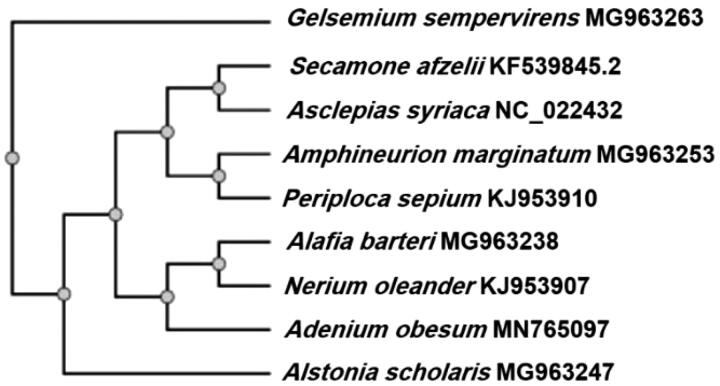
A number of studies based on both morphological as well as DNA sequence data have previously been undertaken to resolve the phylogeny of the family Apocynaceae, the recent classification contains a total of 366 genera, which are recognized and placed within five subfamilies, 25 tribes, and 49 subtribes (Endress et al. 2014), including plastome phylogeny of Apocynaceae (McKain et al. 2018), revealing relationship among the tribe and subtribe. In the present analysis, the BLAST search of the chloroplast genome sequence of A. obesum was performed. Followed by the phylogenetic tree reconstruction with the available representative chloroplast genome sequences [Secamone afzelii (Roem. & Schult.) K. Schum. (KF539845.2, subfam. Secamondoideae), Asclepias syriaca L. (NC_022432 subfam. Asclepiadoideae, tribe Asclepiadeae, subtribe Asclepiadinae), Amphineurion marginatum (Roxb.) D. J. Middleton (MG963253 (subfam. Apocynoideae, tribe Apocyneae, subtribe Amphineruiinae), Periploca sepium Bunge (KJ953910 subfam. Periplocoideae) Alafia barteri Oliv. (MG963238, subfam. Apocynoideae, tribe Nerieae, subtribe Alafiinae), Nerium oleander L. (KJ953907, subfam. Apocynoideae, tribe Nerieae, subtribe Alafiinae), Adenium obesum (Forssk.) Roem. & Schult. MN765097, subfam. Apocynoideae, tribe Nerieae, subtribe Neriinae), Alstonia scholaris (L.) R. Br., MG963247 (subfam. Rauvolioideae), Gelsemium sempervirens (L.) J. St.-Hil. (MG963263, family Gelsemiaceae, outgroup)] using NGPhylogeny.fr (Lemoine et al. 2019) to unravel (a) its phylogenetic relationship within the family and (b) evolution of chloroplast genome in the family Apocynaceae, which resulted in the phylogenetic relationships of the family to be consistent with previous reports (McKain et al. 2018) and its nesting with subfam. Apocynoideae, tribe Nerieae (Figure 1) as per the previous treatment based on morphological and molecular data (Endress et al. 2014).
Figure 1.
The phylogenetic analysis of chloroplast genome of A. obesum with the representative of the subfam. Secamondoideae, Asclepiadoideae, Apocynoideae, Periplocoideae, Rauvolioideae (Family Apocynaceae) outgropup at G. sempervirens (Family Gelsemiaceae). Number next to taxon indicates GenBank accession number.
Funding Statement
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding the work through the research group project [RG-1439-84].
Disclosure statement
No conflict of interest was reported by the author.
References
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Patrick M, Guillaume S. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress ME, Sigrid L-S, Ulrich M. 2014. An updated classification for Apocynaceae. Phytotaxa. 159(3):175–194. [Google Scholar]
- Jung J, Kim JI, Jeong Y-S, Yi G. 2018. AGORA: organellar genome annotation from the amino acid and nucleotide references. Bioinformatics. 34(15):2661–2663. [DOI] [PubMed] [Google Scholar]
- Leebens-Mack JH, Barker MS, Carpenter EJ, Deyholos MK, Gitzendanner MA, Graham SW, Grosse I, Li Z, Melkonian M, Mirarab S, et al. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 574:679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F, Correia D, Lefort V, Doppelt-Azeroual O, Mareuil F, Cohen-Boulakia S, Gascuel O. 2019. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 47(W1):W260–W265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKain MR, Matthew GJ, Simon U‐C, Deren E, Ya Y. 2018. Practical considerations for plant phylogenomics. Appl Plant Sci. 6(3):e1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA, Waterston RH. 2017. DNA sequencing at 40: past, present and future. Nature. 550(7676):345–353. [DOI] [PubMed] [Google Scholar]
- Versiani MA, Ahmed SK, Ikram A, Ali ST, Yasmeen K, Faizi S. 2014. Chemical constituents and biological activities of Adenium obesum (Forsk.) Roem. et Schult. Chem Biodivers. 11(2):171–180. [DOI] [PubMed] [Google Scholar]