Abstract
Platanthera japonica (Thunb. ex A. Marray) Lindl belongs to the genus Platanthera within family Orchidaceae, is an endangered herbal species in the East Asia area. In this study, the complete plastome sequence (cpDNA) of P. japonica was determined by next-generation Illumina sequencing. The cpDNA of this herbal plant is 155,409 bp in size, with a pair of inverted repeat (IR) regions of 26,933 bp that separate a large single-copy (LSC) region of 84,049 bp and a small single-copy (SSC) region of 17,494 bp. The GC content in plastome is 36.9%, and the IR region (43.2%) is higher than that of the LSC and SSC region (34.4% and 29.7%, respectively), which is similar with other Orchidaceae plastomes. The assembled plastome encoded 133 genes, which included 38 tRNA genes, 8 rRNA genes, and 87 protein-coding genes. A total of 24 species were used to construct the phylogenetic relationships among P. japonica and other related species within Orchidaceae. The results showed that P. japonica is closely related to Platanthera chlorantha.
Keywords: Plastome, Platanthera japonica, Illumina sequencing, phylogeny
The family Orchidaceae containing six subfamilies about 25,000 species of 725 genera is one of the biggest families about extant angiosperm groups on earth. The Orchidaceae plants are mainly herbaceous taxa all over the world (Dressler 1981, 1993). Platanthera japonica (Thunb. ex A. Marray) Lindl (Orchidaceae) is a kind of medicinal and ornamental species and has been listed in ‘China Species Red List’ as an endangered plant (Wang and Xie 2004), mainly distributed in China, Korea, and Japan. However, because of some factors (e.g. the unlimited exploiting, the restrictions of their own reproductive mechanism, and habitat destruction), the habitat area of P. japonica is shrinking and makes it an endangered plant (Mendonca and Lins 2007; Ren et al. 2012).
Fresh leaves of P. japonica were collected in Qinling Mountain (34°06′N, 107°54′E, Shaanxi, China), and voucher herbarium specimen (No: XNU0120190311) was deposited at Xianyang Normal University. Total DNA was extracted by CTAB method (Doyle 1987) using the next-generation sequencing with Illumina Hiseq 2500 platform by Sangon Biotech Company (Shanghai, China). After filtering out the low-quality sequence, the high-quality sequences were left as the clean reads. They were assembled by MIRA version 4.0.2 (Chevreux et al. 2004) and MITObim version 1.8 (Hahn et al. 2013) with the plastome of closely related species Habenaria pantlingiana (KJ524104) as the reference. Finally, a total of 278,819 reads have been assembled, and the average coverage is 270.0×. The programs DOGMA (http://dogma.ccbb.utexas.edu/) (Wyman et al. 2004) and Geneious version 8.0.2 (Kearse et al. 2012) were used to annotate the plastome. The annotated plastome sequence of P. japonica has been deposited into GenBank with the accession number MN631092.
The circular complete plastome of P. japonica is 155,409 bp in size, with a pair of IR regions of 26,933 bp that separates a LSC region of 84,049 bp and a SSC region of 17,494 bp. The GC content in plastome is 36.9%, and the IR region (43.2%) is higher than that of the LSC and SSC region (34.4% and 29.7%), which is similar with other Orchidaceae plastomes. The assembled plastome encodes 133 genes, including 38 tRNA genes, 8 rRNA genes, and 87 protein-coding genes. Among them, 14 genes (trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, rpl2, rps12, rps16, rpoC1, ndhA, ndhB, petB, and atpF) contain a single intron, and two genes (ycf3 and clpP) contain two introns.
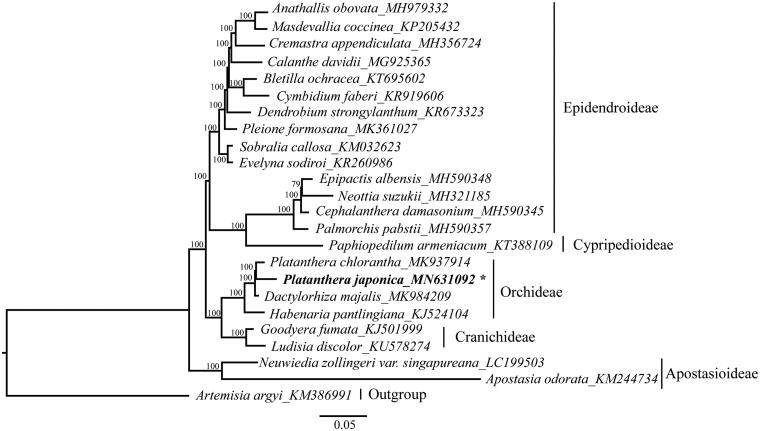
A total of 24 species were used to construct the phylogenetic tree among the main representatives of Orchidaceae species with Artemisia argyi (KM386991) as outgroup (Figure 1). Modeltest version 3.7 (Posada and Crandall 1998) was used to determine the best-fitting model (GTR + G) based on the Akaike information criterion. Maximum-likelihood (ML) analysis was performed using RAxML version 7.2.8 (Stamatakis 2006) with 1000 bootstrap replicates. In the ML tree, the bootstrap values were also high, 20 nodes with 100% bootstrap values. The results indicated that P. japonicais is closely related to P. chlorantha. The complete plastome of P. japonicais can be subsequently used for the systematic study and phylogenetic reconstruction of Orchidaceae.
Figure 1.
Maximum likelihood (ML) tree of 24 complete plastome sequences: Anathallis obovata (MH979332); Masdevallia coccinea (KP205432); Cremastra appendiculata (MH356724); Calanthe davidii (MG925365); Bletilla ochracea (KT695602); Cymbidium faberi (KR919606); Dendrobium strongylanthum (KR673323); Pleione formosana (MK361027); Sobralia callosa (KM032623); Evelyna sodiroi (KR260986); Epipactis albensis (MH590348); Neottia suzukii (MH321185); Cephalanthera damasonium (MH590345); Palmorchis pabstii (MH590357); Paphiopedilum armeniacum (KT388109); Platanthera chlorantha (MK937914); Platanthera japonica (MN631092 in this study); Dactylorhiza majalis (MK984209); Habenaria pantlingiana (KJ524104); Goodyera fumata (KJ501999); Ludisia discolor (KU578274); Neuwiedia zollingeri var. singapureana (LC199503); Apostasia odorata (KM244734); and Artemisia argyi (KM386991).
Funding Statement
This work was supported by the Public health specialty in the Department of Traditional Chinese Medicine [Grant: 2016-44, 2017-66, 2018-43, 2019-68]; Natural Science Basic Research Plan in Shaanxi Province of China [Grant: 2018JM3021]; and State Key Laboratory of Cotton Biology [Grant: CB2018A07].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S.. 2004. Using the mira EST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14(6):1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15. [Google Scholar]
- Dressler RL. 1981. The orchids: natural history and classification. Cambridge (MA): Harvard University Press. [Google Scholar]
- Dressler RL. 1993. Phylogeny and classification of the orchid family. Cambridge (MA): Cambridge University Press. [Google Scholar]
- Hahn C, Bachmann L, Chevreux B.. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucl Acids Res. 41(13):e129–e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Steven SH, Matthew C, Shane S, Simon B, Alex C, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 12:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonca M. P, Lins L. V.. 2007. Revisao das listas das especies da flora eda fauna ameaçadas de extincao do estado de minas gerais. BeloHorizonte (Brazil): Fundacao Biodiversitas. [Google Scholar]
- Posada D, Crandall KA.. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14(9):817–818. [DOI] [PubMed] [Google Scholar]
- Ren ZX, Wang H, Luo YB.. 2012. Deceptive pollination of orchids. Biodivers Sci. 03:270–279. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Wang S, Xie Y.. 2004. China species red list. Beijing (China): Higher Education Press. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]