Abstract
Alopecurus japonicus is a weed in summer crop field, which is harmful to wheat crops. The complete plastome of A. japonicus was reported in this study. The genome was 136,408 bp in length, consisting of an 80,512 bp large single-copy region, a 12,836 bp small single-copy region, and two 21,530 bp inverted repeat regions. The GC content of this plastome was 38.3%. A total of 112 genes were annotated for the plastome of A. japonicus, containing 78 protein-coding genes (PCGs), 30 tRNAs, and 4 rRNAs. Phylogenetic analysis showed that A. japonicus was sister to Alopecurus aequalis.
Keywords: Alopecurus japonicus, plastome, phylogenomics
Alopecurus japonicus is a one-year herb distributed in China, Japan and the Democratic People’s Republic of Korea, mainly in fields and wetlands at low altitudes. It belongs to the Gramineae family as rice, wheat, maize, and sweet sorghum (Bai et al. 2016; Deng et al. 2016; He et al. 2016; Ding et al. 2018; Li et al. 2018). Alopecurus japonicus is a weed in the summer crop field, which is harmful to wheat crops. At present, many studies are focused on its resistance to pesticides (Yang et al. 2007; Mohamed et al. 2012; Wu et al. 2016; Chen et al. 2018). Phylogenetically, A. japonicus belongs to genus Alopecurus, and there is still a big controversy about the systematic position of Alopecurus. Some studies advocated that Alopecurus should be placed in Aveneae or Agrostideae (Hitchcock and Chase 1935; Watson et al. 1986; Hilu and Esen 1990). Tzvelev suggested placing Alopecurus in Phleae (Tzvelev 1989). In this study, we showed the plastome of A. japonicus, which would provide a fundamental genetic resource for studying this important species.
Fresh leaves of A. japonicus were collected from Wanghui Village (Shandong, China; 36°31′N, 115°58′E). Voucher specimen (No.75) has been deposited at College of Life Sciences, Shandong Normal University. Modified CTAB method was used for plant total DNA extraction (Wang et al. 2013). Library preparation and sequencing were performed on the Illumina MiSeq platform at Novogene (Beijing, China). Organelle Genome Assembler (OGA, https://github.com/quxiaojian/OGA) was used to do plastome assembling (Qu 2019). Annotation was performed by using Plastid Genome Annotator (PGA, https://github.com/quxiaojian/PGA) (Qu et al. 2019). Geneious version 9.1.4 was used for manual annotation correction (Matthew et al. 2012). In order to determine the phylogenetic position of A. japonicus, a maximum-likelihood (ML) tree was reconstructed by RAxML version 8.2.10 (Alexandros 2014), using the alignment matrix of 78 protein-coding genes (PCGs) generated by MAFFT version 7.313 (Kazutaka and Standley 2013), the 1000 rapid bootstrap replicates, and the GTRGAMMA substitution model.
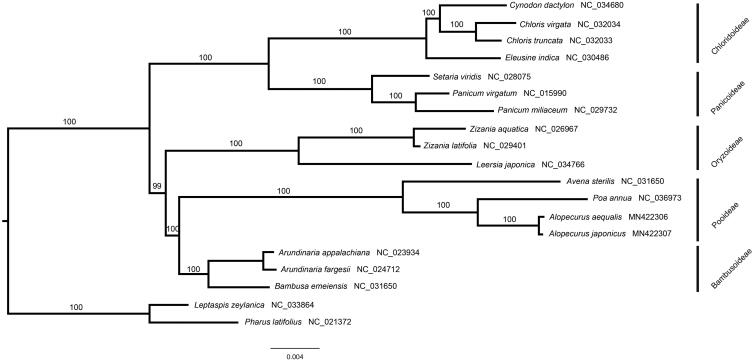
The complete plastome of A. japonicus (GenBank accession number: MN422307) was 136,408 bp in length, consisting of a large single-copy region (80,512 bp), a small single-copy region (12,836 bp), and a pair of inverted repeats regions (21,530 bp). The GC content of this plastome was 38.3%. 112 unique genes were encoded, including 78 PCGs, 30 tRNAs, and 4 rRNAs. The ML phylogenetic tree showed that A. japonicus was closely related to A. aequalis (Figure 1).
Figure 1.
The maximum likelihood (ML) tree was reconstructed by 78 plastome genes. Leptaspis zeylanica and Pharus latifolius are used as out-group. Bootstrap support values are indicated on the branches of the ML tree.
Funding Statement
The study was financially supported by Shandong Provincial Agricultural Elite Varieties Project [2019LZGC017], and Investigation on Undergrowth Herbs of Forestry Department of Shandong Province.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Alexandros S. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Zhao J, Li YP, Zhang F, Zhou JJ, Chen F, Xie XZ. 2016. OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice. Plant Sci. 247:25–34. [DOI] [PubMed] [Google Scholar]
- Chen G, Xu H, Zhang T, Bai C, Dong L. 2018. Fenoxaprop-P-ethyl resistance conferred by cytochrome P450s and target site mutation in Alopecurus japonicus. Pest Manag Sci. 74(7):1694–1703. [DOI] [PubMed] [Google Scholar]
- Deng YQ, Bao J, Yuan F, Liang X, Feng ZT, Wang BS. 2016. Exogenous hydrogen sulfide alleviates salt stress in wheat seedlings by decreasing Na+ content. Plant Growth Regul. 79(3):391–399. [Google Scholar]
- Ding TL, Yang Z, Wei XC, Yuan F, Yin SS, Wang BS. 2018. Evaluation of salt-tolerant germplasm and screening of the salt-tolerance traits of sweet Sorghum in the germination stage. Funct Plant Biol. 45(10):1073–1081. [DOI] [PubMed] [Google Scholar]
- He YA, Li YP, Cui LX, Xie LX, Zheng CK, Zhou GH, Zhou JJ, Xie XZ. 2016. Phytochrome B negatively affects cold tolerance by regulating OsDREB1 gene expression through phytochrome interacting factor-like protein OsPIL16 in rice. Front Plant Sci. 7:1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilu KW, Esen A. 1990. Prolamins in systematics of Poaceae subfam. Plant Syst Evol. 173(1–2):57–70. [Google Scholar]
- Hitchcock AS, Chase A. 1935. Manual of the grasses of the United States. Am J Bot. 21(3):127–139. [Google Scholar]
- Kazutaka K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AQ, Li GH, Zhao YH, Meng ZD, Zhao M, Li CS, Zhang Y, Li PC, Ma CL, Xia H, et al. . 2018. Combined small RNA and gene expression analysis revealed roles of miRNAs in maize response to rice black-streaked dwarf virus infection. Sci Rep. 8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew K, Richard M, Amy W, Steven SH, Matthew C, Shane S, Simon B, Alex C, Sidney M, Chris D. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed IA, Li R, You Z, Li Z. 2012. Japanese foxtail (Alopecurus japonicus) resistance to fenoxaprop and pinoxaden in China. Weed Sci. 60(2):167–171. [Google Scholar]
- Qu XJ. 2019. Complete plastome sequence of an endangered species, Calocedrus rupestris (Cupressaceae). Mitochondrial DNA. 4(1):762–763. [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzvelev NN. 1989. The system of grasses (Poaceae) and their evolution. Bot Rev. 55(3):141–204. [Google Scholar]
- Wang HY, Jiang DF, Huang YH, Wang PM, Li T. 2013. Study on the phylogeny of Nephroma helveticum and allied species. Mycotaxon. 125(1):263–275. [Google Scholar]
- Watson L, Dallwitz MJ, Johnston CR. 1986. Grass genera of the world: 728 detailed descriptions from an automated database. Aust J Bot. 34(2):223–230. [Google Scholar]
- Wu X, Zhang T, Pan L, Wang L, Xu H, Dong L. 2016. Germination requirements differ between fenoxaprop-P-ethyl resistant and susceptible Japanese foxtail (Alopecurus japonicus) biotypes. Weed Sci. 64(4):653–663. [Google Scholar]
- Yang CH, Dong LY, Jun LI, Yang YQ. 2007. Study on resistance of Alopecurus japonicus steud. Populations to haloxyfop-R-methyl in oilseed rape fields. Sci Agric Sin. 9(1):3263–3277. [Google Scholar]