Abstract
Sinojackia huangmeiensis J. W. Ge & X. H. Yao is a member of the genus Sinojackia endemic to central and east China. Here we assembled and annotated the complete chloroplast (cp) genome. It is 158,758 bp in length and encodes 84 protein-coding genes, 37 transfer RNA (tRNA) genes and eight ribosomal RNA (rRNA) genes. The Maximum likelihood phylogenetic analysis confirm that S. huangmeiensis is a closely related but another different species to S. sarcocarpa.
Keywords: Sinojackia huangmeiensis, complete chloroplast genome, Styracaceae, phylogeny
Sinojackia is a small genus with ca. eight species endemic to central and east Asia (Hu 1928; Zhang et al. 2012). S. huangmeiensis is one member of this genus, which is only found in the type locality till now (Yao et al. 2007). Only about 400 individuals are reported in the small area (Luo et al. 2016), but it’s hard to propagate itself. Own white flower and scopperil-shaped fruit, and special systematic position, this species is of important value in horticulture and phylogeny (Luo et al. 2016; Zhao et al. 2016). In present study, the completed chloroplast genome sequence of S. huangmeiensis is reported contributing to conservation of this species and providing significant information for the phylogeny of Styracaceae.
Genomic DNA was extracted from leaves of a seedling of S. huangmeiensis from Qianlin Villadge, Xiaxin Town, Huangmei County in Hubei Province, China (116°00′47.36″E, 29°59′31.91″N; 20 m in altitude, near the lake, Dong et al.-HGNU-0289, 2019. 4. 18; HGTC). Those leaves were stored in the refrigerator at −80 °C. The total genomic DNA was isolated according to a modified CTAB method (Doyle 1987). Total genome DNA of S. huangmeiensis was sequenced by Illumina Hiseq 2500 Sequencing System (Illumina, Hayward, CA) to construct the shotgun library. About 10 Gb pair-end (150 bp) raw short sequence data were obtained. The low quality sequences were filtered out Using CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark) and then reconstructed the chloroplast genome by using MITObim v1.8 (University of Oslo, Oslo, Norway; Kaiseraugst, Switzerland) (Hahn et al. 2013). The complete chloroplast genome of S. huangmeiensis was annotated in Geneious R9 (v9.0.2) (Matthew et al. 2012) and online program Chloroplast Genome Annotation, Visualization, Analysis, and GenBank Submission (CPGAVAS) (Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China) (Liu et al. 2012) and then submitted to GenBank (accession no. MN694844).
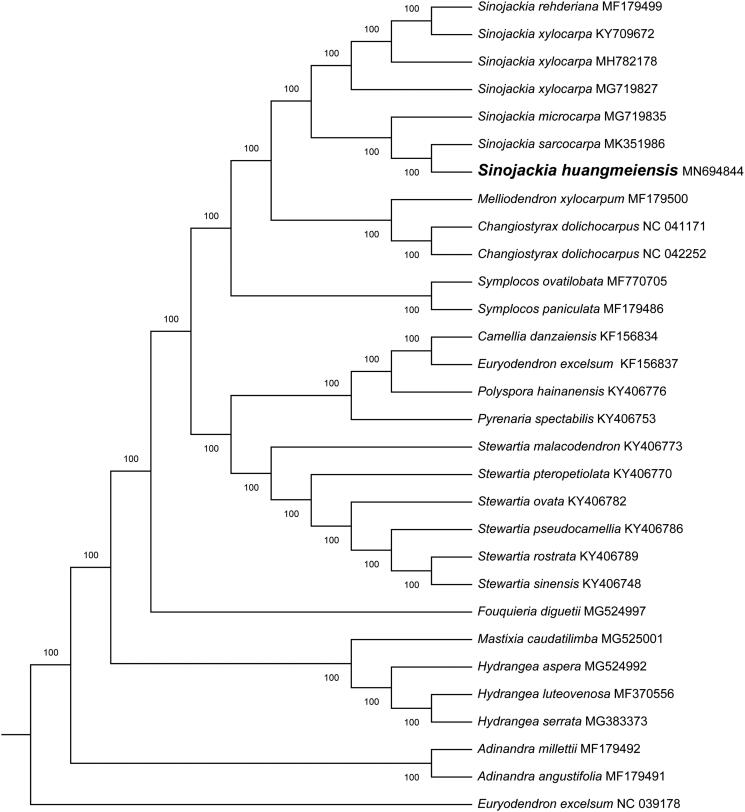
The size of chloroplast genome of S. huangmeiensis is 158,758 bp, including a large single-copy (LSC) region of 88,023 bp and a small single-copy (SSC) region of 18,555 bp separated by a pair identical inverted repeat regions (IRs) of 26,090 bp each. A total of 139 genes were successfully annotated containing 84 protein-coding genes, 37 tRNA genes and 8 rRNA genes. GC content of the whole genome, IRs, LSC and SSC regions are 37.2%, 43.0%, 35.2% and 30.5%, respectively. GC content of IRs region is the highest. 17 genes contain one intron, while 2 genes have two introns. The complete chloroplast genome sequence of S. huangmeiensis and other species from Styracaceae were used to construct phylogenetic tree (Figure 1). Take the plastome of Euryodendron excelsum (NC_039178) as an out-group, a maximum likelihood analysis was performed with RAxML version 8 program (Alexandros 2014) using 1000 bootstrap. The ML tree shows all the published plastome of Sinojackia were grouped into a clade and the phylogenetic relationship was consistent with the previously-published phylogeny (Yao et al. 2008; Cai et al. 2018; Fan et al. 2019). The results show that S. huangmeiensis was most close to S. carcocarpa.
Figure 1.
Maximum likelihood phylogenetic tree for Sinojackia huangmeiensis based on 30 complete chloroplast genomes. The number on each node indicates bootstrap support value.
Funding Statement
This study was financially supported by the Startup Foundation for Doctors of Huanggang Normal University [2042019020 to JJY and 2042019013 to HJD], the High-level Training Program of Huanggang Normal University [204201913203 to JJY], and Foundation for Assessment and Comprehensive Utilization of Characteristic Biological resources in Dabie Mountains [4022019006].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Alexandros S. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai XL, Wang JH, Zhao KK, Zhu ZX, Wang HF. 2018. Complete plastome sequence of Changiostyrax dolichocarpa (Styracaeae): an endangered (EN) plant species endemic to China. Mitochondrial DNA B. 3(2):1031–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Fan J, Fu QC, Liang Z. 2019. Complete chloroplast genome sequence and phylogenetic analysis of Sinojackia sarcocarpa, an endemic plant in Southwest China. Mitochondrial DNA B. 4(1):1350–1351. [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HH. 1928. Sinojackia, a new genus of styracaceae from southeastern China. J Arnold Arbor. 9(2/3):130–131. [Google Scholar]
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Shi QZ, Yang JJ, Fang YP. 2016. Study on natural population of Sinojackia huangmeiensis in Hubei Province. J Anhui Agric Sci. 44(23):67–68. [Google Scholar]
- Matthew K, Richard M, Amy W, Steven SH, Matthew C, Shane S, Simon B, Alex C, Sidney M, Chris D. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao XH, Ye QG, Fritsch PW, Cruz BC, Huang HW. 2008. Phylogeny of Sinojackia (Styracaceae) based on DNA sequence and microsatellite data: implications for taxonomy and conservation. Ann Bot. 101(5):651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao XH, Ye QG, Ge JW, Ming K. 2007. A new species of Sinojackia (Styracaceae) from Hubei, Central China. Novon. 17(1):138–140. [Google Scholar]
- Zhang JJ, Ye QG, Gao PX, Yao XH. 2012. Genetic footprints of habitat fragmentation in the extant populations of Sinojackia (Styracaceae): implications for conservation. Bot J Linn Soc. 170(2):232–242. [Google Scholar]
- Zhao J, Tong YQ, Ge TM, Ge JW. 2016. Genetic diversity estimation and core collection construction of Sinojackia huangmeiensis based on novel microsatellite markers. Biochem Syst Ecol. 64:74–80. [Google Scholar]