Abstract
In this study, we firstly reported the complete chloroplast (cp) genome sequences of the Mangifera sylvatica from Nanning, Guangxi province, China. The complete wild mango cp genome size is 158063 bp with a typical small single-copy region (SSC, 18340 bp), a large single-copy region (LSC, 87008 bp) and a pair of inverted repeats (IRs, 26379 bp and 26379 bp respectively). Out of 112 unique annotated genes in mango cp genome, 78 found to be protein coding, 30 to be tRNA and 4 rRNA genes. Besides, we found 51 microsatellite sequences (SSRs) in the cp genome. Sequence alignment and ML analysis of 29 full plastome data revealed M. sylvatica shares the closest relationship with cultivated mango (M. indica) and form a sister group with Rhus chinensis within Anacardiaceae.
Keywords: Chromoplast, wild mango, Mangifera sylvatica, phylogenetic relationship
Wild mango (Mangifera sylvatica Roxb.), which is sister to the most widely cultivated tropical fruit common mango, is an important medicinal plant belonging to the family Anacardiaceae (Baul et al. 2016). Recent studies have shown that M. sylvatica is of high medicinal and nutrition values as its leaves possess thrombolytic properties and fruits are rich in wild mango butter (Akhter et al. 2016). It is distributed mainly in South-east Asian countries (Baul et al. 2016). As an underutilized wild tree species, it has been threatened in Bangladesh and may go extinct due to forest degradation and climate change (Akhter et al. 2016, 2017). However, genetic resources are still scare to promote domestication and conservation of this species. Here, we assembled the cp genome of M. sylvatica and assessed its phylogenetic position with Illumina sequencing.
One individual plant of M. sylvatica was sampled from Mango Germplasm Resources Protection and Innovation Base, Guangxi Subtropical Crops Research Institute (GSCRI, 22°53′55.5″N, 108°20′33.4″E). Specimen was deposited in the Herbarium of GSCRI (HGSCRI-MS-1). Fresh leaves of the sample were immediately frozen in liquid nitrogen for DNA extraction with the Plant Genomic DNA Kit (TIANGEN, DP305). Adaptors were added to DNA the fragment and an Illumina’s Hiseq2500 sequencer was used for sequencing. After filtering out low quality data, 0.8 Gb clean data were assembled into contigs with SOAPdenovo (v 2.04) (Luo et al. 2012). Then, the assembly results were optimized according to the overlap relationships of reads with GapCloser (v1.12), and redundant segment sequences were removed to obtain the final assembly. Annotations were performed using DOGMA (Wyman et al. 2004) and adjusted according to Wang et al. (2018). Complete genome was submitted to GenBank (MK790101).
Chloroplast genome of M. sylvatica is 158,063 bp in length, with 37.89% GC content. The LSC is 87008 bp in length containing 82 genes, and the SSC is 18340 bp in length containing 13 genes, while two IRs are 26,379 bp containing 18 and 19 genes, respectively. 78 functional genes were annotated with 79,035 bp total length and an average gene length of 1013 bp, which is similar to the M. indica plastome (Azim et al. 2014; Zhao et al. 2019). Among the protein-coding genes, nine genes (rpl2, rpl16, PetB, PetD, rps16, atpF, ropC1, ndhB, and ndhA) had one intron, while rps12, ycf3 and clpP each contained two introns. tRNA genes were situated in all four regions while rRNAs are only found in IR regions. 19 LTRs and 8 DNA transposons were found scattered in the genome, while 51 SSRs were identified including mono- and di-SSR types.
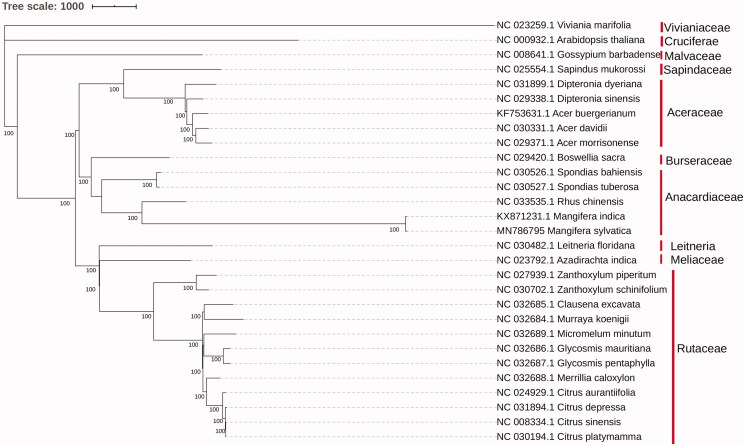
To investigate the phylogenetic position of M. sylvatica, 29 complete chloroplast genome from 10 families were aligned by Clustalw2 (Larkin et al. 2007). ML-analysis and phylogenetic tree plotting were conducted with MEGAX (Kumar et al. 2018). M. sylvatica shares the closest relationship with M. indica and other 3 species in Anacardiaceae with 100% bootstrap support (Figure 1). Anacardiaceae forms a single clade with Burseraceae. These results were consistent with Jo’s study (2017) with only protein-coding and rRNA genes.
Figure 1.
Molecular phylogenetic analysis of 29 plastomes.
The evolutionary history was inferred by using the Maximum Likelihood method based on the Jukes–Cantor model. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. Positions with less than 95% site coverage were eliminated. There was a total of 131,609 positions in the final dataset.
Funding Statement
This study was supported by Guangxi Provincial Natural Science Foundation of China under Grant [2015GXNSFBA139085] and Basic Scientific Research Funding for Special Project of Guangxi Subtropical Crops Research Institute under Grant [201905].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Akhter S, McDonald M A, Marriott R. 2016. Mangifera sylvatica (Wild Mango): a new cocoa butter alternative. Sci Rep. 6(1):32050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter S, McDonald M A, van Breugel P, Sohel S, Kjaer E D, Mariott R. 2017. Habitat distribution modelling to identify areas of high conservation value under climate change for Mangifera sylvatica Roxb. of Bangladesh. Land Use Policy. 60:223–232. [Google Scholar]
- Azim M K, Khan I A, Zhang Y. 2014. Characterization of mango (Mangifera indica L.) transcriptome and chloroplast genome. Plant Mol Biol. 85(1–2):193–208. [DOI] [PubMed] [Google Scholar]
- Baul T K, Alam M J, Nath T K. 2016. Mangifera sylvatica Roxb. in the forests of south-eastern Bangladesh: a potential underutilised tree for small-scale forestry. Small-Scale Forest. 15(2):149–158. [Google Scholar]
- Jo S, Kim H-W, Kim Y-K, Sohn J-Y, Cheon S-H, Kim K-J. 2017. The complete plastome sequences of Mangifera indica L. (Anacardiaceae). Mitochondrial DNA Part B. 2(2):698–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A, Blackshields G, Brown N.P, Chenna R, McGettigan P.A, McWilliam H, Valentin F, Wallace I.M, Wilm A, Lopez R, et al. . 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. . 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chen S, Zhang X. 2018. Whole-genome comparison reveals divergent IR borders and mutation hotspots in chloroplast genomes of herbaceous bamboos (Bambusoideae: Olyreae). Molecules. 23(7):1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S K, Jansen R K, Boore J L. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Gao A, Huang J, Luo R. 2019. The complete sequence of chloroplast genome from mango (Mangifera indica var GuiFei). Mitochondrial DNA Part B. 4(1):1916–1917. [Google Scholar]