Abstract
Bupleurum species are well-known for their antipyretic, analgesic, gallbladder and other functions so that they are very popular in traditional Chinese medicine. However, to our knowledge, there is no completed chloroplast genome of Bupleurum genus in China. In the present study, we determined the complete chloroplast genome sequences for Bupleurum longicaule var. strictum using IIumina sequencing. Bupleurum longicaule var. strictum is 155,578 bp which is composed of two inverted repeats (IR: 26,257bp), a large single copy region (LSC: 86,977 bp), and a small single-copy region (SSC: 16,087 bp). The overall AT content is 65.32%. The chloroplast genome includes a total of 51 functional genes including 15 protein-coding genes and 36 tRNA. A total of 10 genes were duplicated in the IR regions including seven tRNA and three protein-coding genes. Phylogenetic analysis suggested that B. longicaule var. strictum formed a monophyletic clade.
Keywords: Bupleurum, Bupleurum longicaule var. Strictum, Chloroplast genome
The genus Bupleurum, belonging to the Umbelliferae family, encompasses more than 180 species distributed mainly in the Northern Hemisphere. In China, the genus consists of 42 species and 17 varieties, 22 of which are recognized as endemisms (Shan and She 1979). Bupleurum species are well-known for their antipyretic, analgesic, gallbladder, and other functions so that they are very popular in traditional Chinese medicine (Luo and Jin 1991; Pan 2006; Tan et al. 2007). However, to our knowledge, there is no completed chloroplast genome of Bupleurum genus in China. In the present study, we report the completed chloroplast genome of Bupleurum longicaule var. strictum based on the next-generation sequencing method. The chloroplast genome will contribute to molecular phylogeny, genetic information, mechanism of evolution or adoption.
A wild individual of B. longicaule var. strictum was collected from Xinping village (101°37.556′E, 36°34.460′N, 2510 m), Huangzhong country in Qinghai province of China (Voucher specimen: QHGC20190823, HNWP). The Illumina sequencing process was as follows: extraction of Genome DNA, fragmentation of DNA, construction of Illumina pair-end library and sequencing with the Illumina HiSeq 4000 platform. After filtering reads with low-sequencing quality, high-quality clean data were assembled with the SPAdes (Bankevich et al. 2012). And then, annotation was performed with DOGMA (Wyman et al. 2004). The annotated genomic sequence has been submitted to GenBank with the accession number SRR10579389.
The total length of the complete chloroplast genome of B. longicaule var. strictum is 155,578 bp which is composed of two inverted repeats (IR: 26,257bp), a large single-copy region (LSC: 86,977 bp) and a small single-copy region (SSC: 16,087 bp). The overall AT content is 65.32%. The chloroplast genome includes a total of 51 functional genes including 15 protein-coding genes and 36 tRNA. A total of 10 genes were duplicated in the IR regions including seven tRNA and three protein-coding genes. The genome organization, gene/intron content and gene relative positions of the newly sequenced plastid genome were almost identical to other Umbelliferae species.
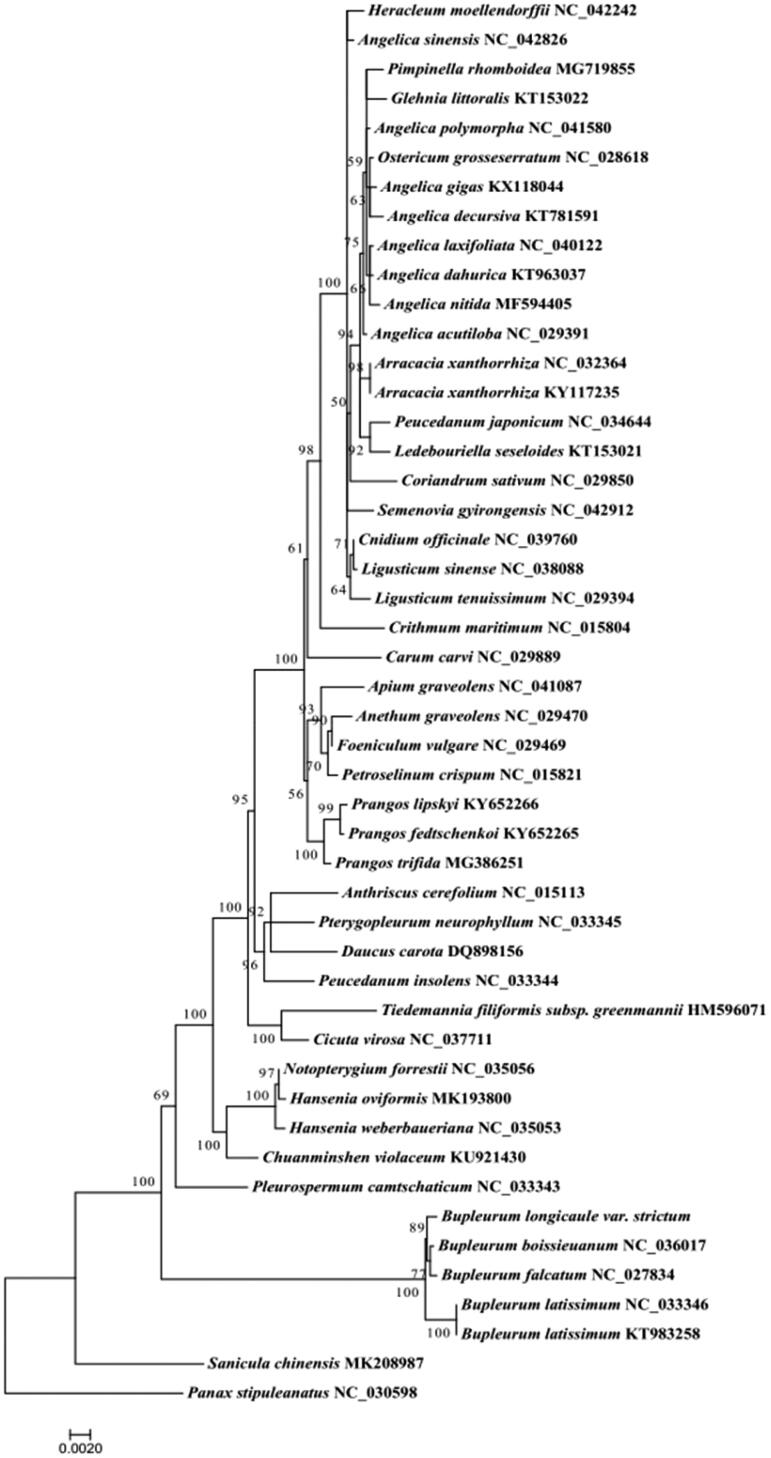
The phylogenetic tree was constructed with the complete chloroplast genomes of B. longicaule var. strictum and 46 other species from Umbelliferae, and Panax stipuleanatus was used as an outgroup. The result showed that B. longicaule var. strictum formed a monophyletic clade (Figure 1).
Figure 1.
Maximum likelihood phylogenetic tree based on 48 complete chloroplast genome sequences.
Funding Statement
This work is supported by Key R&D and Transformation Projects in Qinghai Province; Chinese Academy of Sciences Strategic Leading Science and Technology Projects.
Disclosure statement
The authors are highly grateful to the published genome data in the public database. The authors declare no conflicts of interest and are responsible for the content in the manuscript.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo SQ, Jin HF. 1991. Chemical constituents of the aerial parts of six species of Bupleurum genus medicinally used in south-west region of China. Chin J Chin Mater Med. 16(6):353. [PubMed] [Google Scholar]
- Pan SL. 2006. Bupleurum speices: scientific evaluation and clinical applications. Boca Raton: CRC Press; . [Google Scholar]
- Shan RY, She ML. 1979. Zhong Guo Zhi Wu Zhi (Flora of China). Beijing: Science Press. [Google Scholar]
- Tan LL, Hu ZH, Cai X, Cai X, Chen Y, Shi W. 2007. Histochemical localization and the content compare of main medicinal components of vegetative organs in Bupleurum Chinense DC. J Mol Cell Biol. 40(4):214–220. [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]