Abstract
Semiliquidambar cathayensis is a semi-evergreen broad-leaved tree species distributed in southern China. In 1999, it was approved and published as a national secondary protected plant. We obtained the complete chloroplast genome sequence of S. cathayensis by Illumina sequencing data. The complete chloroplast sequence is 160,430 bp, include large single-copy (LSC) region of 88,991 bp, small single-copy (SSC) region of 18,917 bp, and a pair of invert repeats (IR) regions of 26,261 bp. Plastid genome contain 133 genes, 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that S. cathayensis is closely related to Liquidambar formosana.
Keywords: Semiliquidambar cathayensis, chloroplast genome, phylogeny, Illumina sequencing
Semiliquidambar cathayensis is China’s endemic semi-evergreen tree which belongs to the family Hamamelidaceae. It mainly distributed in subtropical monsoon climate areas in China, such as Fujian, Jiangxi, Guizhou, Guangdong and Guangxi, and it grows well in deep, fertile, loose, moist and well drained acid soils. S. cathayensis is not only a precious timber tree species, but also has medicinal value in its roots, branches and leaves (Ye et al. 2019). In recent years, due to the serious man-made destruction and the difficulty in natural renewal, only a few sporadically distributed tree species remain and the natural community is very few, it has been listed in the red book of endangered plants in China: rare and endangered plants (Fu 1991). S. cathayensis has the comprehensive characters between the Liguidambar and the Altingia (Zhao et al. 2010), which is of great scientific value for studying the phylogeny of Hamamelidaceae. In this study, we report the complete chloroplast genome (cp) of S. cathayensis based on Illumina pair-end sequencing data.
Fresh leaves were collected from one tree of S. cathayensis in FuJian Province, China (Longqi Mountain National Nature Reserve, Jiangle: 117°16′43″E, 26°29′34″N). The voucher specimen is kept at the Herbarium of College of Forestry, Fujian Agriculture and Forestry University (specimen code FAFU0609).
Fresh leaves of S. cathayensis were washed and treated with liquid nitrogen for 10–20 min, and stored in the refrigerator at 80 °C below zero. Then the processed fresh leaves were transported to bgi through dry ice to extract DNA, and the database was built on MGI-seq 2000 platform for sequencing, approximately 2GB data generated. Illumina data were filtered by script in the cluster (default parameter: filter -n 0.02 -l 20 -q 0.4 -i –rmdup). Complete plastid genome of L. formosana (GeneBank accession: KC588388) as reference, plastid genome of S. cathayensis was assembled by GetOrganelle pipe-line (https://github.com/Kinggerm/GetOrganelle), it can get the plastid-like reads, and the reads were viewed and edited by Bandage (Wick et al. 2015). Assembled choroplast genome annotation base on comparison with L. formosana using Geneious v 11.1.5 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. 2012). The annotation result was drawn with the online tool OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. 2013).
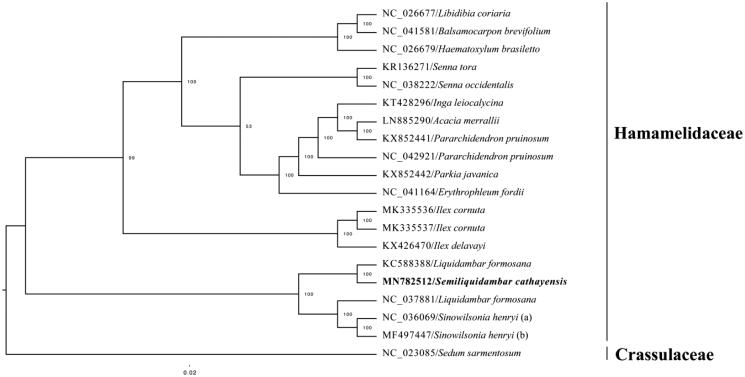
The complete plastid genome sequence of S. cathayensis (GenBank accession: MN782512) was 160,430 bp in length, with a large single-copy (LSC) region of 88,991 bp, a small single-copy (SSC) region of 18,917 bp, and a pair of inverted repeats (IR) regions of 26,261 bp. Complete chloroplastid genome contains 133 genes; there were 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The complete genome GC content was 37.90%. In order to reveal the phylogenetic position of S. cathayensis, a phylogenetic analysis was performed based on 19 complete chloroplast genomes of Hamamelidaceae, one species (Sedum sarmentosum) from Crassulaceae as outgroup. They all downloaded from NCBI GenBank. The sequences were aligned using MAFFT v7.307 (Katoh and Standley 2013), and phylogenetic tree constructed using RAxML (Stamatakis 2014). The phylogenetic tree showed that S. cathayensis was most closely related to L. formosana with strong support (Figure 1).
Figure 1.
Phylogenetic analysis of 19 species of Hamamelidaceae and one species (S. sarmentosum) from Crassulaceae as outgroup based on plastid genome sequences by RAxML, bootstrap support value near the branch.
Acknowledgement
This article can be successfully completed, thanks to my teachers and alumni. I would like to show my deepest gratitude to my supervisor, Professor Liu, who give me the opportunity to write this article, and for his constant guidance to me. Thanks to Professor Chen and Professor Fan for their support and guidance, as well as to Yuting Jiang and Xingzhuang Ye for their technical support. I believe this will be a new starting point for my life. Finally, I would like to thank the experts and professors who participated in the paper evaluation. Your opinions will also be my precious wealth!
Funding Statement
This work was supported by the Seed Industry Innovation and Industrialization Project in Fujian Province, China [ZYCX-LY-2017002]; The Science and Technology Innovation Special Fund Project of Fujian Agriculture and Forestry University [KFA17293A].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Fu LG. 1991. Red book of Chinese plants: rare and endangered plants. Beijing: Science Press. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye XZ, Liu D, Luo JJ, Fan HH, Zhang GF, Liu B, Chen SP. 2019. Transcriptome analysis for rare and endangered plants of Semiliquidambar cathayensis. Bull Botanical Res. 39(02):276–286. [Google Scholar]
- Zhao HT, Song PL, Han GY, Chen Q. 2010. The latest research progress on Semiliquidambar cathayensis H. T. Chang . Northern Horticulture. 21:210–212. [Google Scholar]