Abstract
Distylium macrophyllum H.T.Chang is a critically endangered tree species endemic to southern China. In this study, we assembled and characterized the plastome of D. macrophyllum using Illumina paired-end data. The circular genome is 159,089 bp in size, consisting of two copies of inverted repeat (IR) regions of 26,235 bp, one large single-copy (LSC) region of 87,822 bp, and one small single-copy (SSC) region of 18,797 bp. It encodes a total of 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. Phylogenetic analysis based on 13 cp genome sequences indicated that D. macrophyllum was sister to the clade of D. tsiangii and Parrotia subaequalis.
Keywords: Chloroplast genome, distylium macrophyllum, hamamelidaceae, phylogeny
Distylium macrophyllum H.T.Chang is an evergreen shrub or small tree endemic to southern China. It is mainly distributed in the mountainous areas of northern Guangdong and northwestern Guangxi (Chang 1960). Due to the extremely small population size and narrow range, the species has been classified as critically endangered in the Red List of China Higher Plants (Qin et al. 2017). During the fieldwork, we also found that some natural habitats of the species were degraded or lost due to urban and agricultural development, which would further increase the risk of wild extinction. However, since firstly described by Hung-ta Chang in 1960, there is limited research on the conservation genetics of the species. In this study, we report the complete chloroplast (cp) genome of D. macrophyllum based on Illumina pair-end sequencing data to provide more genomic resources for future conservation.
Fresh young leaves of D. macrophyllum were sampled from a wild individual growing at the Tielongtou Village, Ruyuan, Guangdong Province, China (25.020°N, 113.186°E). The voucher specimen (accession number 19090502) was preserved at the Herbarium of Nanjing Forestry University (HNFU). Total DNA extraction and whole genome sequencing on the Illumina Hiseq X Ten platform were conducted by Nanjing Genepioneer Biotechnologies Inc. (Nanjing, China). A total of 24,105,337 clean reads were produced and then used for the de novo assembly with NOVOplasty 2.7.2 (Dierckxsens et al. 2016). Gene annotation was performed using the CpGAVAS pipeline (Liu et al. 2012).
The complete cp genome of D. macrophyllum (GenBank accession number MN729500) is a circular molecule of 159,089 bp in length, consisting of two copies of IR (26,235 bp) separated by the LSC (87,822 bp) and SSC (18,797 bp) regions. The overall GC content was 38.01%, while the corresponding values of the LSC, SSC, and IR regions were 36.17%, 32.48%, and 43.06%, respectively. The cp genome encoded a total of 132 genes, of which 114 were unique and 18 were duplicated in the IR regions. The 114 unique genes contained 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. Eighteen genes contained introns, 15 of which (nine protein-coding genes and six tRNA genes) contained one intron, and three of which (rps12, ycf3, and clpP) contained two introns.
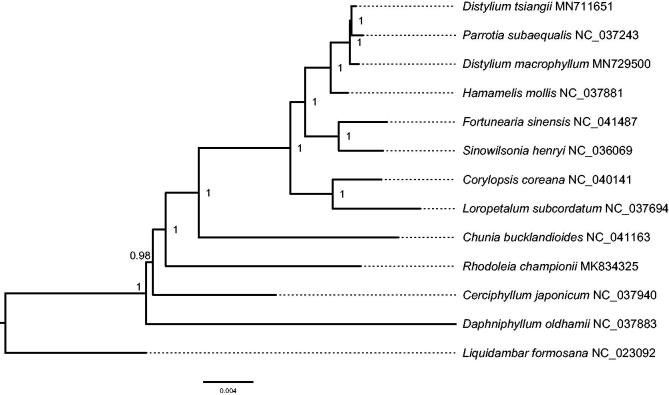
A Bayesian phylogeny was reconstructed using cp genome sequences of 13 species in the woody Saxifragales to determine the phylogenetic position of D. macrophyllum (Figure 1). Two locally collinear blocks were identified and a matrix of 93,936 bp was generated by the HomBlocks pipeline (Bi et al. 2018). Using PartitionFinder 2 (Lanfear et al. 2016), the TVM + I + G model was chosen for the subset of LCBs 1-2. A Markov chain Monte Carlo (MCMC) as implemented in MrBayes 3.2.6 (Ronquist et al. 2012) was run for 1,000,000 generations with two parallel searches using four chains, each starting with a random tree. Trees were sampled every 100 generations and the first 25% was discarded as burn-in. Our results indicated that D. macrophyllum was sister to the clade of D. tsiangii and Parrotia subaequalis with strong support (posterior probability = 1.0).
Figure 1.
Bayesian phylogeny reconstructed by MrBayes 3.2.6 (Ronquist et al. 2012) using cp genome sequences of 13 species in the woody Saxifragales. The posterior probability value is labeled for each node.
Acknowledgments
The authors thank Changteng Yang, Zhifa Liu, and Zhangping You from Nanling National Nature Reserve of Guangdong Province for their help with fieldwork; Lin Chen and Cheng Zhang from Nanjing Forestry University for their assistant with data analysis.
Funding Statement
This work was supported by the Biodiversity Investigation, Observation and Assessment Program of Ministry of Ecology and Environment of China, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22. [DOI] [PubMed] [Google Scholar]
- Chang HT. 1960. Additions to the Hamamelidaceae flora of China, II. Acta Sci Nat Univ Sunyatseni. 5:39–46. [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(18):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Bio Evol. 34:772–773. [DOI] [PubMed] [Google Scholar]
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin HN, Yang Y, Dong SY, He Q, Jia Y, Zhao LN, Yu SX, Liu HY, Liu B, Yan YH, et al. 2017. Threatened species list of China’s higher plants. Biodivers Sci. 25(7):696–744. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]