Abstract
The Dioryctria yiai belongs to Pyralidae in Lepidoptera. The complete mitogenome of D. yiai was described in this study, which is typical circular duplex molecules and 15,430 bp in length, containing the standard metazoan set of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and an A + T-rich region. The gene order is same with other lepidopterans. Except for cox1 started with CGA, all other PCGs started with the standard ATN codons. Most of the PCGs terminated with the stop codon TAA, whereas cox2 has the incomplete stop codon T. The phylogenetic tree showed that D. yiai and other six species belong to Phycitinae, are clustered into a clade.
Keywords: Dioryctria yiai, complete mitogenome, Pyralidae
Dioryctria yiai is a species of snout moth in the genus Dioryctria and is known from Taiwan and China. The larvae feed on Pinus massoniana and damage the branches, cones and shoots of host plant. The species overwinters in the larval stage within the damaged branch, cone or shoot. At present, its mitochondrial genome has not been reported publicly.
In this study, the samples were collected by light trapping in Taiyuan city of China (37°83′33″N, 112°66′61″E) in July 2019, some of these specimens were immediately frozen at −80 °C on board for mitogenome analysis and others were preserved by spreading wings in the Herbarium of Institute of Plant Protection, Shanxi Academy of Agricultural Sciences and their numbers is 2019TYKD1711-1715. Total genomic DNA was extracted from tail tip using the Ezup pillar genomic DNA extraction kit (Sangon Biotech, Shanghai, China). The mitogenome was sequenced by Illumina Hiseq 4000. Gene annotation was performed and circularity was checked using the MITOS2 webserver (Bernt et al. 2013, http://mitos.bioinf.uni-leipzig.de/).
The mitochondrial genome of D. yiai has a total length of 15,430 bp (GenBank accession No. MN658208), consisting of 13 PCGs, 22 tRNA, 2 rRNA genes, and an A + T-rich region. The major strand encodes a larger number of genes (9 PCGs and 14 tRNAs) than the minor strand (4 PCGs, 8 tRNAs, and 2 rRNA genes). Gene content and arrangement are highly conserved and typical of Lepidoptera (Wu et al. 2016). The mitogenome is highly biased toward A/T, contains 42.02% T, 38.99% A, 13.34% C, and 7.65% G, which is a feature commonly present in insects (Boore 1999).
All of the protein-coding genes have ATN as the start codon except for cox1, which starts with CGA. Eleven PCGs have the common stop codon TAA, cox2 has the incomplete stop codon T. All tRNAs exhibit typical clover-leaf secondary structure, except for tRNA-Ser(AGN) lacking the DHU arm, which is common in Lepidoptera insects (Garey and Wolstenholme 1989). The 16S rRNA is 1440 bp in length and the 12S rRNA is 795 bp in length. The A + T-rich region is 329 bp long located between 12S rRNA and tRNA-Met and it is longer than other most Lepidoptera insects. There is a motif ATAGA in downstream of 12S rRNA followed by an 18 bp Poly-T stretch.
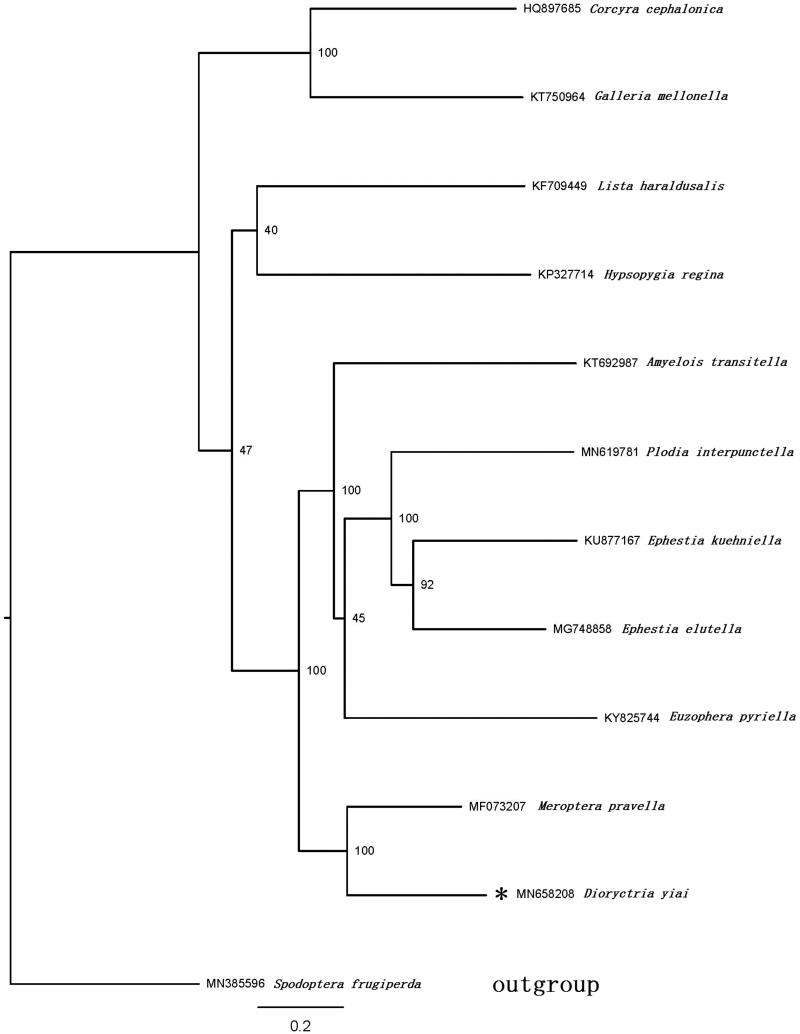
The phylogenetic position of D. yiai was inferred using sequences of the 13 PCGs of 12 species. Eleven of them belong to Pyralidae and a species from Noctuidae (which is used as outgroup) (Figure 1). The sequences were aligned with MAFFT v7.2 software (Katoh and Standley 2013), the evolutionary analyses were conducted with RAxML v8.2.10 (Stamatakis 2014) on the CIPRES Science Gateway (Miller et al. 2010). The result showed that D. yiai and other six species belong to Phycitinae, are clustered into a clade. Galleriinae include two species Corcyra cephalonica and Galleria mellonella and form a monophyly. Epipaschiinae (represented by Lista haraldusalis) is close to Pyralinae ((represented by Hypsopygia regina).
Figure 1.
Maximum-likelihood tree of evolutionary relationships D. yiai based on the complete mitogenomes of 12 Lepidopteran moths. The marked * is the sample sequence in this study.
Nucleotide sequence accession number
The complete mitochondrial genome sequence of D. yiai was deposited in GenBank under the accession number MN658208.
Funding Statement
This work was supported by the National Key Research and Development Program of China [2018YFD0200404-9], the Key R & D projects of Shanxi Province (Social Development) [201903D321058], the Open fund of Shanxi Key Laboratory of Integrated Pest Management in Agriculture [YHSW2019001] and the Special Fund for Outstanding Research Group in Shanxi Academy of Agricultural Sciences [No.YCX2018D2YS08].
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garey JR, Wolstenholme DR. 1989. Platyhelminth mitochondrial DNA: Evidence for early evolutionary origin of a tRNA ser AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol. 28(5):374–387. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14; New Orleans, Louisiana: Institute of Electrical and Electronics Engineers (IEEE). p. 1–8. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YP, Zhao JL, Su TJ, Luo AR, Zhu CD. 2016. The complete mitochondrial genome of Choristoneura longicellana (Lepidoptera: Tortricidae) and phylogenetic analysis of Lepidoptera. Gene. 591(1):161–176. [DOI] [PubMed] [Google Scholar]