Feder, Ali et al. reveal that spatial regulation is built into the heat shock response (HSR) in budding yeast. Heat shock triggers the J-protein Sis1 to relocalize away from the nucleoplasm. Loss of Sis1 in the nucleoplasm releases the transcription factor Hsf1 from Hsp70-mediated repression to activate the HSR.
Abstract
Cells exposed to heat shock induce a conserved gene expression program, the heat shock response (HSR), encoding protein homeostasis (proteostasis) factors. Heat shock also triggers proteostasis factors to form subcellular quality control bodies, but the relationship between these spatial structures and the HSR is unclear. Here we show that localization of the J-protein Sis1, a cofactor for the chaperone Hsp70, controls HSR activation in yeast. Under nonstress conditions, Sis1 is concentrated in the nucleoplasm, where it promotes Hsp70 binding to the transcription factor Hsf1, repressing the HSR. Upon heat shock, Sis1 forms an interconnected network with other proteostasis factors that spans the nucleolus and the surface of the endoplasmic reticulum. We propose that localization of Sis1 to this network directs Hsp70 activity away from Hsf1 in the nucleoplasm, leaving Hsf1 free to induce the HSR. In this manner, Sis1 couples HSR activation to the spatial organization of the proteostasis network.
Introduction
Protein homeostasis (proteostasis) describes a cellular state in which protein synthesis, folding, and degradation are balanced (Sala et al., 2017). When a cell has achieved proteostasis, molecular chaperones, sequestrases, and degradation factors, collectively referred to as the proteostasis network (PN), are expressed at sufficient levels such that nascent, misfolded, and aberrant proteins are efficiently folded, triaged, and degraded and the integrity of the proteome is maintained (Dikic, 2017; Jayaraj et al., 2020; Kim et al., 2013). Environmental fluctuations such as changes in temperature, nutrient availability, and signals from other cells can increase the burden on the PN and overwhelm its capacity. In addition to environmental sources, neurodegenerative disorders such as Parkinson’s disease and amyotrophic lateral sclerosis are characterized by protein aggregates and linked to deficits in PN components (Hipp et al., 2019; Labbadia and Morimoto, 2015; Soto, 2003). By contrast, aggressive human cancers have been shown to usurp the PN to support malignant growth in the presence of high mutational loads (Dai and Sampson, 2016; Dai et al., 2007; Oakes, 2017). Thus, modulation of PN expression has been proposed as a therapeutic avenue to treat both cancer and neurodegenerative diseases (Neef et al., 2011; Whitesell and Lindquist, 2009).
In eukaryotes, heat shock factor 1 (Hsf1) regulates a transcriptional program known as the “heat shock response” (HSR) that activates expression of chaperones such as Hsp70 and Hsp90 along with a suite of other PN genes (Anckar and Sistonen, 2011; Gomez-Pastor et al., 2018). The prevailing model for Hsf1 activation is a “chaperone titration model” in which the accumulation of chaperone clients that result from an overtaxed PN outcompete Hsf1 for access to chaperones, leaving Hsf1 free to induce the HSR (Masser et al., 2019, 2020; Pincus, 2020; Zheng et al., 2016). Several different chaperones have been implicated in Hsf1 repression, including Hsp90, Hsp70, J-proteins, and TRiC/CCT (Anckar and Sistonen, 2011; Brandman et al., 2012; Kim et al., 1999; Neef et al., 2014; Shi et al., 1998; Zou et al., 1998). Recently, multiple studies have pointed to Hsp70 as the primary and direct negative regulator of Hsf1 activity (Kmiecik et al., 2020; Masser et al., 2019; Peffer et al., 2019; Zheng et al., 2016). Since Hsp70 is a major transcriptional target of Hsf1, these two proteins form a negative feedback loop that controls the dynamics of HSR induction (Krakowiak et al., 2018). Although Hsp70 has emerged as a key repressor of Hsf1, roles for the other chaperones have not been ruled out.
While broadly consistent with existing data, the Hsp70 titration model does not account for the spatial organization of the cell in general or the protein quality control machinery in particular. First, the clients that titrate Hsp70 away upon heat shock are thought to be primarily nascent proteins emerging from ribosomes in the cytosol (Masser et al., 2019). Yet, Hsf1 activates target gene transcription in the nucleus in <2 min following heat shock (Kim and Gross, 2013). How do cytosolic clients titrate away nuclear Hsp70 so quickly, especially given that the molecular ratio of Hsp70:Hsf1 in yeast cells exceeds 1,000:1 (Ho et al., 2018)? Second, misfolded reporter proteins and components of the PN, including sequestrases and disaggregases, localize to specific subcellular sites in the cytosol (Escusa-Toret et al., 2013; Ho et al., 2019; Kaganovich et al., 2008; Miller et al., 2015). How does the stress-dependent spatial reorganization of the PN connect to HSR activation? That is, how does the cell biological response coordinate with Hsf1 to modulate the transcriptional response?
In this study, we combine chemical genetics, single-cell reporters, transcriptomics, proteomics, mathematical modeling, and 3D live-cell imaging to investigate the connection between the PN and the HSR in budding yeast. We identify the conserved J-protein Sis1 as a key factor required for Hsp70-mediated repression of Hsf1. J-proteins deliver clients to Hsp70 and activate Hsp70 to bind the clients with high affinity (Craig and Marszalek, 2017). We found that, under nonstress conditions, Sis1 localizes to the nucleoplasm, where it targets Hsp70 to bind and repress Hsf1. Upon heat shock, Sis1 relocalizes to the nucleolar periphery and the cytosolic face of the ER, where it forms a semicontiguous meshwork with other PN factors. Our data support a model in which, during heat shock, Sis1 relocalizes to (1) newly synthesized proteins condensed on the surface of the nucleolus and (2) ribosome-nascent chain complexes on the ER. This depletes Sis1 from the nucleoplasm, reducing the effective affinity of Hsp70 for Hsf1 and ultimately leaving Hsf1 free to activate the HSR. The rapid relocalization of Sis1 to the nucleolar periphery provides a mechanism for near-instantaneous activation of Hsf1 upon heat shock. Sis1 localization dynamics relay the state of the PN to Hsf1 to modulate the HSR accordingly.
Results
Chaperone anchor-away (AA) approach reveals that nuclear Sis1 represses Hsf1 activity
Multiple chaperones have been implicated in regulating Hsf1 to repress the HSR, including Hsp90, Hsp70, and J-proteins (Brandman et al., 2012; Shi et al., 1998; Zou et al., 1998). We and others have recently shown that Hsp70 directly binds and represses Hsf1 in yeast (Krakowiak et al., 2018; Masser et al., 2019; Peffer et al., 2019; Zheng et al., 2016). However, it remains unclear whether the other chaperones are also direct Hsf1 repressors. Since Hsf1 constitutively resides in the nucleus in yeast, the ability of any chaperone to repress Hsf1 should depend on its nuclear localization.
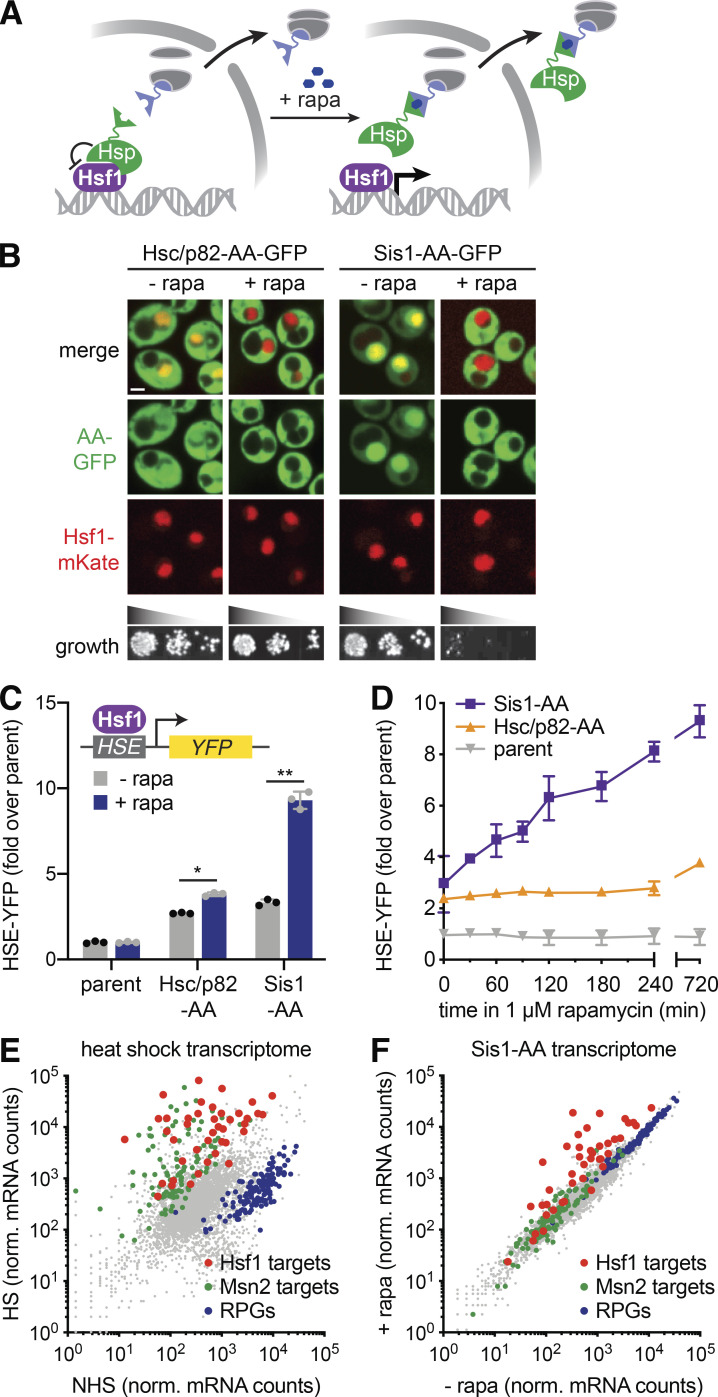
To conditionally remove chaperones from the nucleus and thereby determine whether they could be direct repressors of Hsf1, we used the AA approach to enable rapamycin-inducible nuclear depletion (Fig. 1 A; Haruki et al., 2008). In rapamycin-resistant strains, we AA tagged Hsp70 (Ssa1, Ssa2), Hsp90 (Hsc82, Hsp82), and J-proteins (Ydj1 and Sis1; Fig. S1 B and Table S1). We also doubly tagged Ssa1 and Ssa2 in one strain (Ssa1/2-AA) and Hsc82 and Hsp82 (Hsc/p82-AA) in another strain. Unfortunately, we found that tagging Ssa1/2 at either terminus to enable AA impaired function, rendering AA of Hsp70 unmeaningful, and that rapamycin addition had no effect on Ydj1 (Fig. S1 B). Thus, we focused our analysis on Hsp90 (Hsc/p82) and the J-protein Sis1. We coexpressed GFP-tagged versions of Hsc/p82-AA and Sis1-AA with Hsf1-mKate to visualize the nucleus. In the absence of rapamycin, Hsc/p82-GFP-AA localized equally throughout the cytosol and nucleus, while Sis1-GFP-AA was concentrated in the nucleus (Fig. 1 B). We found that rapamycin selectively depleted Hsc/p82-AA and Sis1-AA from the nucleus while leaving Hsf1-mKate in the nucleus (Fig. 1 C). Rapamycin had no discernable effect on proliferation in Hsc/p82-AA, but it inhibited cell growth in Sis1-AA cells (Fig. 1 C), suggesting either that Sis1 nuclear shuttling is essential or that anchoring Sis1 to the ribosome inactivates it altogether.
Figure 1.
Nuclear depletion of Sis1 rapidly and specifically activates Hsf1. (A) Cartoon of the chaperone AA approach. Upon addition of rapamycin (+ rapa), chaperones of interest are tethered to a ribosomal protein and depleted from the nucleus. (B) Spinning disc confocal images of cells expressing Hsf1-mKate and GFP-tagged versions of Hsp90 (Hsc/p82) and Sis1. Cells were treated with 1 µM rapa for 30 min before imaging. Scale bar is 2 µm. Lower panel: Indicated strains were serially diluted, spotted, and grown at 30°C for 36 h on YPD with or without 1 µM rapa. (C) HSE-YFP reporter assay of AA strains in the presence and absence of rapa normalized to the untagged AA parent strain. Error bars represent the SD of the replicates (n = 3 for each strain and condition), and statistical significance was determined using a two-tailed t test without assuming equal variance (*, P < 0.05; **, P < 0.01). Cells were treated with 1 µM rapa for 8 h before measuring the reporter. (D) Time course of HSE-YFP levels in the Sis1 and Hsp90 AA strains compared with the untagged parent following addition of 1 µM rapa. Error bars represent the SD of the replicates (n = 3 for each strain and time point). (E) RNA-seq of heat-shocked (HS) cells (15 min at 39°C) versus non–heat-shocked (NHS) cells. Hsf1 target genes are shown in red, Msn2 targets in green, and RPGs in blue. (F) RNA-seq analysis of the Sis1 AA strain in the absence and presence of 1 µM rapa for 30 min. Hsf1 target genes are shown in red, Msn2 targets in green, and RPGs in blue. norm., normalized.
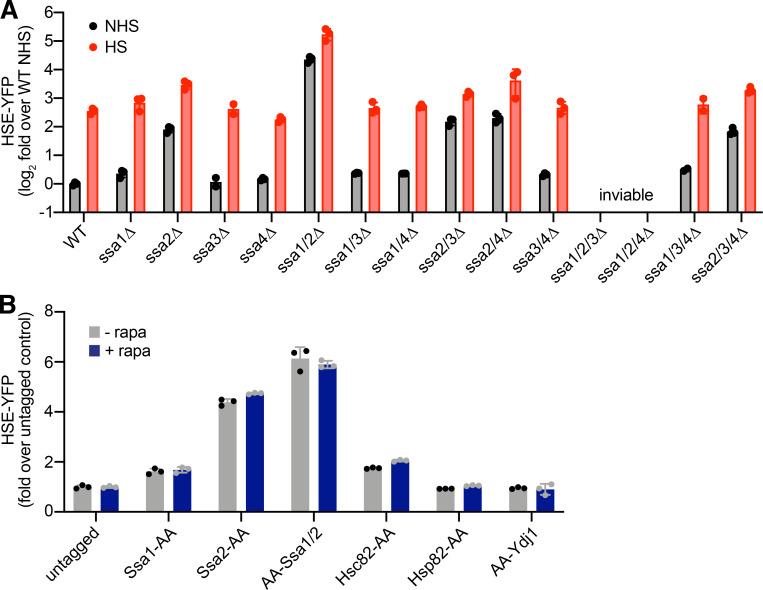
Figure S1.
Hsf1 activity in Hsp70 mutants and chaperone AA strains. (A) HSE-YFP reporter assay for Hsf1 activity in Hsp70 deletion strains. Cells were left untreated or were heat shocked at 39°C for 4 h, and YFP levels were measured by flow cytometry and normalized to untreated WT. Three biological replicates are shown, along with the mean (boxes) and SD (error bars). (B) HSE-YFP reporter assay of AA strains in the presence and absence of rapamycin (rapa) normalized to the untagged AA parent strain. Cells were treated with 1 µM rapa for 8 h before the reporter was measured. These data suggest that C-terminal tagging of Ssa1 and Ssa2 compromises their function nearly as severely as knocking them out. Consistent with this interpretation, individual AA tagging of Ssa1 and Ssa2 resulted in mild and moderate increases HSE-YFP levels, respectively, akin to their respective deletions. N-terminal AA tagging of Ssa2 likewise resulted in a constitutive increase in HSE-YFP signal. Error bars represent the SD of the replicates (n = 3 for each strain and condition). HS, heat shocked; NHS, non–heat shocked.
To assay the effects of anchoring away Hsc/p82 and Sis1 on Hsf1 activity, we expressed a fluorescent reporter of Hsf1 activity in the AA strains. The reporter consists of four repeats of the Hsf1 DNA binding site known as the “heat shock element” (HSE) driving YFP (Fig. 1 C, inset; Krakowiak et al., 2018; Zheng et al., 2016, 2018). The HSE-YFP reporter quantitatively reveals the role of Hsp70 in repressing Hsf1. Consistent with previous high-throughput experiments (Brandman et al., 2012), HSE-YFP levels were significantly increased in cells lacking the highly expressed Hsp70 paralogs (ssa1Δ and ssa2Δ) but not in cells lacking the stress-inducible paralogs (ssa3Δ and ssa4Δ; Fig. S1 A). Double knockout of Hsp70 paralogs (ssa1Δ ssa2Δ) resulted in synergistic activation of the HSE-YFP reporter to >20-fold above WT levels. Triple mutants ssa1Δ ssa2Δ ssa3Δ and ssa1Δ ssa2Δ ssa4Δ were inviable (Fig. S1 A). In the Hsc/p82-AA cells, the HSE-YFP reporter was modestly increased relative to an untagged strain in the absence of rapamycin. Addition of rapamycin resulted in a small but significant further increase in HSE-YFP levels (Fig. 1 C). However, it took multiple hours for nuclear depletion of Hsc/p82-AA to increase HSE-YFP levels (Fig. 1 D). Sis1-AA also displayed increased basal HSE-YFP signal. Yet, addition of rapamycin strongly induced the HSE-YFP reporter in the Sis1-AA strain (Fig. 1 C). AA of Sis1-AA led to an immediate and sustained increase in HSE-YFP signal (Fig. 1 D). The magnitude and immediacy of HSE-YFP induction following Sis1 AA suggested that Sis1 plays a major role in Hsf1 repression under nonstress conditions.
Although nuclear Sis1 is required to repress Hsf1, its nuclear depletion could still activate Hsf1 indirectly by causing general proteostasis collapse. If Sis1 is specifically required to repress Hsf1 in the absence of stress, then its nuclear depletion should activate Hsf1 without triggering simultaneous formation of protein aggregates. To determine whether anchoring away Sis1 results in protein aggregate formation, we performed imaging and fractionation experiments in Sis1-AA cells following heat shock and rapamycin treatment. As a live-cell proxy for protein aggregates, we monitored the localization of Hsp104, a disaggregase that forms clusters during stress that are thought to mark protein aggregates (Tkach and Glover, 2004). Heat shock resulted in significant accumulation of Hsp104-mKate clusters. By contrast, anchoring away Sis1 did not alter Hsp104-mKate localization, suggesting that no aggregates were formed (Fig. S2, A and B). We also performed biochemical fractionation experiments following heat shock and rapamycin treatment. While Ssa1/2 largely moved to the insoluble protein fraction following heat shock, indicative of its association with protein aggregates, Ssa1/2 remained in the soluble fraction following rapamycin treatment, suggesting that aggregates are not forming and Hsp70 is available to repress Hsf1 (Fig. S2, C and D). Thus, while heat shock triggers the formation of protein aggregates as marked by changes in Hsp104 localization and Hsp70 fractionation, anchoring away Sis1 appears to leave proteostasis largely unperturbed.
Figure S2.
AA of Sis1 does not trigger proteostasis collapse. (A) Left: Cells expressing Hsp104-mKate and Sis1-AA-GFP in untreated cells and following the addition of rapamycin (rapa) for 1 h to AA Sis1-AA-GFP. The two dark regions in the rapa-treated cells are the nucleus and the vacuole. Right: Cells expressing Hsp104-mKate were imaged by spinning disc confocal microscopy under non–heat shock (NHS) condition and following 15 min of heat shock (HS) at 39°C. Scale bar is 2 µm. (B) Quantification of the number of Hsp104-mKate clusters in individual cells either left untreated, following heat shock, or following Sis1 AA with rapa. Individual cells are shown, and the total number is indicated in each condition. Lines represent means, and error bars show SEM. Statistical significance was determined using a two-tailed t test without assuming equal variance (****, P < 0.0001). (C) “Total-sup-pellet” assay of Sis1-AA cells either treated with rapa or heat shocked over a 60-min time course. At each time point, cells were harvested, lysed, and fractionated to resolve the soluble (sup.) and aggregate (pellet) protein fractions. Each fraction was blotted and probed with anti-Ssa1/2 antibodies to detect Hsp70. (D) Quantification of the sup. Hsp70 fraction in each sample compared with the untreated sample. The experiment was performed in triplicate, and the error bars represent the 95% confidence bounds on the mean.
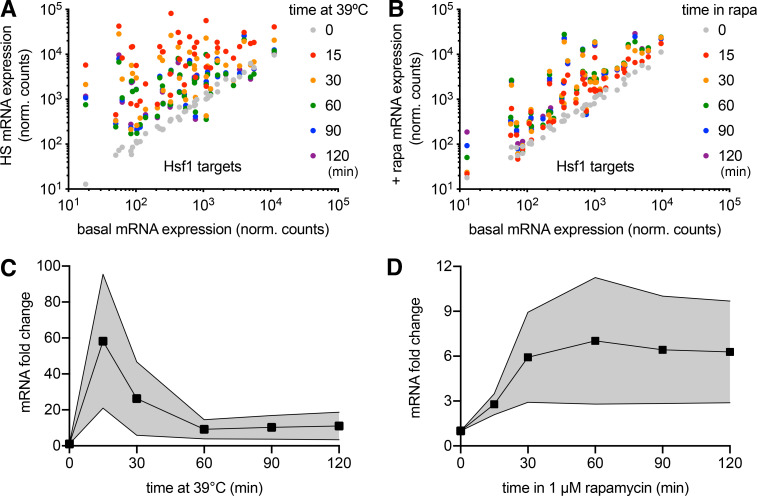
To determine whether anchoring away Sis1 causes other forms of cellular stress, we performed mRNA deep sequencing (RNA sequencing [RNA-seq]) point spread function (PSF) of Sis1-AA cells over time courses following heat shock and rapamycin treatment. In addition to activating the HSR, yeast cells also activate a parallel stress pathway in response to heat shock known as the “general stress response” and repress expression of ribosomal protein genes (RPGs; Solís et al., 2016). Indeed, we observed that heat shock resulted in induction of the full Hsf1 regulon (Pincus et al., 2018) but also induced the general stress response and repressed RPGs (Fig. 1 E). By contrast, Sis1 AA specifically induced the Hsf1 regulon without other changes to the transcriptome (Fig. 1 F). Moreover, while Hsf1 target mRNA expression peaked after 15 min of heat shock and subsequently declined, Sis1-AA resulted in sustained expression of the full Hsf1 regulon, albeit to a lower magnitude than heat shock (Fig. S3). Taken together, the aggregation and transcriptomic experiments demonstrate that anchoring away Sis1 results in specific activation of Hsf1 without triggering proteostasis collapse or other stress responses.
Figure S3.
Comparative transcriptomics following heat shock and Sis1 AA. (A) All 42 Hsf1 target gene expression levels over a heat shock (HS) time course. Most genes peak at the 15-min time point. (B) All 42 Hsf1 target gene expression levels for a time course following Sis1 AA. Levels do not decline over time and peak at later time points for most genes. (C) The average (line and points) and SD (shaded area) of the induction of the 42 Hsf1 target genes over a heat shock time course showing the adaptive response. (D) The same as in E, but for the Sis1 AA time course. There is no adaptation, suggesting the feedback loop has been severed. ns, not significant; rapa, rapamycin.
Nuclear Sis1 drives the interaction between Hsp70 and Hsf1
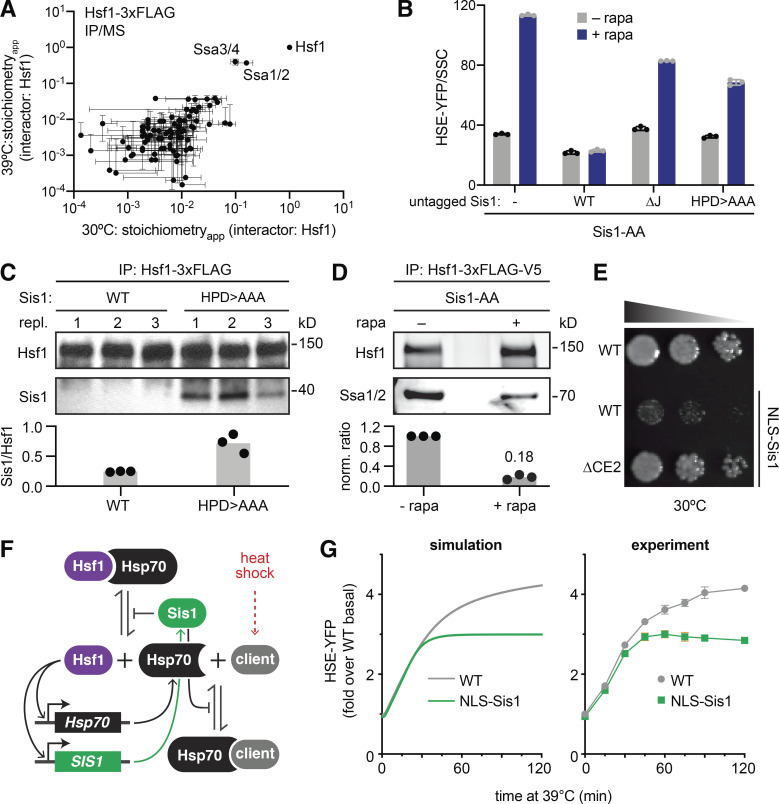
We next tested if Hsf1 forms a protein complex with Sis1. We performed anti-FLAG immunoprecipitation (IP) of Hsf1-3xFLAG from unstressed cells and from cells that had been heat shocked for 60 min to induce high-level expression of Sis1. While we efficiently pulled down Hsf1, mass spectrometry (MS) analysis failed to identify Sis1 as an interactor in either condition (Table S2). Only Ssa1/2/3/4 (Hsp70 paralogs) associated specifically with Hsf1, consistent with our previous results (Fig. 2 A; Zheng et al., 2016).
Figure 2.
Sis1 promotes interaction between Hsf1 and Hsp70. (A) Anti-FLAG IP of Hsf1-3xFLAG-V5 followed by MS at 30°C and at 39°C for 60 min. IPs were performed in biological triplicates. Levels of interacting proteins were plotted relative to the amount of Hsf1 measured in each replicate to generate a stoichiometryapp value. Mean and SD of the replicates are shown. (B) Sis1 functional complementation assay. HSE-YFP levels were measured in Sis1-AA cells expressing additional copies of untagged Sis1 from the SIS1 promoter: WT Sis1, ΔJ, a mutant of Sis1 in which the HPD motif residues are mutated to alanine (HPD>AAA), or no additional Sis1 (−). Cells were treated with 1 µM rapamycin (+ rapa) for 8 h before the reporter was measured by flow cytometry. YFP levels in each cell were normalized by each cell’s side scatter value (SSC; a proxy for size), and the medians of the resulting distributions are plotted. Error bars represent the SD of the replicates (n = 3 for each strain and condition). (C) Anti-FLAG IP of Hsf1-3xFLAG in cells expressing V5-tagged Sis1, either WT or HPD>AAA. Cells were grown at 30°C in log phase before harvesting. IP eluates were blotted and probed with anti-FLAG (Hsf1) and anti-V5 (Sis1) antibodies. Values plotted below are the ratios of Sis1:Hsf1 in each replicate (repl.); gray boxes show the mean of the replicates. (D) Anti-FLAG/V5 serial IP of Hsf1-3xFLAG-V5 followed by anti-FLAG and anti-Ssa1/2 Western blots. Cells were treated in the absence and presence of 60 min of 1 µM rapa treatment to AA Sis1. Plotted values are normalized ratios of Ssa1/2:Hsf1 in each of three biological replicates; gray boxes show the mean values of the replicates. (E) Dilution series spot assay. Ectopic expression of NLS-Sis1 impairs cells growth; deletion of the CE2 region of Hsf1 rescues growth in NLS-Sis1 cells. Cells were grown for 36 h on YPD + 20 nM estradiol to induce NLS-Sis1. (F) Schematic of the mathematical model of the Hsf1 regulation by the Sis1 and Hsp70. During heat shock, Hsp70 clients accumulate and compete for Sis1 and Hsp70 and release Hsf1. Active Hsf1 then induces expression of Sis1 and Hsp70. (G) Simulations of the mathematical model and corresponding experiments of heat shock time courses of HSE-YFP reporter strains with and without ectopic expression of NLS-Sis1. NLS-Sis1 was induced with 20 nM estradiol for 1 h before the time course. Error bars represent the SD of the replicates (n = 3 for each strain and condition).
As a J-protein, Sis1 is thought to deliver clients to Hsp70 and activate high-affinity Hsp70 binding (Craig and Marszalek, 2017). Thus, since we found no evidence that Sis1 forms a stable complex with Hsf1, we hypothesized that Sis1 promotes the interaction between Hsf1 and Hsp70 but does not remain part of the mature complex. To test this, we (1) tested the role of the Sis1 J-domain in repressing Hsf1, (2) used an Sis1 mutant to trap the putative interaction with Hsf1, and (3) assessed the role of Sis1 in promoting the interaction of Hsf1 with Hsp70.
To determine whether the J-domain participates in Hsf1 repression, we generated a mutant of Sis1 that completely lacks the J-domain (ΔJ) and another mutant that harbors alanine substitutions at the conserved HPD motif required for Sis1 to transfer clients to Hsp70 (HPD>AAA; Kityk et al., 2018). In the Sis1-AA strain expressing the HSE-YFP reporter, we integrated an additional, untagged allele of Sis1—WT, ΔJ, or HPD>AAA—into the genome under control of the SIS1 promoter. We assayed these additional alleles for functional complementation by measuring HSE-YFP levels following addition of rapamycin to AA WT Sis1. While untagged WT perfectly complemented, addition of rapamycin induced the HSE-YFP reporter in both ΔJ and HPD>AAA cells (Fig. 2 B). Thus, the J-domain and HPD motif are required for Sis1 to fully repress Hsf1.
While the J-domain is required for full repression of Hsf1, HSE-YFP levels after adding rapamycin were reduced in ΔJ and HPD>AAA cells relative to cells without any untagged Sis1, suggesting these alleles are able to partially complement (Fig. 2 B). Since Sis1 is a dimer, it could be that the mutants are binding to the tagged WT Sis1 and thereby retaining some residual activity. However, even in this case, one tagged protein may be sufficient to result in anchoring away the dimer. Alternatively, in the absence of the ability to activate Hsp70, the mutants of Sis1 could still be able to bind to clients but not transfer them to Hsp70, thus trapping substrates in a complex that is typically a transient intermediate. In the case of Hsf1, this could result in partial repression of transcription for steric reasons. To test whether the HPD>AAA mutant could trap the interaction with Hsf1, we performed IPs of Hsf1-3xFLAG in cells expressing V5-tagged versions of either WT Sis1 or HPD>AAA. Indeed, while we were unable to detect WT Sis1, the HPD>AAA mutant coprecipitated with Hsf1 in lysates from unstressed cells (Fig. 2 C). These data suggest that Hsf1 functions as a canonical client for the J-protein/Hsp70 system: Sis1 and Hsf1 directly interact in a transient complex before Hsf1 is transferred to Hsp70.
To test whether Sis1 is required to promote the interaction of Hsf1 with Hsp70, we performed serial IPs of Hsf1-3xFLAG-V5 in the Sis1-AA strain in the presence and absence of rapamycin and blotted for Hsp70 (Ssa1/2). Anchoring away Sis1 resulted in greater than a fivefold decrease in the amount of Ssa1/2 that coprecipitated with Hsf1-3xFLAG-V5 (Fig. 2 D). Thus, in the absence of nuclear Sis1, the interaction between Hsp70 and Hsf1 is reduced, indicating that Sis1 promotes the interaction of Hsf1 with Hsp70 in the absence of stress.
As an additional piece of evidence that Sis1 mediates its repressive effect on Hsf1 by promoting Hsp70 binding, we identified a genetic interaction relating Sis1 to Hsf1 and Hsp70. We found that ectopic expression of Sis1 fused to a nuclear localization signal (NLS-Sis1) impairs growth in WT cells under nonstress conditions. However, this growth phenotype is suppressed in cells expressing Hsf1ΔCE2 as the only copy of Hsf1. Hsf1ΔCE2 lacks a binding site for Hsp70 that represses Hsf1 activity (Fig. 2 E; Krakowiak et al., 2018; Peffer et al., 2019). Thus, relieving Hsf1 repression by Hsp70 is sufficient to rescue growth in cells with too much Sis1 in the nucleus. This result implies that overexpression of Sis1 in the nucleus leads to hyperrepression of Hsf1 by Hsp70 to the point that Hsf1 ceases to perform its essential basal transcriptional function. Without the CE2 binding site for Hsp70, NLS-Sis1 cannot completely inactivate Hsf1. This genetic result further supports a role for Sis1 in repressing Hsf1 via Hsp70.
Mathematical modeling of the Sis1–Hsp70–Hsf1 regulatory circuit
To formalize the role of Sis1 in the Hsf1 regulatory circuit, we generated a mathematical model of the HSR. Like our previous versions (Krakowiak et al., 2018; Zheng et al., 2016), the model is based on a negative feedback loop in which Hsf1 activates Hsp70 expression and Hsp70 represses Hsf1 activity. Heat shock generates “clients” (proteins recognized by chaperones) that accumulate and titrate away Hsp70, releasing active Hsf1 (Fig. 2 F). We incorporated Sis1 into the model by having the dissociation rate of the Hsf1–Hsp70 complex depend inversely on the concentration of Sis1; that is, the more Sis1, the greater the stability of the Hsf1–Hsp70 complex (see Materials and methods). Since Sis1 is a transcriptional target of Hsf1, we also modeled Sis1 induction in response to heat shock using the same Hill function describing induction of Hsp70 and the HSE-YFP reporter. The output of the model is expression of HSE-YFP, enabling comparison with the cellular response.
We first confirmed that the model was able to recapitulate the dynamics of the HSR by comparing a simulation of the HSE-YFP reporter over a heat shock time course in WT cells with experimental data (Fig. 2 G). Next, we tested if the model was capable of capturing the effect of overexpression of Sis1. The model only simulates the cell nucleus, so overexpression of Sis1 in the model is equivalent to an increase in nuclear Sis1 in cells. The model predicted that increased expression of Sis1 in the nucleus should reduce the maximum HSE-YFP output over a heat shock time course and attenuate the response faster than WT (Fig. 2 G). To test this prediction experimentally, we generated a strain with the HSE-YFP reporter in which we could induce ectopic expression of Sis1 with an appended NLS-Sis1 (Park et al., 2013). We performed a heat shock time course from 25°C to 39°C for 2 h in cells with and without induction of ectopic NLS-Sis1. In agreement with the model, cells expressing NLS-Sis1 failed to reach the maximal HSE-YFP level achieved by WT and deactivated more quickly (Fig. 2 G). In the model, the reason that the initial increase in Hsf1 activity during heat shock is the same in WT and NLS-Sis1 is that Hsp70 becomes limiting immediately after heat shock, not Sis1. Immediately following heat shock, clients greatly outnumber Hsp70, so no matter how much Sis1 is present, all the Hsp70 is bound by clients. Once Hsf1 induces more Hsp70, the extra Sis1 serves to increase the affinity of newly available Hsp70 for Hsf1, enabling rapid deactivation. These results demonstrate that a model based on the Sis1–Hsp70–Hsf1 regulatory axis is consistent with experimental data, and such a model can quantitatively recapitulate the effects of perturbations to Sis1.
The Sis1 interactome during acute heat shock
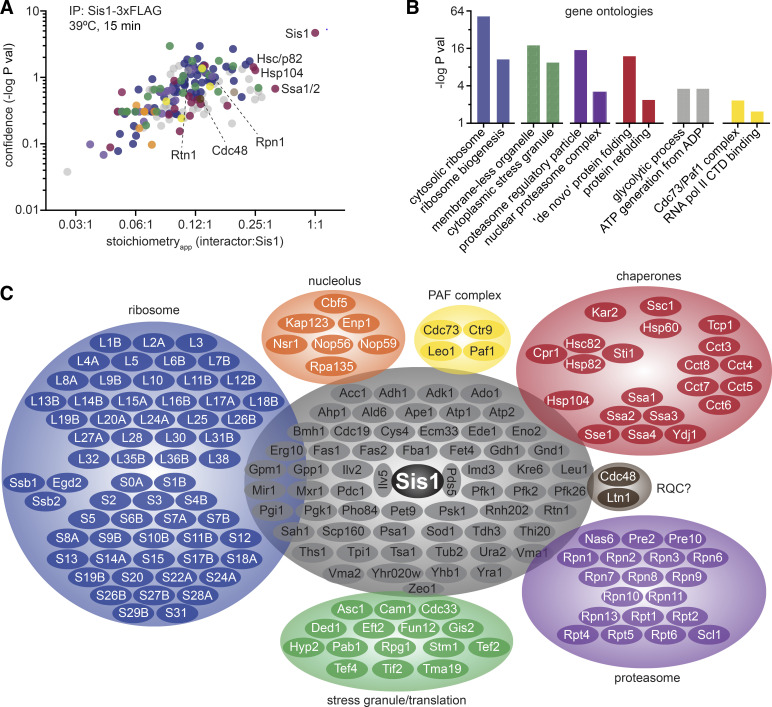
Our chemical genetic experiments and mathematical modeling suggest that Sis1 is a key regulator of the HSR, but they provide little insight into the physiological role of Sis1 during heat shock. To identify endogenous proteins that interact with Sis1, we performed IP-MS of Sis1-3xFLAG following a 15-min heat shock. We identified 192 proteins with >99% confidence in 3 biological replicates (Table S3), and Sis1 was both the most abundant and most significantly enriched protein in the set. In Fig. 3 A, we plot the significance with which each of these proteins was enriched over an untagged control as a function of the relative abundance of each protein with respect to Sis1 (“apparent stoichiometry” [stoichiometryapp]). Gene ontology analysis of these proteins revealed functional enrichment for ribosomal proteins and biogenesis factors, stress granule components, proteasome subunits, chaperones, glycolytic enzymes, and the Paf transcription elongation complex (Fig. 3 B). In addition to these categories, we also observed interactions with nucleolar factors and the ER structural protein Rtn1 (Fig. 3, A and C). The heat shock–dependent Sis1 interaction network thus appears to link major cellular chaperone systems (Hsp70, Hsp90, and TRiC/CCT) with other major proteostasis systems, including stress granules and the proteasome. The mix of nucleolar, cytosolic, and ER factors among the Sis1 interactors suggests that Sis1 localizes in a complex subcellular pattern during heat shock.
Figure 3.
The Sis1 interactome during heat shock. (A) Anti-FLAG IPs were performed from an untagged strain and a strain expressing Sis1-3xFLAG, both heat shocked for 15 min at 39°C. The significance over background is plotted as a function of the stoichiometryapp—the ratio of the background-subtracted quantitative value of each of the 192 interacting proteins to the value of Sis1. Proteins are color coded to match the categories in C. (B) Gene ontology associations enriched among the Sis1 interactors during heat shock. (C) Sis1 interactors grouped by functional category. Ribosomal proteins are abbreviated with their subunit designation (L or S) and their identifier. Proteins in gray are all those that do not belong to the other groups, enriched for highly expressed cytosolic enzymes. RNA pol II CTD, RNA polymerase II C-terminal domain.
Sis1 localizes to the nucleolar periphery and cytosolic clusters during heat shock
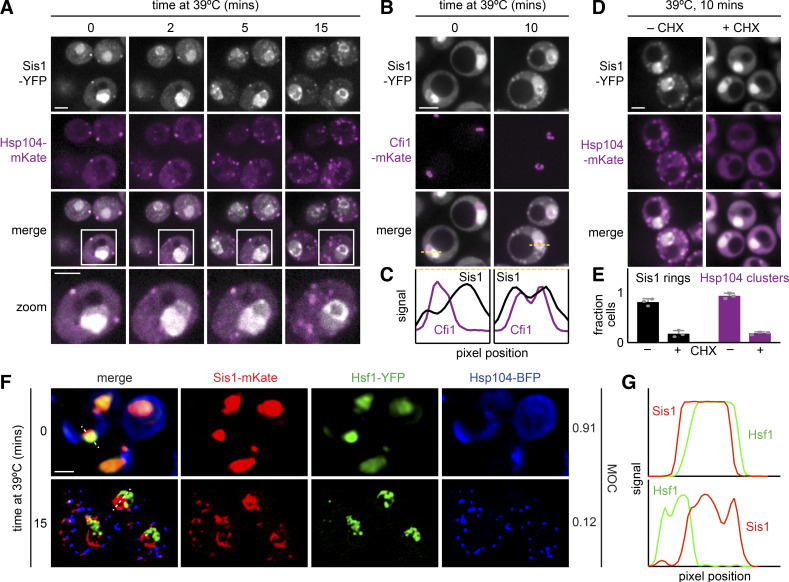
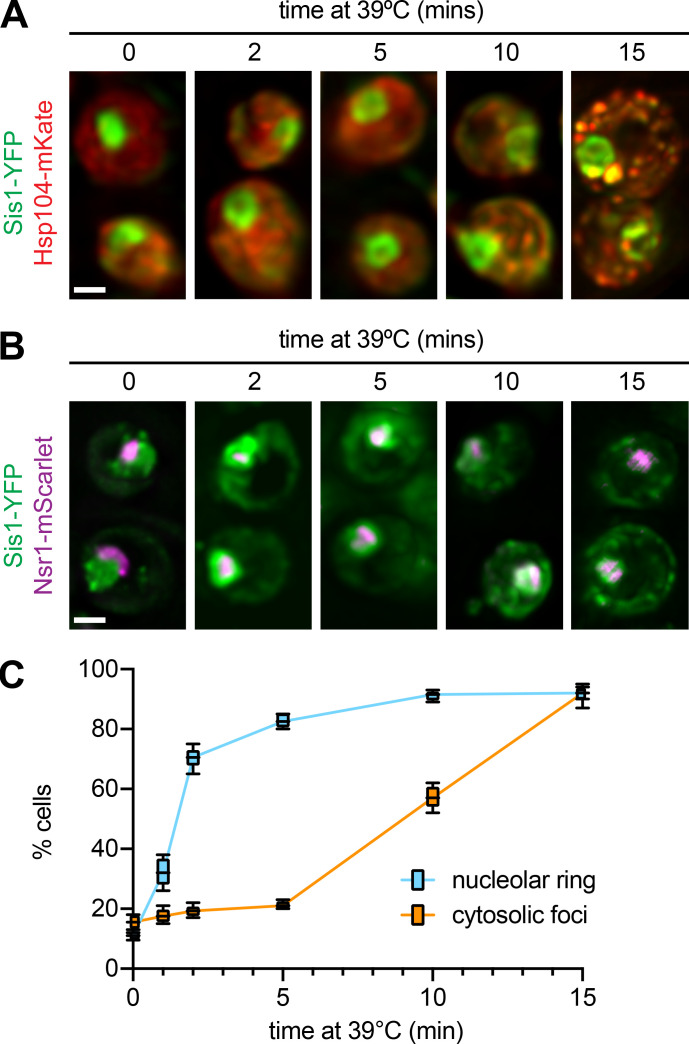
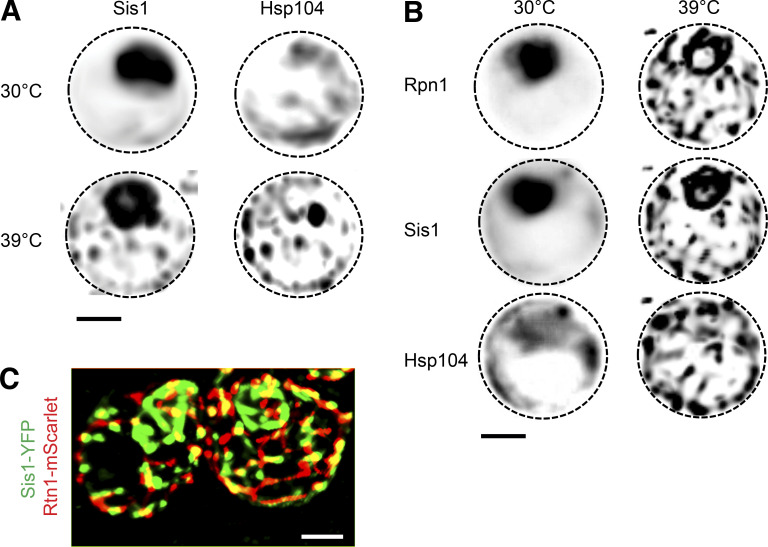
To investigate the localization of Sis1 during heat shock, we tagged Sis1 with YFP to enable live-cell fluorescence imaging over a heat shock time course. In the Sis1-YFP cells, we tagged the disaggregase chaperone Hsp104 with mKate. Hsp104 is among the strongest Sis1 interactors (Fig. 3 A) and is known to form cytosolic clusters during heat shock. Hsp104 clusters are an indicator that a given cell is stressed. Prior to heat shock, Sis1-YFP was concentrated in the nucleus in all cells but also showed diffuse cytosolic signal along with a few cytosolic clusters that colocalized with Hsp104-mKate (Fig. 4 A). Rapidly upon heat shock, Sis1-YFP relocalized to form a ring in the nucleus and, in concert with Hsp104-mKate, coalesced into multiple cytosolic clusters (Fig. 4 A and Video 1). Quantification of a time course of fixed cells revealed that Sis1 formed the subnuclear ring in most cells in <2 min after heat shock, while the formation of cytosolic clusters that colocalize with Hsp104 takes >5 min (Fig. S4, A and C).
Figure 4.
Sis1 localizes to a perinucleolar ring and cytosolic clusters during heat shock. (A) Live-cell heat shock time course of cells expressing endogenously tagged Sis1-YFP and Hsp104-mKate. Cells were imaged by spinning disc confocal microscopy. Scale bar is 2 µm. (B) Cells expressing endogenously tagged Sis1-YFP and the nucleolar marker Cfi1-mKate imaged under nonstress and heat shock conditions. Scale bar is 2 µm. (C) Scans of fluorescence signal for Sis1-YFP (black) and Cfi1-mKate (purple) along the dashed lines in the merged images in B. (D) Cells expressing endogenously tagged Sis1-YFP and Hsp104-mKate following 10 min of heat shock after either no pretreatment or pretreatment with cycloheximide (CHX) to arrest protein synthesis. Scale bar is 2 µm. (E) Quantification of the fraction of cells with an Sis1 nuclear ring (black) and Hsp104 clusters (purple) after either no pretreatment or pretreatment with CHX. Experiments were performed with >50 cells in each replicate. Error bars represent the SD of the replicates (n = 3 for each strain and condition). (F) Live cells expressing endogenously tagged Sis1-mKate, Hsf1-YFP, and Hsp104-BFP were imaged in a lattice light-sheet microscope under nonstress conditions and following heat shock at 39°C for 15 min. MOC is a fraction of Sis1 that colocalizes with Hsf1. Scale bar is 2 µm. (G) Scans of fluorescence signal for Sis1-mKate (red) and Hsf1-YFP (green) along the dashed lines in the merged images in F.
Video 1.
Live cell imaging on a spinning disc confocal microscope of cells growing at 25°C expressing Sis1-YFP (white) and Hsp104-mKate (magenta) shifted to 39°C and imaged over time. 45 s elapse between frames.
Figure S4.
Sis1 perinucleolar localization is an early event following heat shock. (A) Images of fixed cells expressing Sis1-YFP and Hsp104-mKate over a series of early time points following heat shock. (B) Images of fixed cells expressing Sis1-YFP and Nsr1-mScarlet over a series of early time points following heat shock. Nsr1 is a nucleolar protein identified in the Sis1-3xFLAG IP/MS experiment (Fig. 3). Scale bar is 2 μm. (C) Quantification of the fraction of cells with Sis1 in nuclear rings and cytosolic clusters that colocalize with Hsf1. Experiments were performed in triplicate: Line shows the mean; box shows the SD; and error bars show the range.
The nuclear ring encompassed a subregion of the nucleus that was relatively depleted of Sis1-YFP under nonstress conditions. Based on the crescent-shaped morphology of this area in unstressed cells (Figs. 4 A and S4 B) and the IP-MS data showing nucleolar proteins among the Sis1 interactors (Fig. 3 C), we hypothesized this nuclear subregion to be the nucleolus (Aris and Blobel, 1988). To monitor Sis1 localization relative to the nucleolus, we imaged Sis1-YFP in a strain in which we tagged the nucleolar resident protein Cfi1 with mKate (Visintin et al., 1999). Under nonstress conditions, Cfi1-mKate localized to the expected subregion of the nucleus. Following heat shock, Sis1-YFP formed a ring around Cfi1-mKate (Fig. 4 B). Fluorescence intensity line scans showed that the signal peaks for Cfi1-mKate and Sis1-YFP are adjacent before heat shock; upon heat shock, the Cfi1-mKate signal becomes surrounded by the Sis1-YFP signal (Fig. 4 C). In addition to Cfi1-mKate, we also imaged Nsr1, a nucleolar protein that we identified among the Sis1 interactors (Fig. 3 C). Nsr1-mScarlet displayed a crescent-shaped localization pattern adjacent to Sis1-YFP under nonstress conditions and became immediately surrounded by Sis1-YFP upon heat shock (Fig. S4, B and C). These data demonstrate that Sis1 encircles the nucleolus during heat shock.
The nucleolus is the site of ribosome biogenesis. Nascent ribosomal proteins traffic from the cytosol, where they are synthesized to the nucleolus, where they are incorporated with ribosomal RNA into large and small ribosomal subunits. Unincorporated “orphan” ribosomal proteins (oRPs) are known to be potent activators of Hsf1 in the absence of stress (Albert et al., 2019; Tye et al., 2019), and ribosomal proteins constituted >30% (59 of 192) of the Sis1-interacting proteins (Table S3). We hypothesize that heat shock results in the accumulation of recently synthesized oRPs that recruit Sis1 to form the perinucleolar ring.
A prediction of this hypothesis is that ongoing protein synthesis, a prerequisite for ribosome biogenesis, should be required for Sis1 to relocalize during heat shock. Notably, ongoing protein synthesis has recently been shown to be required for Hsp104 to form clusters and for full Hsf1 activation during heat shock (Masser et al., 2019; Tye et al., 2019). To block translation in general and the production of ribosomal proteins in particular, we added cycloheximide 5 min before heat shocking cells expressing Sis1-YFP and Hsp104-mKate, allowing time for recently translated ribosomal proteins to be incorporated into ribosomal subunits. In the presence of cycloheximide, we observed strong reduction in formation of both Sis1 cytosolic clusters and nucleolar rings upon heat shock (Fig. 4, D and E). This suggests that ongoing translation is required to trigger Sis1 relocalization during heat shock and is consistent with a role for newly synthesized ribosomal proteins in Sis1 localization to the nucleolus. However, whether oRPs are the biochemical entities that recruit Sis1 to the nucleolus remains to be determined.
Regardless of the species involved, an implication of Sis1 relocalization to the nucleolar periphery is that it may be depleted from the nucleoplasm, where Hsf1 resides. To determine if Sis1 localizes away from Hsf1 during heat shock, we tagged Sis1 with mKate in a strain expressing Hsf1-YFP and Hsp104-BFP. Under nonstress conditions, Sis1-mKate and Hsf1-YFP both show diffuse nuclear localization patterns, and Hsp104-BFP is diffuse in the cytosol (Fig. 4 F). Quantification revealed >90% signal overlap between Sis1-mKate and Hsf1-YFP (Mander’s overlap coefficient [MOC] = 0.91; Manders et al., 1992). Upon heat shock, consistent with previous observations, Sis1-mKate and Hsp104-BFP formed puncta in the cytosol; Sis1-mKate formed a subnuclear ring; and Hsf1-YFP formed subnuclear clusters (Fig. 4 F; Chowdhary et al., 2019). Despite both being in the nucleus, Sis1-mKate and Hsf1-YFP showed nearly mutually exclusive spatial patterns and a greater than sevenfold reduction in signal overlap during heat shock (MOC = 0.12; Fig. 4 G). Taken together, these data demonstrate that Sis1 localizes to a perinucleolar ring and cytosolic clusters during heat shock and away from Hsf1, provided that cells are actively translating.
Sis1 and Hsp104 colocalize with the proteasome on the ER
In addition to nucleolar factors, the IP/MS dataset revealed Sis1 interactions with cytosolic and ER factors (Fig. 3 C). To resolve the subcellular localization of Sis1 with respect to these extranuclear interactors with high spatial resolution, we employed lattice light sheet imaging to generate 3D reconstructions of cells under nonstress and heat shock conditions (Chen et al., 2014). We constructed tricolor yeast strains expressing Sis1-YFP, Hsp104-BFP, and a third factor of interest tagged with mScarlet, all integrated in the genome. We investigated connections between Sis1, the proteasome, and the ER-associated factors.
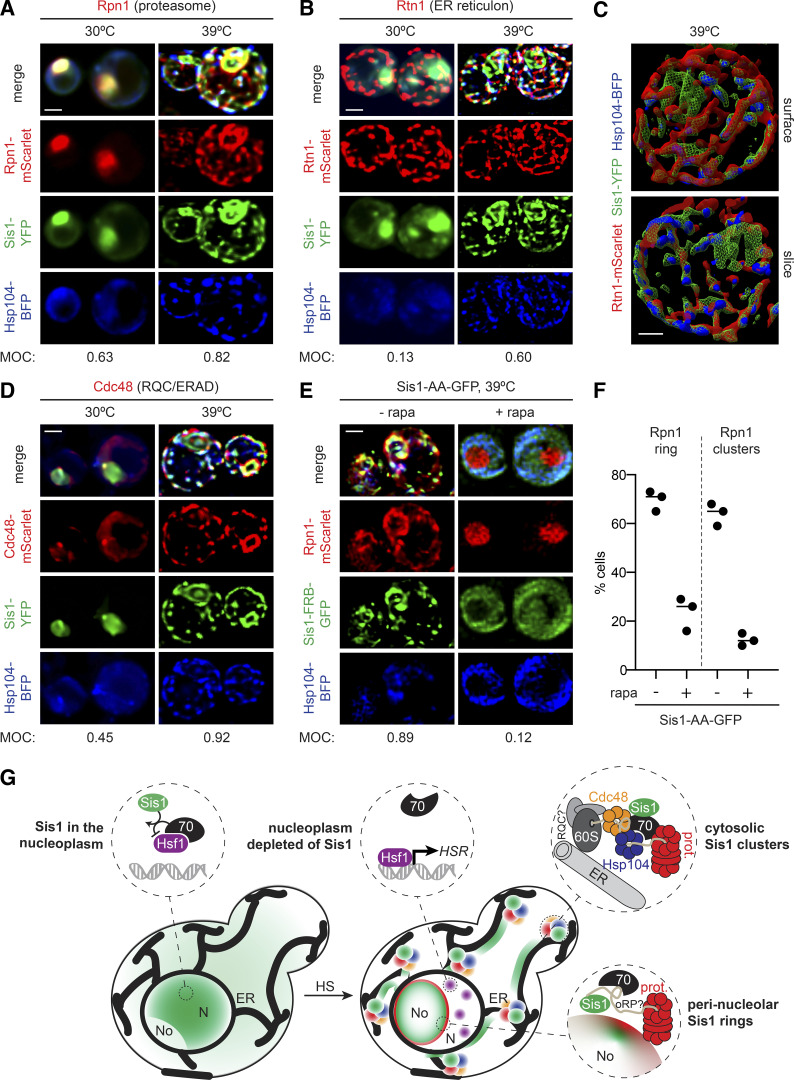
To image the proteasome, we tagged the subunit Rpn1 with mScarlet. Prior to heat shock, Rpn1-mScarlet localized diffusely to the nucleus and showed a high level of overlap with Sis1-YFP (MOC = 0.63; Fig. 5 A). Upon heat shock, Sis1-YFP and Rpn1-mScarlet colocalized in a nuclear ring and cytosolic clusters (MOC = 0.82; Fig. 5 A, Fig. S5 A, and Video 2). The colocalization of Rpn1-mScarlet and Sis1-YFP suggests that Sis1 localizes to subcellular sites of proteasomal degradation during heat shock. The cytosolic clusters, but not the nuclear ring, also colocalized with Hsp104-BFP (Fig. 5 A). Moreover, rather than forming a series of discrete puncta, the cytosolic clusters containing Sis1-YFP, Hsp104-BFP, and Rpn1-mScarlet appeared to be organized into a semicontiguous network (Fig. 5 A; and Fig. S5, A and B).
Figure 5.
Sis1 forms an interconnected spatial network with other proteostasis factors during heat shock (HS). (A and B) Deconvolved lattice light-sheet 3D reconstructions of live cells under nonstress conditions and 15 min of HS at 39°C expressing endogenously tagged Sis1-YFP and Hsp104-BFP with Rpn1-mScarlet (A), a subunit of the proteasome; or Rtn1-mScarlet (B), a reticulon protein component of the ER membrane. (C) Top: Space-filling 3D surface rendering of a cell at 39°C showing the association of Sis1-YFP (green) and Hsp104-BFP (blue) with the inside surface of the reticulated ER as marked by Rtn1-mScarlet (red). Bottom: Coronal slice at the midpoint of the cell above. The Sis1-YFP subnuclear ring (green) can be seen and is disconnected from the cytosolic Rtn1 network. Scale bar is 1 µm. (D) Cdc48-mScarlet, an AAA ATPase involved in ERAD and RQC. (E) AA of Sis1 precludes formation the spatial PN. Cells expressing Hsp104-BFP and Rpn1-mScarlet to mark the proteasome in an Sis1-AA background with Sis1-AA-GFP were imaged using a lattice light-sheet microscope following 15 min of HS at 39°C following no pretreatment or pretreatment with rapamycin (rapa) to AA Sis1. In A, B, D, and E, scale bar is 2 µm, and MOC is the fraction of YFP that overlaps with mScarlet. (F) The fraction of cells showing Rpn1 subnuclear rings and cytosolic clusters was quantified during HS in cells with no pretreatment or with rapa pretreatment to AA Sis1. Experiments were performed in triplicate with >20 cells per replicate; the line shows the mean of the replicates. (G) Cartoon model of how the HS-dependent spatial reorganization of the protein homeostasis network is coupled to the regulation of the HSR. Left: In the absence of stress, Sis1 is diffuse throughout the cell and concentrated in the nucleus. In the nucleus, it activates Hsp70 to repress Hsf1. Right: Upon HS, Sis1 relocalizes to the periphery of the nucleolus (No) and the surface of the ER in the cytosol. In the nucleoplasm (N), Hsf1 is now free of Hsp70 and can cluster and activate the HS transcriptional response. Sis1 colocalizes with the proteasome (prot.) at the nucleolar periphery. On the ER, we propose that Sis1 interacts with stalled 60S ribosomes with nascent chains, Cdc48 and possibly the RQC complex, Hsp104, and the proteasome to participate in the resolution of HS-induced cotranslational misfolding.
Figure S5.
The proteostasis machinery forms a contiguous network on the ER. (A) Maximum-intensity negative projections of Sis1-YFP and Hsp104-BFP signal in cells under nonstress and heat shock conditions (39°C for 15 min). The signal is enhanced to show the cytosolic interconnections between the nodes. Scale bar is 2 µm. (B) The same as in A, but for the proteasome component Rpn1. (C) Overlay of Sis1-YFP and Rtn1-mScarlet from Fig. 5 C. Scale bar is 2 µm.
Video 2.
3D rotation of a lattice light-sheet image of a cell expressing Sis1-YFP (green), Rpn1-mScarlet (red), and Hsp104-BFP (blue) at 39°C for 15 min. Rpn1 marks the proteasome.
We wondered whether the network-like organization of the cytosolic clusters could be due to an association with the ER. The ER has previously been implicated as an organizational hub for the spatial arrangement of protein quality control factors, including Hsp104 (Escusa-Toret et al., 2013; Zhou et al., 2014). Moreover, we found that the ER structural protein Rtn1 and the ER/ribosome-associated quality control (RQC) factor Cdc48 coprecipitated with Sis1-3xFLAG during heat shock (Fig. 3 C). To test whether the Sis1 cytosolic network forms in proximity to the ER, we imaged Rtn1-mScarlet in cells expressing Sis1-YFP and Hsp104-BFP. Indeed, Sis1-YFP increased its signal overlap with Rtn1-mScarlet from MOC = 0.13 under nonstress conditions to MOC = 0.60 during heat shock (Fig. 5 B), suggesting increased association with the ER. The remaining Sis1 signal is largely composed of the perinucleolar ring (Fig. S5 C). Space-filling 3D cell projections reveal the orientation of the interaction network, with Rtn1 toward the periphery and Sis1 and Hsp104 lining the interior (Fig. 5 C and Video 3). These data suggest that Sis1 forms a highly connected network with Hsp104 and the proteasome on the surface of the ER.
Video 3.
3D rotation of a lattice light-sheet image of a cell expressing Sis1-YFP (green), Rtn1-mScarlet (red), and Hsp104-BFP (blue) heat shocked at 39°C for 15 min. This video shows colocalization of Sis1 and Hsp104 with the reticulated ER during heat shock.
To determine whether these ER-associated clusters could be the sites of protein quality control processes upstream of proteasomal degradation, we imaged Cdc48-mScarlet. Cdc48, another protein we found to coimmunoprecipitate with Sis1-3xFLAG during heat shock (Fig. 3 C), is an AAA-ATPase that mediates the handoff of ubiquitylated nascent chains to the proteasome in multiple quality control processes, including ER-associated degradation (ERAD) and RQC (Brandman et al., 2012; Nakatsukasa et al., 2008). In the absence of stress, Cdc48-mScarlet displayed diffuse cytosolic signal and overlapped at the nuclear periphery with Sis1-YFP (MOC = 0.45; Fig. 5 B). Heat shock triggered Cdc48-mScarlet relocalization to a network at the cell cortex that largely overlapped with Sis1-YFP (MOC = 0.92; Fig. 5 B and Video 4). Taken together, the imaging results demonstrate that Sis1 localizes to the surface of the ER along with Cdc48, Hsp104, and Rpn1, suggesting that these clusters may harbor multiple protein quality control processes.
Video 4.
3D rotation of a lattice light-sheet image of a cell expressing Sis1-YFP (green), Cdc48-mScarlet (red), and Hsp104-BFP (blue) at 39°C for 15 min. Cdc48 functions in RQC and ERAD.
Last, we tested if Sis1 is required for the proteasome to relocalize during heat shock. To this end, we anchored away Sis1 before heat shocking cells and monitored localization of Rpn1-mScarlet. Following Sis1 AA, we found that Rpn1-mScarlet remained nucleoplasmic during heat shock and failed to form a nuclear ring or cytosolic clusters (Fig. 5, E and F). This suggests either that unanchored Sis1 is required to recruit the proteasome to the nucleolar ring and cytosolic clusters or that anchoring away Sis1 before heat shock, which is sufficient to activate Hsf1 and the HSR (Fig. 1 F), preadapts the cell such that the effects of heat shock do not overwhelm the PN. However, RNA-seq indicates that anchoring away Sis1 for 15 min results in less than threefold induction for the average Hsf1 target gene (compared with >50-fold induction after 15 min of heat shock; Fig. S3, C and D), suggesting preadaptation is likely to be limited. Thus, Sis1 relocalization may be an upstream event required for the relocalization of the proteasome during heat shock.
Discussion
Here, we show that localization of the J-protein Sis1 is a key determinant of Hsf1 activity and transcriptional induction of the HSR. Nuclear depletion of Sis1 in the absence of stress triggers Hsf1 dissociation from Hsp70 and specifically activates the HSR. During physiological heat shock, Sis1 leaves the nucleoplasm and relocalizes to a subnuclear ring and cytosolic clusters. The Sis1 subnuclear ring surrounds the nucleolus. The cytosolic clusters form a semicontiguous network adjacent to the ER and colocalize with the disaggregase Hsp104, the proteasome, and Cdc48 (Fig. 5). By spatially collaborating with proteostasis machinery at the surfaces of the nucleolus and ER during heat shock, Sis1 relocalizes away from Hsf1 and thereby ceases to promote Hsp70-mediated Hsf1 repression. Thus, Sis1 couples the spatial remodeling of the proteostasis machinery to regulation of the HSR. Whether Sis1 relocalization represents a common mechanism to activate Hsf1 in response to other stresses remains to be explored. However, complementary to our results, Sis1 was recently identified as the strongest Hsf1 repressor in the genome in a CRISPR interference–based screen for Hsf1 regulators conducted under a battery of environmental conditions (Alford et al., 2020 Preprint).
Identification of a role for Sis1 in promoting Hsp70-mediated repression of Hsf1 addresses two conceptual challenges in the current “chaperone titration model” of Hsf1 regulation. In the current model, heat shock triggers cotranslational protein misfolding on ribosomes in the cytosol, and these nascent misfolded proteins titrate Hsp70 away from Hsf1 in the nucleus (Masser et al., 2019, 2020). The first conceptual challenge is based on the observation that Hsp70 outnumbers Hsf1 by three orders of magnitude in the cell (Ho et al., 2018). How is it possible for all the Hsp70 to be titrated away from Hsf1, given such lopsided stoichiometry? The second conceptual challenge is based on the requirement for ongoing translation in order for heat shock to activate Hsf1 (Masser et al., 2019). If the clients that titrate Hsp70 away from Hsf1 are nascent misfolded proteins, how can these ribosome-associated nascent chains in the cytosol titrate away Hsp70 from the nucleus to enable near-instantaneous Hsf1 activation? Sis1 helps to resolve both of these issues.
First, Sis1 renders the stoichiometric imbalance between Hsp70 and Hsf1 irrelevant. We found that Sis1 requires its J-domain and HPD motif in order to repress Hsf1 (Fig. 2 B); that is, Sis1 must be able to deliver substrates to Hsp70 and activate ATP hydrolysis. This implies that Hsp70 binding to Hsf1 is a nonequilibrium process that cannot be explained by simple mass action. The fraction of Hsf1 bound to Hsp70 thus does not depend on the total concentration of Hsp70 in the cell; rather, it depends on the concentration of Sis1 in the nucleoplasm: Sis1 is the limiting factor. As Sis1 moves away from the nucleoplasm during heat shock and its local concentration near Hsf1 drops, the effective affinity of Hsp70 for Hsf1 drops precipitously, and Hsf1 dissociates. Thus, Hsf1 is derepressed, regardless of the local Hsp70 concentration.
Second, the rapid relocalization of Sis1 in response to heat shock means that Hsp70 does not need to be titrated out of the nucleus. Rather, Sis1 is depleted from the nucleoplasm and rapidly moves to the nucleolar periphery following heat shock, thereby releasing Hsf1 from repression by Hsp70. Subsequently, Sis1 also forms a network of cytosolic clusters (Figs. 4 and S4). Formation of both localization patterns—the perinucleolar ring and the cytosolic clusters—is blocked if cells are pretreated with cycloheximide to inhibit translation 5 min before heat shock (Fig. 4, D and E). Thus, it is likely that newly synthesized proteins that localize to the nucleus are responsible for recruiting Sis1 to the nucleolar periphery. The requirement for newly synthesized proteins to drive rapid Sis1 relocalization within the nucleus reconciles seemingly contradictory requirements of the chaperone titration model: Near-instantaneous activation of Hsf1 following heat shock is achieved by Sis1 relocalization, obviating the need to invoke the slower process of titration of nuclear Hsp70 by cytosolic misfolded proteins.
Sis1 relocalization to the nucleolar periphery during heat shock is consistent with an emerging view of the nucleolus as a protein quality control compartment during stress (Frottin et al., 2019). The nucleolus is the site of ribosome biogenesis, and yeast cells must generate >105 nascent ribosomal proteins per minute to support cell division (Woolford and Baserga, 2013). Until they are embedded in the ribosome, ribosomal proteins are unstable and aggregation prone (Albert et al., 2019; Tye et al., 2019). Moreover, ribosomal proteins contain many positively charged and hydrophobic residues, precisely the amino acids enriched in Hsp70 substrate binding sites (Rüdiger et al., 1997). Indeed, oRPs have been shown to be potent Hsf1 activators in the absence of other stresses (Albert et al., 2019; Tye et al., 2019). Future experiments will be required to establish whether oRPs accumulate at the nucleolar periphery during heat shock and directly interact with Sis1, as well as whether the nucleolus is itself performing a quality control function in yeast.
In the cytosol, Sis1 also displays stress-dependent relocalization. 3D reconstructions revealed that, rather than forming a series of discrete foci, Sis1 and Hsp104 form a semicontiguous network along with the proteasome and Cdc48 that appears to associate with the ER (Fig. 5, A–D; and Fig. S5 C). We propose that this network of proteostasis factors serves to organize chaperones in two dimensions to triage ribosomes with misfolded nascent chains and facilitate proteasomal degradation of the nascent chains (Fig. 5 G). This reorganization of Sis1 and other proteostasis components during stress demonstrates that, in addition to being a network of coregulated genes, the PN forms a dynamic and spatially organized physical structure. Stress-dependent relocalization of Sis1 establishes a general spatial mechanism that enables direct communication between the PN throughout the cell and Hsf1 in the nucleus to regulate the HSR according to need.
Materials and methods
Strain construction and cell growth
Yeast strains used in this study are listed in Table S1. All strains are derivatives of W303, and fluorescent protein and epitope tags are integrated into the genome. Cells were cultured in SDC media (synthetic media with dextrose and complete amino acids) for confocal imaging and flow cytometry experiments, and they were cultured in SDC-riboflavin and folic acid for low autofluorescence for lattice light-sheet imaging. Cells were cultured in yeast extract peptone dextrose (YPD) media for IP and RNA-seq experiments. Heat shocks were performed at 39°C. AA experiments were conducted using 1 µM rapamycin.
HSE-YFP flow cytometry assays
Reporter levels in untreated, endpoint, and time-course assays from heat-shocked and rapamycin-treated cells were measured in the Whitehead Flow Cytometry Facility or the University of Chicago Cytometry and Antibody Technology Facility. These measurements were performed using a BD Fortessa flow cytometer equipped with a high-throughput sampler, and the results were analyzed using FlowJo.
RNA-seq sample preparation and analysis
Total RNA was purified from yeast using hot acid phenol extraction and ethanol precipitation (Solís et al., 2016), and polyadenylation-positive RNA-seq libraries were constructed using the NEB Next Ultra RNA Kit (E7770; New England Biolabs). Sequencing was performed on an Illumina HiSeq 2500 at the Whitehead Institute Genome Technology Core Facility, and reads were aligned and quantified as previously described (Solís et al., 2016). Raw sequence files and processed data were deposited in the Gene Expression Omnibus (accession no. GSE145936).
IP
Hsf1-3xFLAG-V5 was serially immunoprecipitated using anti-FLAG (M8823; Sigma-Aldrich) and anti-V5 (M167-11; MBL) magnetic beads, and eluates were blotted for IP/Western blot analysis (Zheng et al., 2016; Zheng and Pincus, 2017). For IP/MS of Sis1-3xFLAG and Hsf1-3xFLAG, the protocol was modified to only perform the anti-FLAG IP using a short incubation of the anti-FLAG beads with total lysate for 15 min before washing and eluting with 3xFLAG peptide. The short incubation was designed to increase the likelihood of capturing transient interactions while reducing nonspecific interactions. All IP experiments were performed in biological triplicate. Triplicate IPs were also performed on an untagged strain exposed to the same conditions to subtract background and calculate significance. Immunoblots were performed with anti-FLAG (M2, F1804; Sigma-Aldrich), anti-V5 (V5-10, V8012; Sigma-Aldrich), or anti-Ssa1/2 (rabbit polyclonal; gift from Elizabeth A. Craig, Department of Biochemistry, University of Wisconsin, Madison, WI; Lopez-Buesa et al., 1998) antibodies as indicated.
Total/soluble/pellet fractionation
Cells were collected by filtration and immediately cryopreserved. Lysate was prepared using cryomilling, and differential centrifugation was performed to separate total, supernatant, and pellet fractions (Wallace et al., 2015). Sample fractions were analyzed by immunoblotting for the presence of Hsp70 (anti-Ssa1/2; gift from Elizabeth A. Craig).
MS
For the Sis1-3xFLAG-V5 untreated samples and the Hsf1-3xFLAG-V5 samples, MS analysis was performed as described in the subsections below. Additional MS analysis was performed at the Whitehead Proteomics Core Facility (Zheng et al., 2016).
In-solution trypsin digestion
30 µl eluate was in-solution digested with trypsin by first reducing in 50 mM ammonium bicarbonate with 6 µl RapiGest surfactant (Waters) and 10% 200 mM Tris(2-carboxyethyl)phosphine, alkylated with 50 mM iodoacetamide (33 µl) in the dark for 30 min at RT, and digested with 1:50 vol/vol trypsin (Promega) at 37°C overnight. Detergent was removed with 1 µl trifluoroacetic acid at 37°C for 45 min. Digested peptides were cleaned up on a C18 column (Pierce), speed vacuumed, and sent to the Proteomics Core at Mayo Clinic for liquid chromatography–tandem MS (LC-MS/MS).
HPLC for MS
All samples were resuspended in Burdick and Jackson HPLC-grade water containing 0.2% formic acid (Fluka), 0.1% trifluoroacetic acid (Pierce), and 0.002% ZWITTERGENT 3-16 (Calbiochem), a sulfobetaine detergent that contributes the following distinct peaks at the end of chromatograms: MH+ at 392 m/z, and in-source dimer [2M + H+] at 783 m/z, and some minor impurities of ZWITTERGENT 3-12 seen as MH+ at 336 m/z. The peptide samples were loaded to a 0.25-µl C8 OptiPak trapping cartridge custom packed with Michrom Magic (Optimize Technologies) C8, washed, then switched in line with a 20-cm × 75-µm C18 packed spray tip nano column packed with Michrom Magic C18AQ for a two-step gradient. Mobile phase A was water/acetonitrile/formic acid (98/2/0.2), and mobile phase B was acetonitrile/isopropanol/water/formic acid (80/10/10/0.2). Using a flow rate of 350 nl/min, a 90-min, two-step LC gradient was run from 5% B to 50% B in 60 min, followed by 50–95% B over the next 10 min, holding for 10 min at 95% B, brought back to starting conditions, and reequilibrated.
LC-MS/MS data acquisition and analysis
The samples were analyzed via data-dependent electrospray LC-MS/MS on a Thermo Q-Exactive Orbitrap mass spectrometer using a 70,000 resolving power survey scan in profile mode, mass-to-charge ratio 360–2,000 D, with lock masses, followed by 20 high-energy collisional dissociation fragmentation scans at 17,500 resolution on doubly and triply charged precursors. Single-charged ions were excluded, and ions selected for MS/MS were placed on an exclusion list for 60 s. An inclusion list was used that consisted of expected prototypic peptide ions in the 2+ and 3+ charge state for the yeast proteins ySIS1 and yHSF1. All LC-MS/MS RAW data files were analyzed with MaxQuant version 1.5.2.8, searching against the Swiss-Prot yeast database (downloaded May 23, 2019, with isoforms; 12,154 entries) *.fasta sequence, using the following criteria: label-free quantification (LFQ) was selected for quantitation with a minimum of one high-confidence peptide to assign LFQ intensities. Trypsin was selected as the protease, with maximum missing cleavage set to 2. Carbamidomethyl (C) was selected as a fixed modification. Variable modifications were set to oxidization (M), formylation (N-term), deamidation (NQ), and phospo (STY). The orbitrap mass spectrometer was selected using an MS error of 20 ppm and an MS/MS error of 0.5 D. A 1% false discovery rate cutoff was selected for peptide, protein, and site identifications. Ratios were reported on the basis of LFQ intensities of protein peak areas determined by MaxQuant (version 1.5.2.8) and reported in the proteingroups.TXT file. The proteingroups.TXT file was processed in Perseus (version 1.6.7). Proteins were removed from this results file if they were flagged by MaxQuant as “Contaminants,, “Reverse,” or “Only identified by site.” Three biological replicates were performed. Samples were filtered to require hits to have been seen in at least two replicates per condition. LFQ peak intensities were log2 transformed and median normalized, and missing values were imputed via default settings in Perseus.
Mathematical modeling
To model the Sis1-Hsp70-Hsf1 circuit, we have expanded the differential equation model from Zheng et al. (2016) to include the effects of Sis1. The model consists of five different protein species—Hsf1, Hsp70 (HSP), client protein (UP), Sis1, reporter protein YFP—and two protein complexes:
where denotes the nuclear concentration of each respective species. We refer the reader to Zheng et al. (2016) for modeling details, assumptions, and parametric values. The rate denotes the binding of Hsp70 to Hsf1 to create an inactive complex , and the complex dissociates with rate Sis1 enhances the repression of Hsf1 by Hsp70, and we phenomenologically capture this by assuming that is dependent on Sis1 levels as per the following Hill equation:
with and The rate is the binding of Hsp70 to client proteins to create the complex that dissociates with rate The degradation of UP by Hsp70 is captured via the rate The activation of both YFP and Hsp70 by Hsf1 is modeled by a Hill equation with and The activation of Sis1 is similarly modeled, but with activation rate that depends on the nuclear export signal.
The above differential equation model was run with the following initial values (in a.u.) at time
The effect of NLSs was modeled by changing a single parameter, with for WT and for NLS.
Confocal imaging
Confocal imaging was performed at the Nikon Imaging Center at the Whitehead Institute for Biomedical Research using a spinning disc system (Krakowiak et al., 2018) with the following parameters: Nikon Ti-E base, 1.49 NA, 100× objective, Yokogawa CSU-X1 spinning disc, Andor iXon 897E EM charge-coupled device camera, and MetaMorph acquisition software. Cells were grown and indicated fluorophores were imaged in SDC media, and heat shock was performed on live cells from 25°C to 39°C using an objective heater (Chowdhary et al., 2019). Images were autocontrasted using ImageJ, and single z-slices are shown.
Lattice light-sheet imaging and analysis
Lattice light-sheet imaging was performed at the University of Chicago Integrated Light Microscopy Core using a phase 2 system designed by Intelligent Imaging Innovations and run in SlideBook 6.0 software (Intelligent Imaging Innovations). The design is a modification of the original (Chen et al., 2014) with greater automation and stability. Optics were aligned daily, and bead PSFs were collected before cells were imaged. The imaging camera used was a Hamamatsu Fusion chilled sCMOS run at default speed/quality. The annulus mask was set for a 20-µm beam length (outer NA, 0.55; inner NA, 0.493) with 400-nm thickness, with dither set at 9 µm. Laser intensities were set to balance signal and bleaching rates. Temperature was controlled by a built-in Peltier device (empirically set to indicated temperatures). Sample scan image stacks were deskewed in SlideBook, and the TIF series images were processed using National Institutes of Health ImageJ (Fiji version). Graphics processing unit–based Richardson-Lucy deconvolution used measured PSFs or theoretical PSFs via Brian Northan’s “Ops” implementation (https://github.com/imagej/ops-experiments). Reconstructions and videos were assembled using ClearVolume (Royer et al., 2015).
Online supplemental material
Figs. S1, S2, and S3 are associated with Fig. 1 and show control experiments and additional data. Fig. S4 is related to Fig. 4, and Fig. S5 is related to Fig. 5. These figures show additional imaging results. The videos are related to Figs. 4 and 5. Video 1 shows cells over time during heat shock, while Video 2, Video 3, and Video 4 are 3D projections of single time points during heat shock. Table S1, Table S2, and Table S3 list the yeast strains and MS data related to Figs. 2 and 3.
Supplementary Material
lists yeast strains used in this study.
contains Hsf1-3xFLAG IP/MS results.
contains Sis1-3xFLAG IP/MS results.
Acknowledgments
We thank H. Yoo and D.A. Drummond for providing important insights into the biochemical role of Sis1 and for technical assistance with the fractionation assay. We thank J. Greenberg, E. Ferguson, G. Bushkin, and the other members of the Pincus laboratory for insightful discussions and comments on the manuscript. We are grateful to T. Volkert and S. Gupta at the Whitehead Genome Technology Core and E. Spooner at the Whitehead Proteomics Core Facility for technical assistance.
The initial phase of this work was funded by an Early Independence Award from the National Institutes of Health Office of the Director (DP5 OD017941 to D. Pincus), while the later phase was supported by institutional startup funding provided by the Department of Molecular Genetics and Cell Biology in the Biological Sciences Division at the University of Chicago (to D. Pincus).
The authors declare no competing financial interests.
Author contributions: conceptualization: D. Pincus; investigation: Z.A. Feder, A. Ali, J. Krakowiak, X. Zheng, D. Wolfgeher, V.P. Bindokas, and D. Pincus; methodology: V.P. Bindokas, D. Wolfgeher, and D. Pincus; formal analysis: A. Singh; writing – original draft: D. Pincus; writing – review and editing: all authors; visualization: D. Pincus; supervision: S.J. Kron and D. Pincus.
References
- Albert B., Kos-Braun I.C., Henras A.K., Dez C., Rueda M.P., Zhang X., Gadal O., Kos M., and Shore D.. 2019. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. eLife. 8:e45002 10.7554/eLife.45002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford B.D., Valiant G., and Brandman O.. 2020. Genome-wide, time-sensitive interrogation of the heat shock response under diverse stressors via ReporterSeq. bioRxiv. (Preprint posted March 30, 2020). 10.1101/2020.03.29.014845 [DOI] [Google Scholar]
- Anckar J., and Sistonen L.. 2011. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 80:1089–1115. 10.1146/annurev-biochem-060809-095203 [DOI] [PubMed] [Google Scholar]
- Aris J.P., and Blobel G.. 1988. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107:17–31. 10.1083/jcb.107.1.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.W., Zhou S., King D., Shen P.S., Weibezahn J., et al. 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 151:1042–1054. 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.C., Legant W.R., Wang K., Shao L., Milkie D.E., Davidson M.W., Janetopoulos C., Wu X.S., Hammer J.A. III, Liu Z., et al. 2014. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 346:1257998 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary S., Kainth A.S., Pincus D., and Gross D.S.. 2019. Heat shock factor 1 drives intergenic association of its target gene loci upon heat shock. Cell Rep. 26:18–28.e5. 10.1016/j.celrep.2018.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E.A., and Marszalek J.. 2017. How do J-proteins get Hsp70 to do so many different things? Trends Biochem. Sci. 42:355–368. 10.1016/j.tibs.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., and Sampson S.B.. 2016. HSF1: guardian of proteostasis in cancer. Trends Cell Biol. 26:17–28. 10.1016/j.tcb.2015.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Whitesell L., Rogers A.B., and Lindquist S.. 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 130:1005–1018. 10.1016/j.cell.2007.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I. 2017. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86:193–224. 10.1146/annurev-biochem-061516-044908 [DOI] [PubMed] [Google Scholar]
- Escusa-Toret S., Vonk W.I., and Frydman J.. 2013. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol. 15:1231–1243. 10.1038/ncb2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin F., Schueder F., Tiwary S., Gupta R., Körner R., Schlichthaerle T., Cox J., Jungmann R., Hartl F.U., and Hipp M.S.. 2019. The nucleolus functions as a phase-separated protein quality control compartment. Science. 365:342–347. 10.1126/science.aaw9157 [DOI] [PubMed] [Google Scholar]
- Gomez-Pastor R., Burchfiel E.T., and Thiele D.J.. 2018. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19:4–19. 10.1038/nrm.2017.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., and Laemmli U.K.. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 31:925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Hipp M.S., Kasturi P., and Hartl F.U.. 2019. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 20:421–435. 10.1038/s41580-019-0101-y [DOI] [PubMed] [Google Scholar]
- Ho B., Baryshnikova A., and Brown G.W.. 2018. Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 6:192–205.e3. 10.1016/j.cels.2017.12.004 [DOI] [PubMed] [Google Scholar]
- Ho C.T., Grousl T., Shatz O., Jawed A., Ruger-Herreros C., Semmelink M., Zahn R., Richter K., Bukau B., and Mogk A.. 2019. Cellular sequestrases maintain basal Hsp70 capacity ensuring balanced proteostasis. Nat. Commun. 10:4851 10.1038/s41467-019-12868-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraj G.G., Hipp M.S., and Hartl F.U.. 2020. Functional modules of the proteostasis network. Cold Spring Harb. Perspect. Biol. 12:a033951 10.1101/cshperspect.a033951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., and Frydman J.. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature. 454:1088–1095. 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.R., Kang H.S., and Kim H.D.. 1999. Geldanamycin induces heat shock protein expression through activation of HSF1 in K562 erythroleukemic cells. IUBMB Life. 48:429–433. 10.1080/713803536 [DOI] [PubMed] [Google Scholar]
- Kim S., and Gross D.S.. 2013. Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and mediator tail subunits Med15 and Med16. J. Biol. Chem. 288:12197–12213. 10.1074/jbc.M112.449553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.E., Hipp M.S., Bracher A., Hayer-Hartl M., and Hartl F.U.. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82:323–355. 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- Kityk R., Kopp J., and Mayer M.P.. 2018. Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Mol. Cell. 69:227–237.e4. 10.1016/j.molcel.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Kmiecik S.W., Le Breton L., and Mayer M.P.. 2020. Feedback regulation of heat shock factor 1 (Hsf1) activity by Hsp70-mediated trimer unzipping and dissociation from DNA. EMBO J. 39:e104096 10.15252/embj.2019104096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakowiak J., Zheng X., Patel N., Feder Z.A., Anandhakumar J., Valerius K., Gross D.S., Khalil A.S., and Pincus D.. 2018. Hsf1 and Hsp70 constitute a two-component feedback loop that regulates the yeast heat shock response. eLife. 7:e31668 10.7554/eLife.31668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J., and Morimoto R.I.. 2015. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84:435–464. 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Buesa P., Pfund C., and Craig E.A.. 1998. The biochemical properties of the ATPase activity of a 70-kDa heat shock protein (Hsp70) are governed by the C-terminal domains. Proc. Natl. Acad. Sci. USA. 95:15253–15258. 10.1073/pnas.95.26.15253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders E.M., Stap J., Brakenhoff G.J., van Driel R., and Aten J.A.. 1992. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 103:857–862. [DOI] [PubMed] [Google Scholar]
- Masser A.E., Ciccarelli M., and Andréasson C.. 2020. Hsf1 on a leash - controlling the heat shock response by chaperone titration. Exp. Cell Res. 396:112246 10.1016/j.yexcr.2020.112246 [DOI] [PubMed] [Google Scholar]
- Masser A.E., Kang W., Roy J., Mohanakrishnan Kaimal J., Quintana-Cordero J., Friedländer M.R., and Andréasson C.. 2019. Cytoplasmic protein misfolding titrates Hsp70 to activate nuclear Hsf1. eLife. 8:e47791 10.7554/eLife.47791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.B., Ho C.T., Winkler J., Khokhrina M., Neuner A., Mohamed M.Y., Guilbride D.L., Richter K., Lisby M., Schiebel E., et al. 2015. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 34:778–797. 10.15252/embj.201489524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Huyer G., Michaelis S., and Brodsky J.L.. 2008. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 132:101–112. 10.1016/j.cell.2007.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef D.W., Jaeger A.M., and Thiele D.J.. 2011. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat. Rev. Drug Discov. 10:930–944. 10.1038/nrd3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef D.W., Jaeger A.M., Gomez-Pastor R., Willmund F., Frydman J., and Thiele D.J.. 2014. A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep. 9:955–966. 10.1016/j.celrep.2014.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes S.A. 2017. Endoplasmic reticulum proteostasis: a key checkpoint in cancer. Am. J. Physiol. Cell Physiol. 312:C93–C102. 10.1152/ajpcell.00266.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Kukushkin Y., Gupta R., Chen T., Konagai A., Hipp M.S., Hayer-Hartl M., and Hartl F.U.. 2013. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 154:134–145. 10.1016/j.cell.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Peffer S., Gonçalves D., and Morano K.A.. 2019. Regulation of the Hsf1-dependent transcriptome via conserved bipartite contacts with Hsp70 promotes survival in yeast. J. Biol. Chem. 294:12191–12202. 10.1074/jbc.RA119.008822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus D. 2020. Regulation of Hsf1 and the heat shock response. Adv. Exp. Med. Biol. 1243:41–50. 10.1007/978-3-030-40204-4_3 [DOI] [PubMed] [Google Scholar]
- Pincus D., Anandhakumar J., Thiru P., Guertin M.J., Erkine A.M., and Gross D.S.. 2018. Genetic and epigenetic determinants establish a continuum of Hsf1 occupancy and activity across the yeast genome. Mol. Biol. Cell. 29:3168–3182. 10.1091/mbc.E18-06-0353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer L.A., Weigert M., Günther U., Maghelli N., Jug F., Sbalzarini I.F., and Myers E.W.. 2015. ClearVolume: open-source live 3D visualization for light-sheet microscopy. Nat. Methods. 12:480–481. 10.1038/nmeth.3372 [DOI] [PubMed] [Google Scholar]
- Rüdiger S., Buchberger A., and Bukau B.. 1997. Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 4:342–349. 10.1038/nsb0597-342 [DOI] [PubMed] [Google Scholar]
- Sala A.J., Bott L.C., and Morimoto R.I.. 2017. Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 216:1231–1241. 10.1083/jcb.201612111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Mosser D.D., and Morimoto R.I.. 1998. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 12:654–666. 10.1101/gad.12.5.654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís E.J., Pandey J.P., Zheng X., Jin D.X., Gupta P.B., Airoldi E.M., Pincus D., and Denic V.. 2016. Defining the essential function of yeast Hsf1 reveals a compact transcriptional program for maintaining eukaryotic proteostasis. Mol. Cell. 63:60–71. 10.1016/j.molcel.2016.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. 2003. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 4:49–60. 10.1038/nrn1007 [DOI] [PubMed] [Google Scholar]
- Tkach J.M., and Glover J.R.. 2004. Amino acid substitutions in the C-terminal AAA+ module of Hsp104 prevent substrate recognition by disrupting oligomerization and cause high temperature inactivation. J. Biol. Chem. 279:35692–35701. 10.1074/jbc.M400782200 [DOI] [PubMed] [Google Scholar]
- Tye B.W., Commins N., Ryazanova L.V., Wühr M., Springer M., Pincus D., and Churchman L.S.. 2019. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. eLife. 8:e43002 10.7554/eLife.43002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Hwang E.S., and Amon A.. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 398:818–823. 10.1038/19775 [DOI] [PubMed] [Google Scholar]
- Wallace E.W., Kear-Scott J.L., Pilipenko E.V., Schwartz M.H., Laskowski P.R., Rojek A.E., Katanski C.D., Riback J.A., Dion M.F., Franks A.M., et al. 2015. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell. 162:1286–1298. 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L., and Lindquist S.. 2009. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin. Ther. Targets. 13:469–478. 10.1517/14728220902832697 [DOI] [PubMed] [Google Scholar]
- Woolford J.L. Jr., and Baserga S.J.. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 195:643–681. 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., and Pincus D.. 2017. Serial immunoprecipitation of 3xFLAG/V5-tagged yeast proteins to identify specific interactions with chaperone proteins. Bio Protoc. 7:e2348 10.21769/BioProtoc.2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Beyzavi A., Krakowiak J., Patel N., Khalil A.S., and Pincus D.. 2018. Hsf1 phosphorylation generates cell-to-cell variation in Hsp90 levels and promotes phenotypic plasticity. Cell Rep. 22:3099–3106. 10.1016/j.celrep.2018.02.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Krakowiak J., Patel N., Beyzavi A., Ezike J., Khalil A.S., and Pincus D.. 2016. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife. 5:e18638 10.7554/eLife.18638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Slaughter B.D., Unruh J.R., Guo F., Yu Z., Mickey K., Narkar A., Ross R.T., McClain M., and Li R.. 2014. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 159:530–542. 10.1016/j.cell.2014.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Guo Y., Guettouche T., Smith D.F., and Voellmy R.. 1998. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 94:471–480. 10.1016/S0092-8674(00)81588-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists yeast strains used in this study.
contains Hsf1-3xFLAG IP/MS results.
contains Sis1-3xFLAG IP/MS results.