Abstract
Epimedium elachyphyllum, which belongs to Berberidaceae, is only distributed in Guizhou province of China. In this study, the complete chloroplast (cp)genome of E. elachyphyllum was sequenced and assembled. The circular genome is 157,201 bp in length, which comprises of a large single-copy region (LSC, 88,519 bp), a small single-copy region (SSC, 17,042 bp), and a pair of inverted repeat regions (IRa and IRb, 25,820 bp). The genome of E. elachyphyllum contains 112 unique genes, of which 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. Phylogenetic analysis based on 18 complete cp genome sequences indicated that E. elachyphyllum was closely related to E. dolichostemon.
Keywords: Chloroplast genome, Epimedium elachyphyllum Stearn, Berberidaceae
Epimedium elachyphyllum, an endangered species endemic to China, is only distributed in a small area in the Guizhou province of China. Due to habitat loss, E. elachyphyllum has been classified as endangered in “China Species Red List” (Ministry of Environment Protection of the People’s Republic of China 2013). Epimedium L. family contains about 60 species intermittently distributed in the Eurasia between Europe and eastern Asia. (Xu et al. 2019). The major bioactive components are flavonoid glycosides in Epimedii Folium (Wang et al. 2007; Ma et al. 2011). The therapeutic effects of E. elachyphyllum was the same as E. sagittatum (sieb. Et zucc.) Maxim (He and Zhang 1994). Previous studies showed that the classification and phylogeny were not completely clear in the Epimedium family (Guo et al. 2018). It was reported that the chloroplast genome has a highly conserved sequence ranging approximately 150k bp, which proves more variation information to discriminate closely related plants (Li et al. 2015). Up to now, the complete chloroplast (cp) genome of five Epimedium species have been reported (Zhang et al. 2016; Liu et al. 2019). So far, no data are available regarding the chloroplast genome of E. elachyphyllum. In this study, we assembled and characterized the complete chloroplast (cp) genome of E. elachyphyllum for the first time. The complete chloroplast (cp) genome sequence of E. elachyphyllum is a valuable resource for further studies, including species identification and genetic evolution in the family Berberidaceae.
In this study, E. elachyphyllum sample was collected from the Songtao County of Guizhou province in China (28°26′N, 108°42′E). A voucher specimen (Zhang218) was deposited at the Herbarium of the Institute of Medicinal Plant (IMPLAD), Beijing, China. Total genomic DNA was extracted from the fresh leaves of E. elachyphyllum using the modified CTAB method (Doyle and Doyle 1987). The high-quality DNA was sheared to the size of 300 bp for the shotgun library construction. The sequencing was performed on an Illumina Novaseq PE150 platform (Illumina Inc, San Diego, CA), and 150 bp paired-end reads were generated. The filtered reads were assembled into the complete chloroplast (cp) genome using the program GetOrganelle v1.5 (Jin et al. 2018) with E. acuminatum chloroplast genome (GenBank accession number: NC_029941) as a reference. The annotation of chloroplast genome was conducted through the online program CPGAVAS 2 (Shi et al. 2019), followed by manual correction if required. The annotated genomic sequence has been registered in GenBank with an accession number (MN873562).
The complete chloroplast (cp) genome of E. elachyphyllum is 157,201 bp in length and shows a typical quadripartite structure, which consists of a large single-copy region (LSC, 88,519 bp), a small single-copy region (SSC, 17,042 bp), and a pair of inverted repeat regions (IRa and IRb, 25,820 bp). The genome of E. elachyphyllum contains 112 unique genes, of which 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The total GC content of E. elachyphyllum chloroplast genome is 38.77%, while the corresponding GC content of LSC, SSC, IR regions are 37.36%, 32.78%, and 43.16%, respectively. The intron-exon structure analysis indicated that eleven protein-coding genes and seven tRNA genes contained one intron, while four genes (ycf3, clpP and rps12) had two introns.
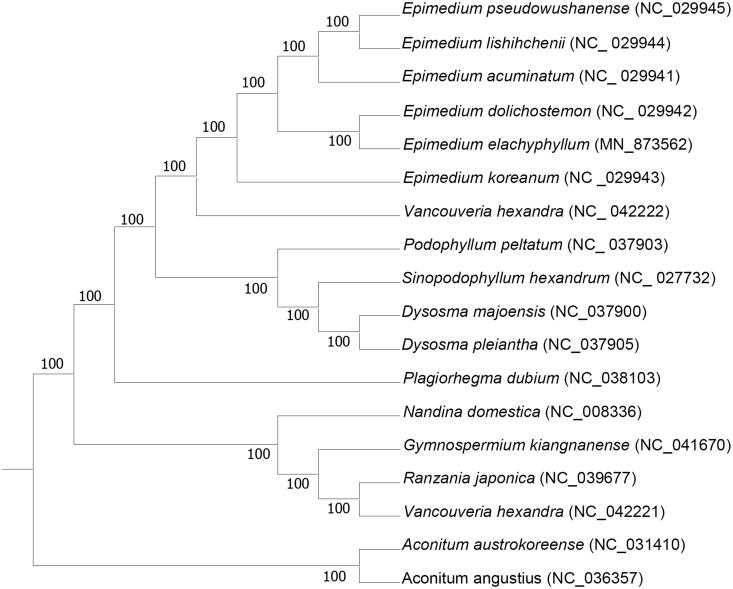
To identify the phylogenetic relationship of E. elachyphyllum, 15 complete chloroplast (cp) genomes of Berberidaceae species were used to reconstruct a maximum-likelihood (ML) phylogenetic tree using RAxML v8.2.10 (Stamatakis 2014), with Aconitum angustius and Aconitum austrokoreenseas as the outgroup. Phylogenetic analysis indicated that E. elachyphyllum is closely related to E. dolichostemon (Figure 1). The published E. elachyphyllum chloroplast genome will provide useful information for phylogenetic and evolutionary studies in Berberidaceae.
Figure 1.
ML phylogenetic tree inferred from 18 complete cp genomes.
Funding Statement
This work was supported by the CAMS Innovation Fund for Medical Sciences (CIFMS) [2017-12 M-3-013], the National Natural Science Foundation of China [81473302], the Chongqing Science and Technology Burea [cstc2018jcyjAX0316, cc-cstc-CA-19-2], and the Traditional Chinese Medicine Science and the Technology Projects of Chongqing Health and Family Planning Commission [ZY201802117].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Guo M, Xu Y, Ren L, He S, Pang X. 2018. A systematic study on DNA barcoding of medicinally important genus Epimedium L. (Berberidaceae). Genes. 9(12):637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SZ, Zhang TL. 1994. A new species of medicinal plants of Epimedium L. from GUIZHOU. Guihaia. 14:25–26. [Google Scholar]
- Jin JJ, Yu WB, Yang JB. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479. [Google Scholar]
- Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. 2015. Plant DNA barcoding: from gene to genome. Biol Rev. 90(1):157–166. [DOI] [PubMed] [Google Scholar]
- Liu X, Yang Q, Zhang C, Shen G, Guo B. 2019. The complete chloroplast (cp) genome of Epimedium sagittatum (Sieb. Et Zucc.) Maxim. (Berberidaceae), a traditional Chinese herb. Mitochondrial DNA Part B. 4(2):2572–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, He X, Yang Y, Li M, Hao D, Jia Z. 2011. The genus Epimedium: an ethnopharmacological and phytochemical review. J Ethnopharmacol. 134(3):519–541. [DOI] [PubMed] [Google Scholar]
- Ministry of Environment Protection of the People’s Republic of China. 2013. China species red list: higher plants. http://www.zhb.gov.cn/gkml/hbb/bgg/.201309/t20130912_260061.htm.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyser. Nucleic Acids Res. 47:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GJ, Tsai TH, Lin LC. 2007. Prenylflavonol, acylated flavonol glycosides and related compounds from Epimedium sagittatum. Phytochemistry. 68(19):2455–2464. [DOI] [PubMed] [Google Scholar]
- Xu Y, Liu L, Liu S, He Y, Li R, Ge F. 2019. The taxonomic relevance of flower colour for Epimedium (Berberidaceae), with morphological and nomenclatural notes for five species from China. PK. 118:33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Du L, Liu A, Chen J, Wu L, Hu W, Zhang W, Kim K, Lee SC, Yang TJ, et al. 2016. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 7:306–308. [DOI] [PMC free article] [PubMed] [Google Scholar]