Abstract
We describe the mitogenome sequence of alpine butterfly Aglais ladakensis (Lepidoptera: Nymphalidae: Nymphalinae) collected from the Qilianshan Mountain, Gansu province, China. The molecule is 15,222 bp in length, containing 37 typical insect mitochondrial genes and one AT-rich region. All protein-coding genes (PCGs) start with ATN codons, except for COI gene with CGA, which is often found in other butterflies. In addition, seven PCGs harbor the typical stop codon TAA, whereas six PCGs terminate with TA or T. The rrnL and rrnS genes are 1316 bp and 735 bp in length, respectively. The AT-rich region is 394 bp in size and harbors several features characteristic of the lepidopterans, including the motif ATAGA followed by a 19 bp poly-T stretch and a microsatellite-like (TA)8 element. Phylogenetic analysis shows that the Qinghai-Tibet Plateau (QTP) distributed A. ladakensis of this study is closely related to the A. milberti, which is the only Aglais species that occurs in the alpine caves of North America.
Keywords: Mitochondrial genome, phylogeny, Nymphalinae, Nymphalini, Aglais ladakensis
The tribe Nymphalini is one of the most diverse butterfly groups distributed all around the world and this diversity (e.g. wing patterns, behaviors, and host-plant associations) have led to many Nymphalini species as model organisms in evolutionary and ecological studies (Janz et al. 2001; Wahlberg et al. 2005). However, the taxonomy and phylogeny of Nymphalini are still standing as a controversial issue (Harvey 1991; Wahlberg 2006). In recent decades, mitogenome have been widely used as an informative molecular marker in evolutionary study areas (Boore 1999; Galtier et al. 2009).
In this study, we newly determined the complete mitogenome sequence of the Aglais ladakensis, and reconstructed the phylogenetic tree of the species with other related Nymphalini species. Aglais ladakensis (distributed at about 4200 meters high) was collected from Qilianshan Mountain, Qinhai province, China (coordinates: E100.39, N38.50) in July 2016. After morphological identification, a voucher specimen (ANUH-20160710) was kept in the Herbarium of Anhui Normal University (Wuhu, China). Total genomic DNA was extracted from thorax muscle using a DNA extraction kit (Shanghai, China) and the PCR amplication and sequencing were conducted after Hao et al. (2013). The resultant reads were assembled and annotated using the BioEdit 7.0 (Hall 1999) and MEGA 7.0 (Kumar et al. 2016).
The whole mitogenome is a circular molecule of 15,222 bp in size (GenBank accession No. MN732892) with a AT bias of 80.5%, containing 13 protein-coding genes (PCGs), 22 transfer tRNA genes, 2 ribosomal rRNA genes, and a noncoding AT-rich control region. All PCGs are initiated by typical ATN codons, except for COI gene, which starts with the unusual CGA as observed in most of the other sequenced nymphalids (Gan et al. 2016; Timmermans et al. 2016). Seven PCGs have a complete stop codon TAA, whereas six PCGs (COI, COII, COIII, ND1, ND4, and ND5) stop with incomplete TA or T. All tRNAs harbor the typical predicted secondary cloverleaf structures except for the tRNASer(AGN), as seen in other determined butterflies (Hao et al. 2013; Shi et al. 2018). The rrnL and rrnS genes are 1316 bp and 735 bp in length, respectively. The AT-rich region is 394 bp long with several structures characteristic of lepidopterans, including the motif ATAGA followed by a 19 bp poly-T stretch and a microsatellite-like (TA)8 element.
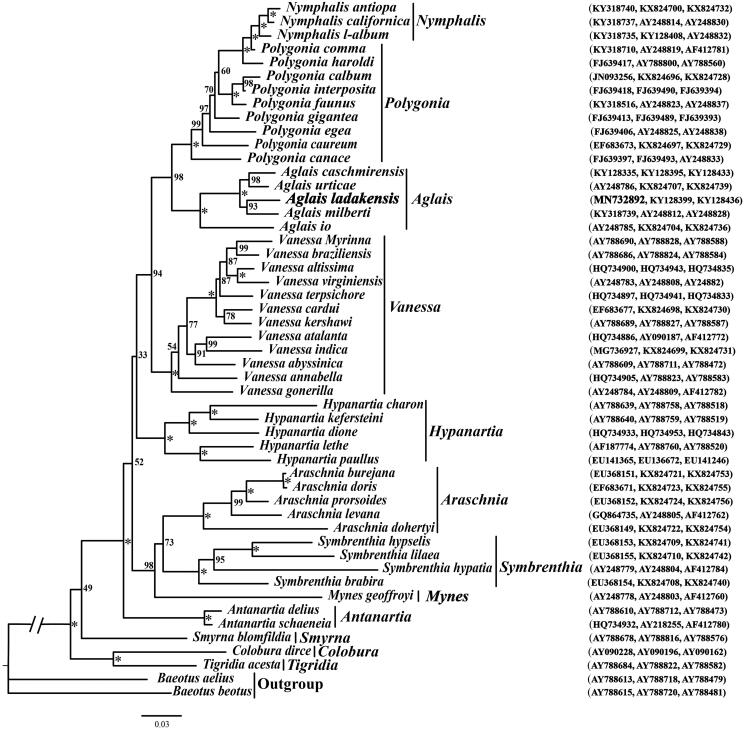
Using two Baeotus nymphalid species as the outgroups, the maximum-likelihood (ML) phylogenetic analysis were performed with IQ-TREE v.1.6.8 (Nguyen et al. 2015) based on the concatenated mitochondrial COI and two nuclear gene (EF-1a, and wingless) sequence data to clarify the phylogenetic relationship of A. ladakensis with other 50 nymphalids available from GenBank (Figure 1). The resultant phylogenetic tree showed that the Nymphalini of this study includes 12 genera, the genus Aglais is sister to the grouping of two genera, namely, the Nymphalis and Polygonia; in addition, the alpine A. ladakensis of this study that is distributed Qinghai-Tibet Plateau (QTP) is closely related to the Milbert's Tortoiseshell A. milberti, which is the only Aglais species that occurs in the alpine caves of North America.
Figure 1.
The maximum-likelihood (ML) phylogenetic tree of Nymphalini inferred from nucleotide sequence data of mitochondrial CO1, nuclear EF-1a and wingless genes. The numbers beside the nodes are percentages of 1000 bootstrap values (*BP = 100%). The alphanumeric characters in parentheses represent the GenBank accession numbers.
Funding Statement
This work was supported by the National Natural Science Foundation of China [41472028].
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this paper.
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Nabholz B, Glémin S, Hurst G. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 18(22):4541–4550. [DOI] [PubMed] [Google Scholar]
- Gan SS, Chen YH, Zuo N, Xia CC, Hao JS. 2016. The complete mitochondrial genome of Tirumala limniace (Lepidoptera: Nymphalidae: Danainae). Mitochondr DNA A. 27(2):1096–1098. [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98. [Google Scholar]
- Hao JS, Sun ME, Shi QH, Sun XY, Shao LL, Yang Q. 2013. Complete mitogenomes of Euploea mulciber (Nymphalidae: Danainae) and Libythea celtis (Nymphalidae: Libytheinae) and their phylogenetic implications. ISRN Genom. 2013:1–14. [Google Scholar]
- Harvey DJ. 1991. Higher classification of the Nymphalidae, Appendix B. In: Nijhout HF, editor. Higher classification of the Nymphalidae, Appendix B. Washington (DC): Smithsonian Institution Press; p. 255–273. [Google Scholar]
- Janz N, Nyblom K, Nylin S. 2001. Evolutionary dynamics of host-plant specialization: a case study of the tribe Nymphalini. Evolution. 55(4):783–796. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi QH, Xing JH, Liu XH, Hao JS. 2018. The complete mitochondrial genome of comma, polygonia c-aureum (Lepidoptera: Nymphalidae: Nymphalinae). Mitochondr DNA B. 3(1):53–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans M, Lees DC, Thompson MJ, Sáfián S, Brattström O. 2016. Mitogenomics of 'Old World Acraea' butterflies reveals a highly divergent 'Bematistes. Mol Phylogenet Evol. 97:233–241. [DOI] [PubMed] [Google Scholar]
- Wahlberg N. 2006. That awkward age for butterflies: insights from the age of the butterfly subfamily Nymphalinae (Lepidoptera: Nymphalidae). Syst Biol. 55(5):703–714. [DOI] [PubMed] [Google Scholar]
- Wahlberg N, Brower AV, Nylin S. 2005. Phylogenetic relationships and historical biogeography of tribes and genera in the subfamily Nymphalinae (Lepidoptera: Nymphalidae). Biol J Linn Soc. 86(2):227–251. [Google Scholar]