Abstract
Monte Carlo (MC) track structure simulation tools are commonly used for predicting radiation induced DNA damage by modeling the physical and chemical reactions at the nanometer scale. However, the outcome of these MC simulations is particularly sensitive to the adopted parameters which vary significantly across studies. In this study, a previously developed full model of nuclear DNA was used to describe the DNA geometry. The TOPAS-nBio MC toolkit was used to investigate the impact of physics and chemistry models as well as three key parameters (the energy threshold for direct damage, the chemical stage time length, and the probability of damage between hydroxyl radical reactions with DNA) on the induction of DNA damage. Our results show that the difference in physics and chemistry models alone can cause differences up to 34% and 16% in the DNA double strand break (DSB) yield, respectively. Additionally, changing the direct damage threshold, chemical stage length, and hydroxyl damage probability can cause differences of up to 26%, 51%, and 71% in predicted DSB yields, respectively, for the configurations in this study.
Introduction
With the recognition that DNA is a significant target for causing biological effects after ionization irradiation (Hall and Giaccia, 2018), modeling DNA damage mechanistically is a critical research topic. Different Monte Carlo (MC) track structure simulation tools as well as detailed DNA models have been developed to predict levels of DNA damage (Tang et al., 2019; Sakata et al., 2019; Lampe et al., 2018b; Meylan et al., 2017; Friedland et al., 2017; Friedland et al., 2003; Nikjoo et al., 2001; Nikjoo et al., 2016; Nikjoo et al., 2006) by modeling the direct damages caused by initial physical interactions in the physical stage and indirect damages caused by subsequent chemical interactions of radiation induced reactive oxygen species in the physico-chemical and the chemical stages. The simulation of DNA damage allows researchers to further their understanding of the induction of cellular damage after ionizing radiation in order to interpret, analyze or design experiments (Campa et al., 2005).
Differences in the simulation tool and geometry structure intrinsically affect the DNA damage result. Furthermore, different studies adopted different parameter values, which could also significantly alter simulation results. For instance, some studies (Mokari et al., 2018; Lampe et al., 2018b; Meylan et al., 2017; Nikjoo et al., 2001) assumed that a direct strand break (SB) is formed with a fixed threshold of 17.5 eV, i.e., a direct SB will be formed if the accumulated energy deposition within one sugar phosphate or hydration shell exceeds this threshold. In contrast, other studies (Friedland et al., 2017; Friedland et al., 2011; Friedland et al., 2003; Sakata et al., 2019) used a linear acceptance threshold ranging from 5 to 37.5 eV, i.e., the probability to form a direct SB linearly increases from 0 when less than 5 eV energy deposition was accumulated to 1 when more than 37.5 eV energy deposition was accumulated in the target DNA volume. Many other parameters (see Table 1), such as the length of the chemical stage and the probability of chemical interactions to cause DNA damages also vary over different studies.
Table 1.
Simulation parameters adopted in different MC studies
| Reference | Code | Physics constructor | Chemical stage length | ·OH damage probability(c) | Direct damage threshold |
|---|---|---|---|---|---|
| (Nikjoo et al., 2001) | PITS | —— | 1 ns | 0.13(a) | 17.5 eV |
| (Friedland et al., 2017) | PARTRAC | —— | 100 ns | 0.65(b) | 5–37.5 eV |
| (Friedland et al., 2011) | PARTRAC | —— | 2.5 ns | 0.65(b) | 5–37.5 eV |
| (Friedland et al., 2003) | PARTRAC | —— | 10 ns | 0.13(a) | 5–37.5 eV |
| (Sakata et al., 2019) | Geant4-DNA | option4 | 2.5 ns | 0.4(b) | 5–37.5 eV |
| (Mokari et al., 2018) | Geant4-DNA | Default | 1 ns | 0.65(b) | 17.5 eV |
| (Lampe et al., 2018a) | Geant4-DNA | option4 | 1 ns | 0.4(b) | 17.5 eV |
| (Meylan et al., 2017) | Geant4-DNA | default | 2.5 ns | 0.4(b) | 17.5 eV |
Defined between ·OH and whole DNA volume
Defined between ·OH and deoxyribose (“backbone”).
The ·OH damage probability of P·OH-DNA = 0.13 is equivalent to P·OH-backbone = 0.65 (Friedland et al., 2003)
Differences can also be found in handling of interactions among studies. Friedland et al. assumed hydroxyl radicals were scavenged after an encounter with histones using a radius of 4.5 nm as the interaction distance, while other chemical species were assumed to not interact with the DNA or with histones by preventing these species from diffusing into the volumes occupied by these molecules (Friedland et al., 2003). Sakata et al. and Lampe et al. used a parameter of radical kill radius (rkill) in the chemical stage, which defines a radius from the DNA volume beyond which chemical molecules will be terminated and would not be simulated to avoid modeling radical tracks that will not interact with the DNA; this subsequently speeds up the simulation runtime (Lampe et al., 2018a; Sakata et al., 2019).
Differences in the parameters as well as the handling of interactions can lead to significant uncertainties in the simulation of DNA damages. Lampe et al. studied the impact of physics models, direct damage threshold, the hydroxyl damage probability and the radical kill distance on the DNA damage after irradiating a test geometry filled with DNA segments with electrons (Lampe et al., 2018a).
In this study, TOPAS-nBio was used for a sensitivity study of physics and chemistry models as well as three key parameters that could affect direct and indirect DNA damage results. A previously developed full nucleus model (Zhu et al., 2019) was used for the description of the DNA geometry. DNA damages were simulated with different physics constructors, chemistry models, direct damage threshold values, chemical stage time length, and the hydroxyl damage probability. The impacts of these parameters on the DNA damages were quantified and compared with the ratio of DNA damage yield. In addition to the differences in the above listed parameters, the nucleus geometry is another important aspect when studying DNA damage. Different research groups have developed their own nucleus geometries and implemented them in their own respective codes (Tang et al., 2019; Meylan et al., 2017; Friedland et al., 2017; Nikjoo and Girard, 2012; Sakata et al., 2019). However, the uncertainties due to the chosen DNA geometry were not considered in this work.
Methods
Simulation tool and geometry setup
TOPAS-nBio (Schuemann et al., 2019a) is a MC track structure simulation application layered on Geant4-DNA (Incerti et al., 2018; Incerti et al., 2010a), which extends the functionality of TOPAS (Perl et al., 2012), an advanced and user-friendly MC tool based on Geant4 (Agostinelli et al., 2003; Allison et al., 2006; Allison et al., 2016) to support simulation studies at the patient scale. TOPAS-nBio offers an extensive library of advanced biological geometries ranging from the micrometer scale (e.g. cells and organelles) down to the nanometer scale (e.g. DNA molecules and proteins) to support studies at the cellular and sub-cellular level (McNamara et al., 2018).
In this study, a previously developed full nucleus model (Figure 1) (Zhu et al., 2019), which was built using a fractal pattern based on a 3D Hilbert space-filling curve (Lieberman-Aiden et al., 2009; McNamara et al., 2018), was used to describe the DNA geometry. The spherical nucleus model has a diameter of 9.3 μm, and it was organized in a hierarchical pattern in the order of DNA double helix, nucleosomes, chromatin fibers, and chromosomal territories. The double helix structure was formed with a half-cylindrical base volume with a radius of 0.5 nm and a quarter-cylindrical sugar-phosphate backbone volume with a radius of 1.15 nm (Henthorn et al., 2017). Considering that ionizations in the hydration shell could also lead to DNA damage (Meylan et al., 2017; Friedland et al., 2017; Friedland et al., 2003), a 0.16 nm thick quarter-cylindrical shell surrounding the backbone volume was modeled as the hydration shell. The DNA double helix wraps around the cylindrical histone volume to form the nucleosome (Figure 1 (a)), which is the unit of chromatin fibers (Figure 1 (b)). Each chromatin fiber has 51 nucleosomes and contains 15150 base pairs (bp) of DNA. The nucleus was voxelized and consists of 14328 voxels; each voxel was filled with chromatin fibers folded according to a continuous 3D Hilbert space-filling curve. The resulting nucleus (Figure 1 (c)) includes 46 chromosomes and was filled with 6.08 Gbp (Giga base pairs) of DNA; the DNA density of the nucleus was 14.4 Mbp/μm3 and agrees with the typical DNA density of a mammalian nucleus of 15 Mbp/μm3 (Ghosh and Jost, 2018).
Figure 1.
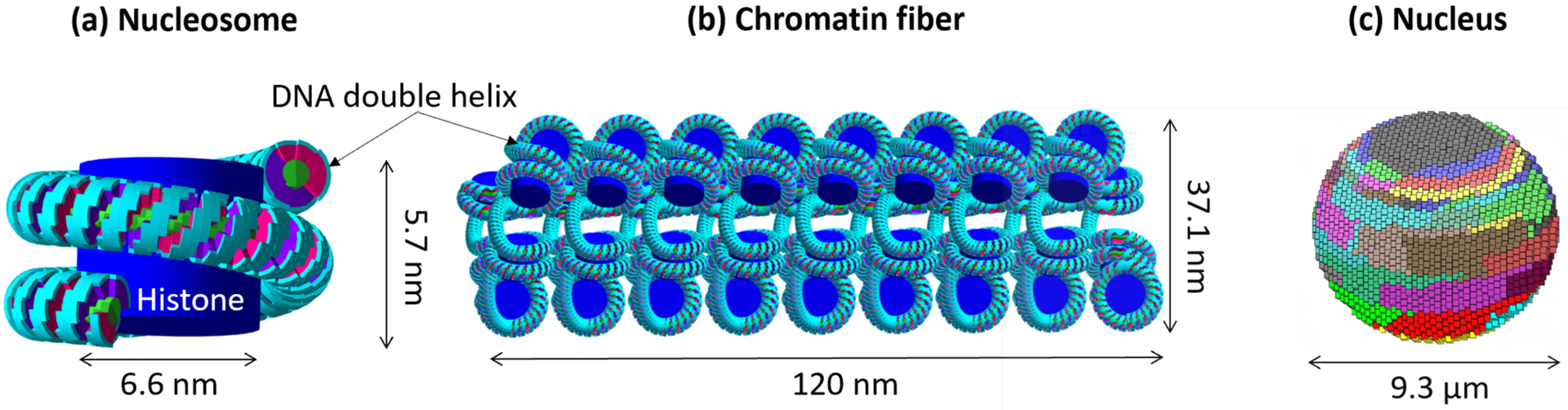
The nuclear DNA model. (a). The nucleosome with 200 base pairs (bp) of DNA; (b). The chromatin fiber with 15.15 kbp DNA; (c). The whole nucleus with 6.08 Gbp DNA consists of 46 chromosomes; voxels of the same color represent a chromosome.
DNA damage induction after irradiation by 0.5–50 MeV monoenergetic protons were simulated with the full nucleus model and scored in the SDD (Standard for DNA Damage) format for analysis (Schuemann et al., 2019b). The nucleus was placed at the center of a cubic world volume with a side length of 14 μm, and the primary protons were sampled on the surface of the nucleus and directed inside the nucleus with a random direction to mimic the random relative orientation between the incident particle and chromatin fiber in experimental irradiations. If particles or radicals stepped outside the world boundary, they were considered to be terminated and were not reflected back into the world volume.
To achieve acceptable statistical accuracy, 100 runs were performed for each simulated particle energy. Each simulation run deposited a dose of ~1 Gy within the nucleus and the average number of histories to deposit a dose of 1 Gy for different energies was listed in Table 2. The statistical uncertainties of DSB yields were smaller than 2% (except for simulations with 10 ns chemical stage length, which were very time consuming, and the statistical uncertainty was limited to 5% for these simulations).
Table 2.
Average number of primary protons to deposit a dose of 1 Gy within the nucleus.
| Proton energy (MeV) | Average number of primary protons |
|---|---|
| 0.5 | 6.3 |
| 0.6 | 7.5 |
| 0.8 | 9.9 |
| 1 | 12.1 |
| 1.5 | 16.9 |
| 2 | 21.1 |
| 5 | 43 |
| 10 | 76 |
| 20 | 139.4 |
| 50 | 312 |
Physics and chemistry models in TOPAS-nBio
Most of the interaction models and cross sectional data used in MC track structure tools were derived based on water, and were considered as a near accurate description for biological tissue which is mostly water. The simulation of DNA damage includes three stages, starting from interactions in the physical stage, which lasts about 1 femtosecond. In this stage, primary and secondary particles interact with the DNA volume according to predefined interaction processes within the limits of the energy thresholds of each modeled physics process. The Geant4 (Geant4-DNA) release (version 10.5) used for this analysis provides 9 physics constructors: G4EmDNAPhysics (default) and G4EmDNAPhysics_option1–8 for simulations at the DNA scale. These physics constructors are based on three main constructors (G4EmDNAPhysics_option2 (opt2), G4EmDNAPhysics_option4 (opt4), and G4EmDNAPhysics_option6 (opt6)) that are recommended for simulations in liquid water (Incerti et al., 2018). Opt2 is an accelerated version of the default physics constructor; it uses a cumulated version of differential cross sections to describe the physical interactions while the default constructor uses non-cumulated differential cross sections. Opt4 includes an updated set of cross sections for electron-impact excitation and ionization in liquid water and provides an alternative elastic scattering model. Opt6 is an implementation of the CPA100 track structure code to Geant4-DNA. More details about different physics constructors in Geant4-DNA can be found elsewhere (Bordage et al., 2016; Bernal et al., 2015; Tran et al., 2015; Incerti et al., 2010b; Incerti et al., 2018). All of these physics constructors are available in TOPAS-nBio.
Besides the physics constructors available in Geant4-DNA, TOPAS-nBio provides a flexible physics constructor, TsEmDNAPhysics, which uses the same physics processes and associated models as G4EmDNAPhysics. However, users can easily modify the interaction models in TsEmDNAPhysics constructor with a few command lines in the parameter file.
The pre-chemical stage follows the physical stage with a time scale of a few picoseconds and is followed by the chemical stage that can last up to microseconds. In the pre-chemical stage, the chemical species are generated by ionized or excited water molecules via dissociative decay or auto-ionization, and then the initial chemical species diffuse and react in the chemical stage. As mentioned before, potential reflections of chemical species at the geometry boundary have not been considered as this process is not currently supported in Geant4-DNA and TOPAS-nBio. Geant4-DNA provides a chemistry model (G4EmDNAChemistry) to describe the production, diffusion, and interaction of all chemical species (Karamitros et al., 2014). TOPAS-nBio provides a chemistry model (TsEmDNAChemistry) to simulate the production of water radiolysis and the subsequent chemical interactions in the pre-chemical and chemical stage with reviewed and updated parameters, including the electron solvated thermalization distance, diffusion coefficients, and reaction rates in the G4EmDNAChemistry model. The updated values of parameters were adopted from RITRACKS (Plante and Devroye, 2017), and this update was recently included in Geant4-DNA as G4EmDNAChemistry_option1 (Shin et al., 2019). The TsEmDNAChemistry model was validated by calculating time-dependent G-values (number of molecular species per 100 eV of energy deposit) for electrons, protons and α particles covering a range of LET (0.05–230 keV/μm) (Ramos-Méndez et al., 2018).
Induction of DNA damage
Initial DNA damages caused by the physical and chemical interactions within the nucleus were quantified as SBs in this work. SBs may result from the direct interactions of primaries and secondaries with the backbone volume and neighboring hydration shell or the indirect interactions of the chemical species with the backbone volume. Following our previous work (Zhu et al., 2019), all radiolysis products and their interactions were simulated. The table of considered chemical species and their reactions can be found in the reference (Ramos-Méndez et al., 2018). When chemical species diffuse into DNA volumes (base, backbone, hydration shell, and histone), they were considered to react and be scavenged by the DNA-related volumes. However, only interactions between the hydroxyl (·OH) and the DNA backbone were regarded to be able to cause an indirect SB. No radical kill radius was applied in this work.
If two SBs were located on the opposite sides of the DNA double helix strand and separated by less than 10 base pairs, it was assumed to be a double strand break (DSB). Otherwise, the two SBs were classified as two single strand breaks (SSBs). DSBs were further classified as direct, indirect or hybrid DSBs according to the process that initiated SBs comprising the DSB. Following other studies, the DNA damages were counted after each history, i.e., inter-track damages were not considered assuming that proton tracks act independently (Meylan et al., 2017; Henthorn et al., 2017). The complexity of DSBs was not considered in this analysis.
Sensitivity study for parameters involved in DNA damage simulations
Differences in physics and chemistry models as well as the selection of other relevant parameter values across different studies can cause uncertainties in the DNA damage modeling. In this investigation, sensitivity studies were performed with TOPAS-nBio for physics and chemistry models, and three key parameters as listed in Table 3. For each parameter, DNA damages were simulated with the values in the second column in Table 3 while values for other parameters were fixed at the default values (third column in Table 3). For instance, for the sensitivity study for physics constructors, DNA damages were simulated with opt2, opt4, and opt6, while for all of these simulations, the chemistry model was TsEmDNAChemistry, and values of direct damage threshold, chemical stage length and ·OH damage probability were fixed at thresholds of 17.5 eV, 1 ns and P·OH-backbone = 0.4, respectively.
Table 3.
Parameter set investigated in this study
| Parameter type | Value for sensitivity study | Default value |
|---|---|---|
| Physics constructors | G4EmDNAPhysics_option2/4/6 | TsEmDNAPhysics |
| Chemistry model | G4EmDNAChemistry, TsEmDNAChemistry | TsEmDNAChemistry |
| Direct damaged threshold | 17.5 eV, 5–37.5 eV | 17.5 eV |
| Chemical stage length | 1 ns, 2.5 ns, 10 ns | 1 ns |
| ·OH damage probability | P·OH-backbone = 0.4, P·OH-backbone = 0.65 | P·OH-backbone = 0.4 |
Results and Discussion
DNA damage results were simulated with three recommended physics constructors (opt2, opt4, and opt6) and default physics constructors in Geant4-DNA (G4EmDNAPhysics) as well as TOPAS-nBio (TsEmDNAPhysics). Figure 2 shows that TsEmDNAPhysics (Ts), G4EmDNAPhysics (G4), and opt2 give statistically equivalent damage yields. Changing to other physics constructors resulted in different total damage yields and different contributions from direct or indirect damage. Opt4 gave the highest total SB yield while opt6 gave the highest direct SB yield but the lowest indirect SB yield. Figure 3 (b) shows that opt4 and opt6 caused up to 34% and 23% more DSBs, respectively, than opt2, and Figure 4 shows that contributions of direct, indirect and hybrid DSBs to the total DSBs were also different when simulating with different physics constructors.
Figure 2.
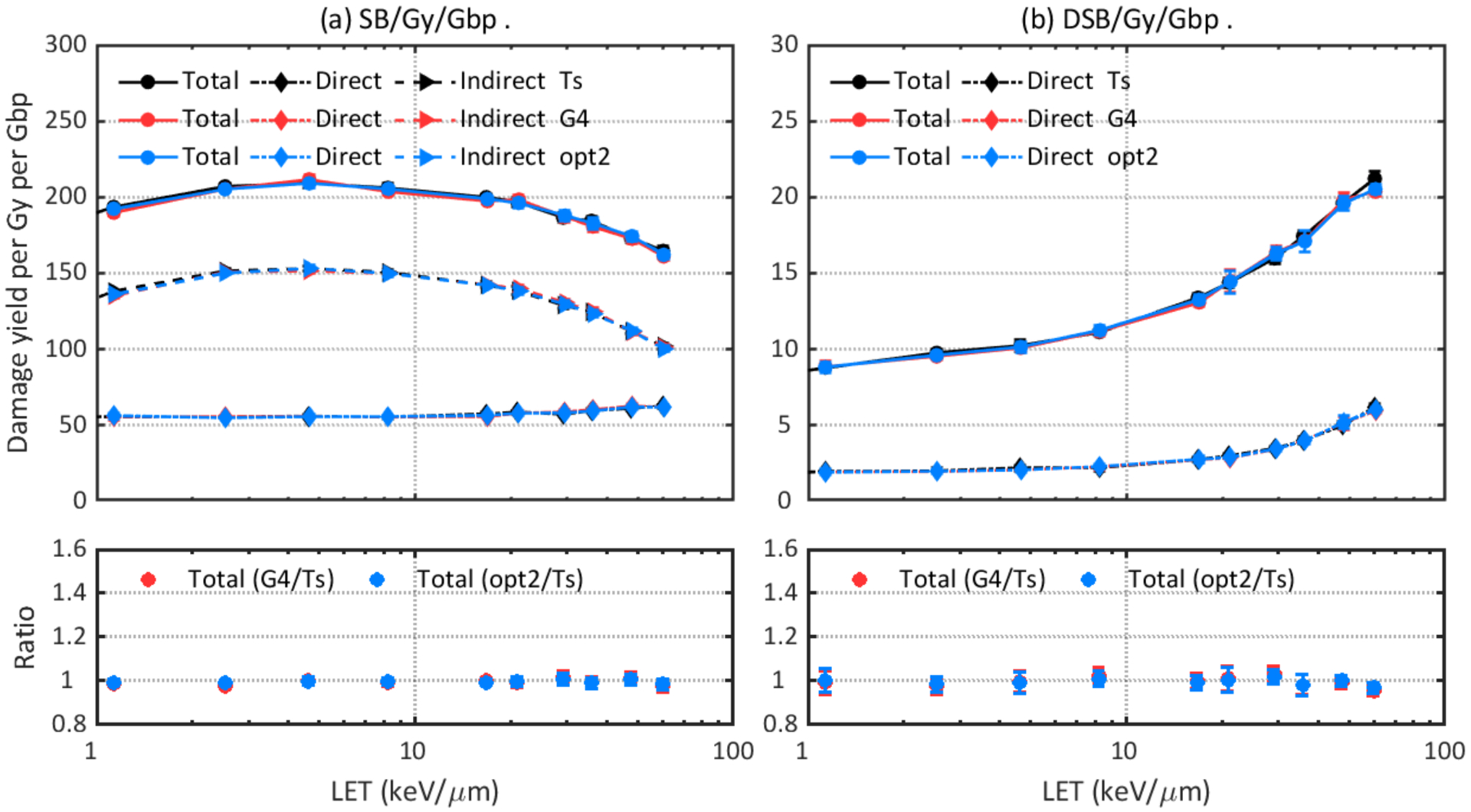
Comparison of DNA damage yields simulated with default physics constructors in TOPAS-nBio (Ts: TsEmDNAPhysics) and Geant4-DNA (G4: G4EmDNAPhysics) as well as the accelerated Geant4-DNA default constructor (opt2: G4EmDNAPhysics_option2).
Figure 3.
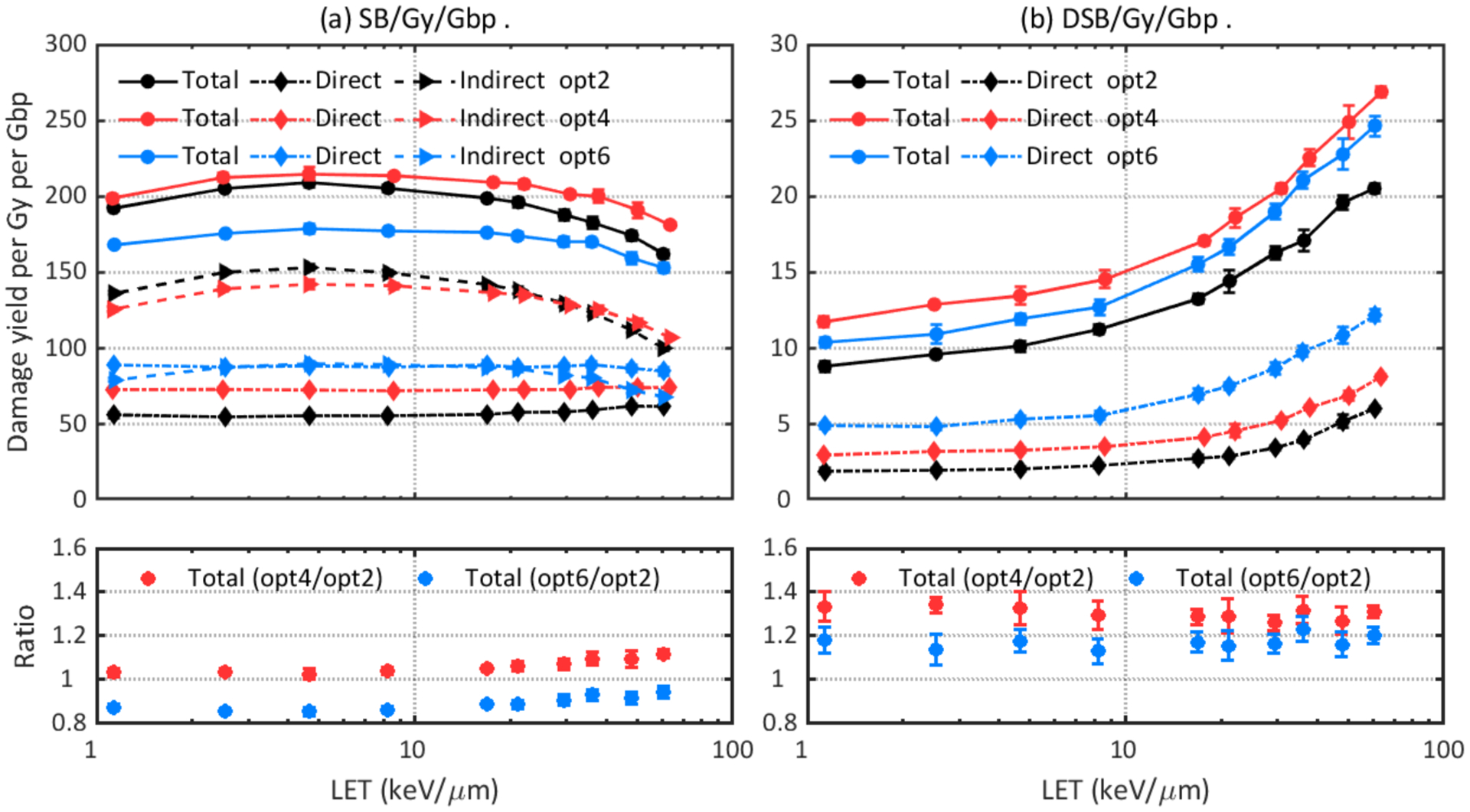
Comparison of DNA damage yields simulated with different physics constructors: opt2, opt4, and opt6. Upper left: total, direct, and indirect SB yields. Lower left: ratio of total SB yields. Upper right: total and direct DSB yields. Lower right: ratio of total DSB yields.
Figure 4.
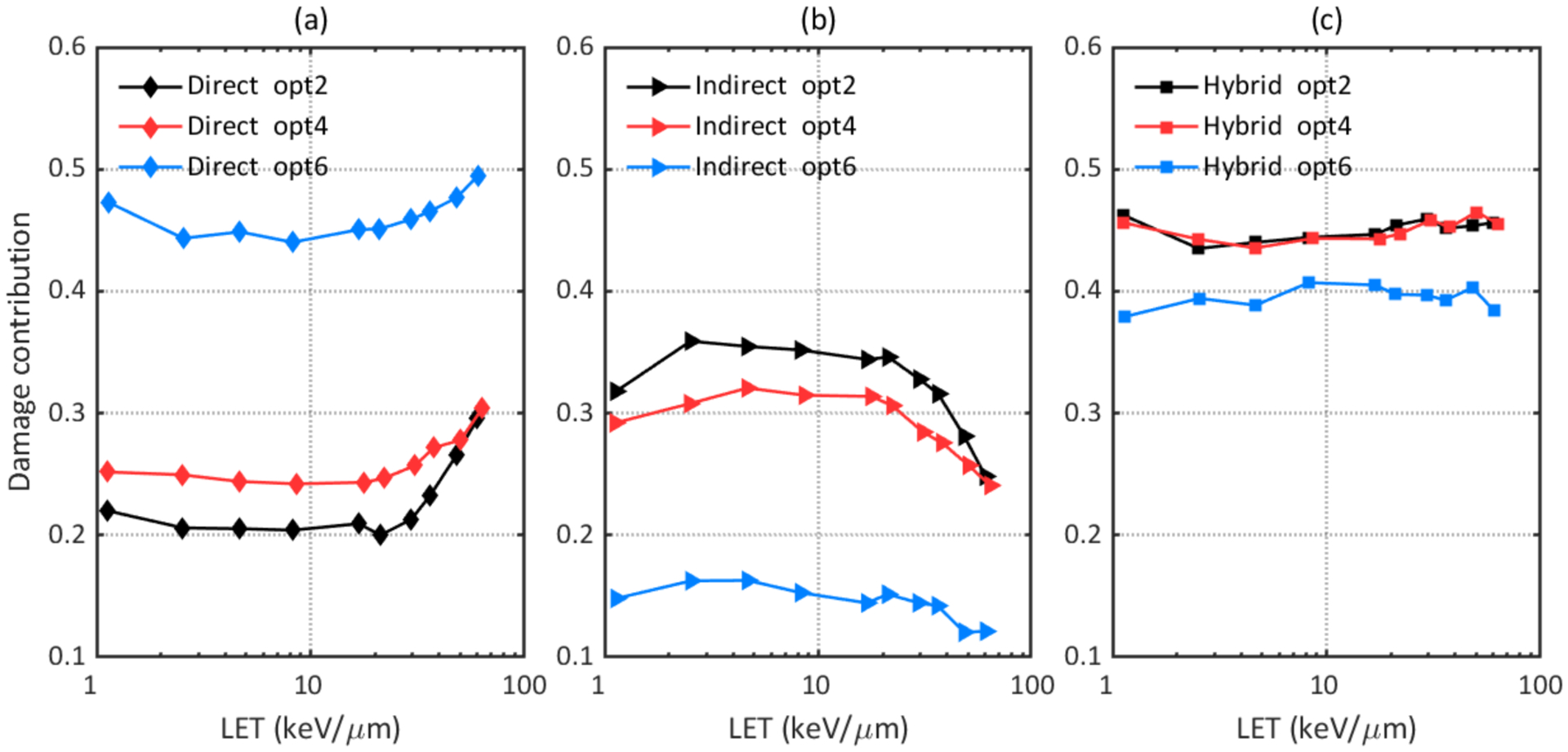
Comparison of contributions of different types of DSB to the total DSB yield simulated with different physics constructors. (a) Contribution of direct DSB; (b) Contribution of indirect DSB; (c) Contribution of hybrid DSB.
The main reason for the differences is the adopted process models and cross section data for electrons (an overview of these model differences can be found in (Incerti et al., 2018; Bernal et al., 2015)). Dose point kernel (DPK) simulations with different physics constructors were considered as benchmarks for the accuracy of electron elastic and inelastic scattering models (Incerti et al., 2018). DPK is the average energy deposited per unit distance along the radius of a sphere with its center being the starting point of a particle source, and it describes spatial distribution of energy transferred from the particle source to the medium. The comparison of DPK obtained with different physics constructors showed opt2 is the most diffusive model (i.e. it has longer tail toward large radius values) while opt4 and opt6 are more concentrated models (Bordes et al., 2017; Shin et al., 2018; Kyriakou et al., 2016). A more diffuse model results in less instant recombination of initial pre-chemical and chemical species. Shin et al (Shin et al., 2019) showed that opt2 simulates the more spread distribution of electron interactions. This produces ˙OH radicals sufficiently separated to lead to higher G-values at the initiation of the chemical stage, hence more interactions between ˙OH and the DNA structure are expected, and our results show opt2 indeed gave higher indirect SB yield.
Figure 5 shows the total DSB yield to total SB yield ratios for different constructors. and opt2 gave the lowest DSB yield to SB yield ratios for the whole LET region, which is consistent with the diffusive characteristic of opt2 as SBs should be located close enough to form a DSB.
Figure 5.
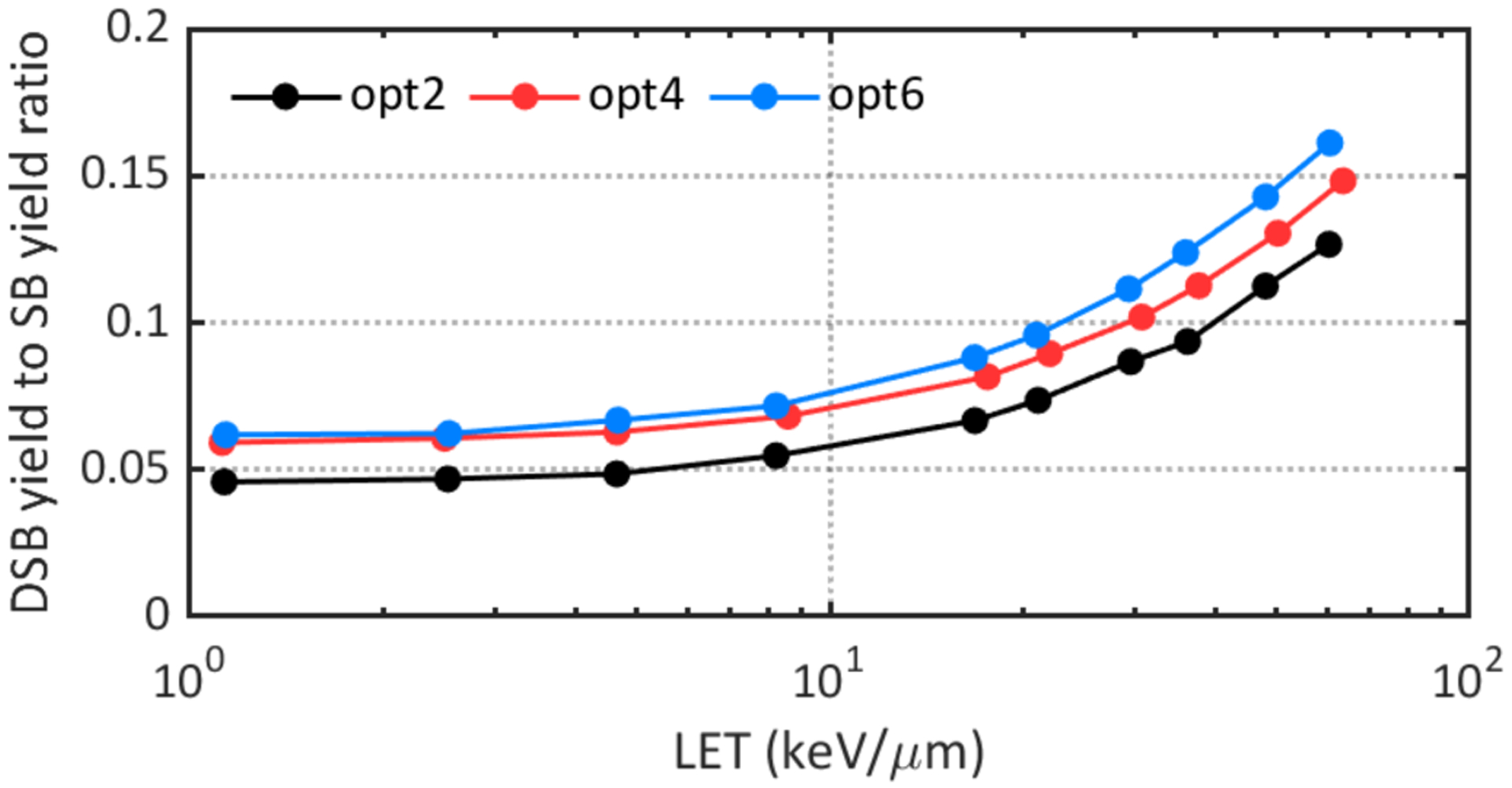
Comparison of DSB yield to SB yield ratios obtained with different physics constructors.
Our previous calculations (Zhu et al., 2019) showed that more than 80% of the direct DNA damages after proton irradiation were caused by secondary electrons. Figure 6 and Table 4 show counts of energy depositions of secondary electrons from 0.5 MeV primary protons in the backbone and hydration shell simulated with different physics constructors. In this case, opt6 resulted in the highest number of energy deposition events beyond the 17.5 eV damage threshold, which leads to the highest direct SB yield. On the other hand, opt2 and opt4 gave higher total counts of energy depositions, which then led to more excited water molecules (H2O*) and ionized water molecules (H2O+) in the pre-chemical stage, subsequently resulting in more radiolysis products that could lead to indirect damages.
Figure 6.
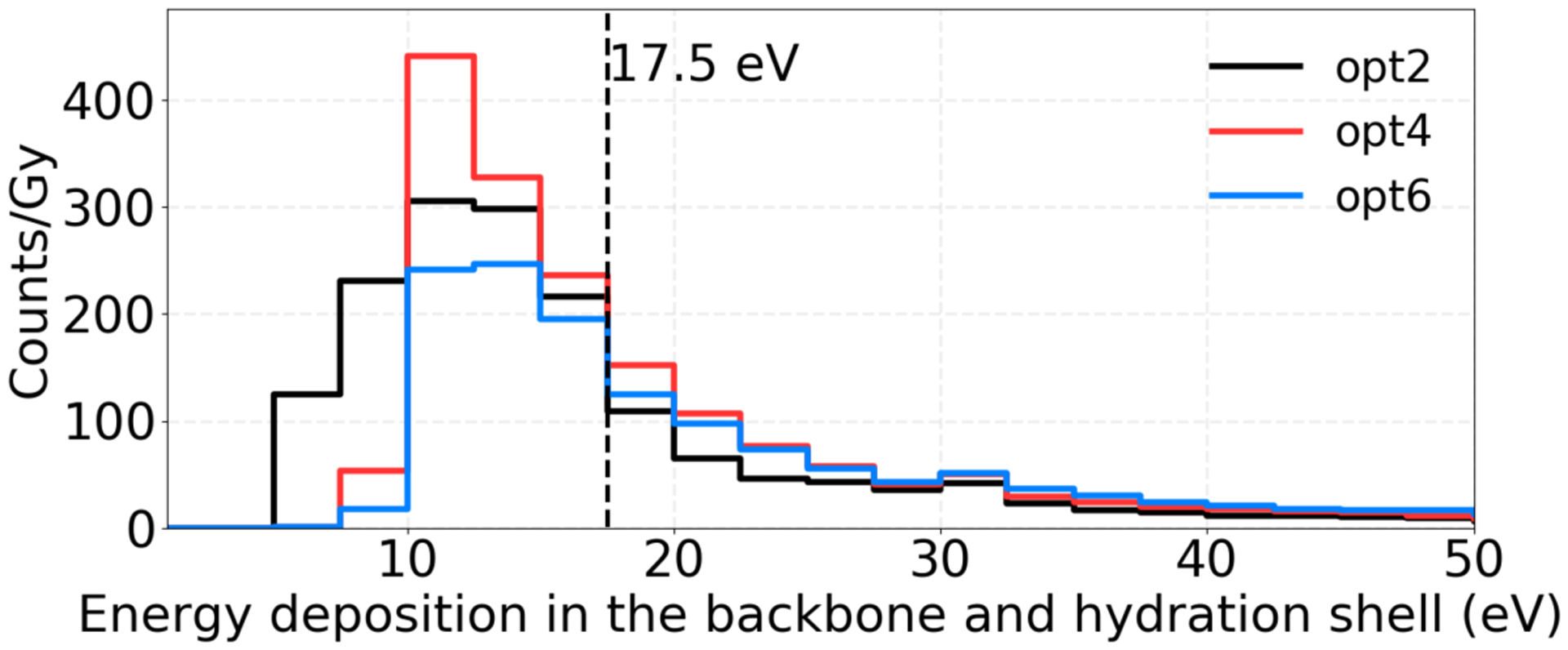
Energy deposition histograms of secondary electrons from 0.5 MeV primary protons in the backbone and hydration shell simulated with different physics constructors. The counts were normalized to the dose to the nucleus.
Table 4.
Counts of energy depositions of secondary electrons from 0.5 MeV primary protons simulated with different physics constructors. The counts were normalized to the dose to the nucleus.
| Opt2 | Opt4 | Opt6 | |
|---|---|---|---|
| Counts/Gy (> 0 eV) in the whole nucleus | 1650.6 | 1729.8 | 1401.4 |
| Counts/Gy (>17.5 eV) in the backbone and hydration shell | 474.9 | 671 | 700.1 |
Accordingly, another critical parameter that could affect the DNA damage results in the physical stage is the energy threshold to produce direct damage. Figure 7 compares the DNA damage results simulated with different energy thresholds: a fixed threshold of 17.5 eV and a linear acceptance threshold ranging from 5–37.5 eV. In the configuration of this work, up to 12% more SBs and 26% more DSBs were observed with the linear acceptance threshold. This trend is consistent with other works (Lampe et al., 2018a; Meylan et al., 2017; Sakata et al., 2019).
Figure 7.
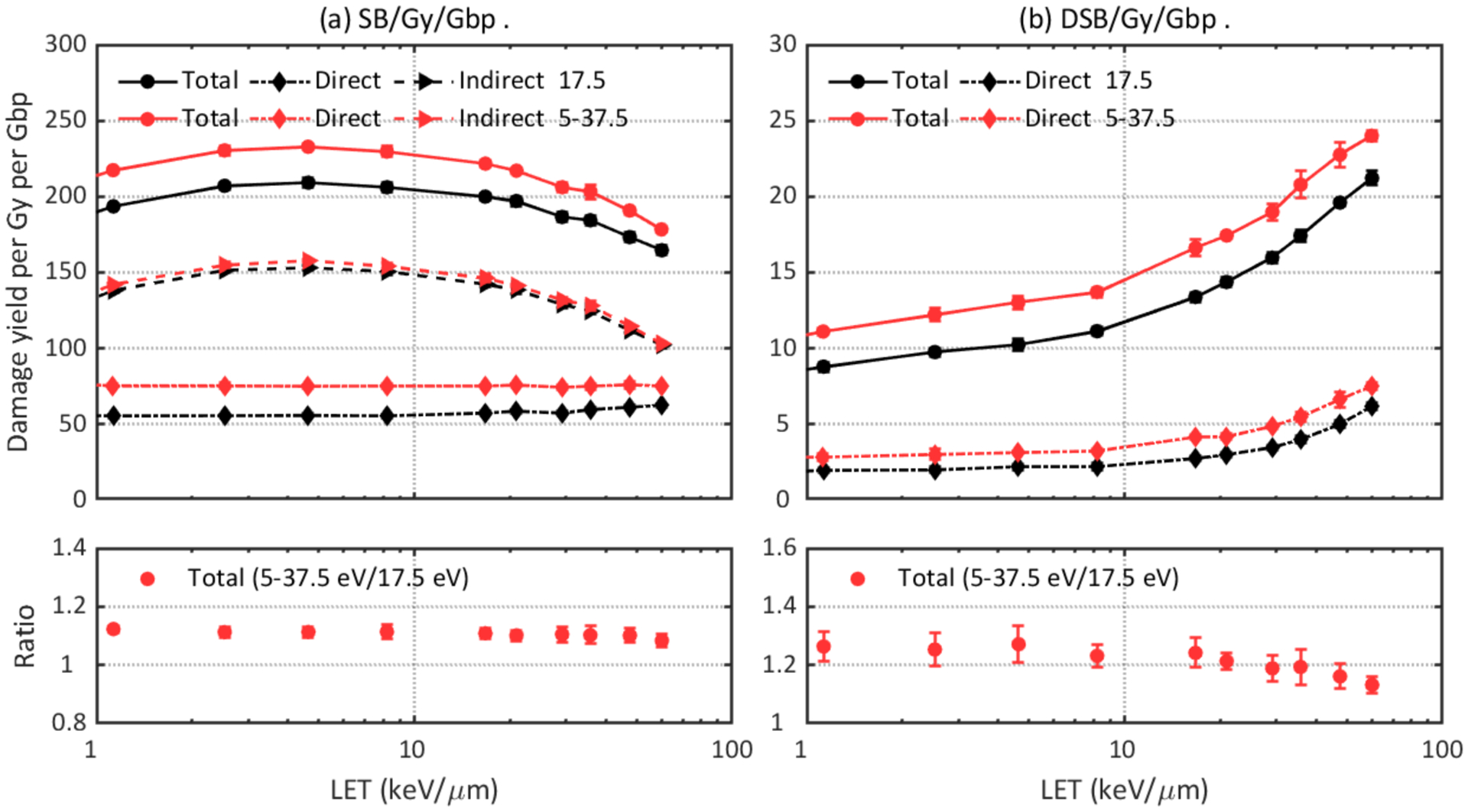
Comparison of DNA damage yields simulated with different direct damage thresholds: a fixed threshold of 17.5 eV or a linear acceptance threshold ranging from 5 to 37.5 eV. Upper left: total, direct, and indirect SB yields. Lower left: ratio of total SB yields. Upper right: total and direct DSB yields. Lower right: ratio of total DSB yields.
Following the physical stage, the ionized and exited water molecules will produce initial chemical species through dissociative decay or auto-ionization leading to chemical interactions. The intrinsic difference between the Geant4-DNA and TOPAS-nBio chemistry models results in different G-values and hence indirect damage yields. Figure 8 shows the difference of DNA damage simulated with G4EmDNAChemistry and TsEmDNAChemistry. The G4EmDNAChemistry model results in ~10% more SBs and DSBs than the TsEmDNAChemistry model in the investigated LET region. This is because only hydroxyl radicals were assumed to be able to cause indirect damages and the yield of hydroxyl radicals was affected by the diffusion and reaction rates in the two models. TsEmDNAChemistry produced ~10% fewer hydroxyl molecules than G4EmDNAChemistry at 1 ns (Ramos-Méndez et al., 2018), thus resulting in fewer indirect damages.
Figure 8.
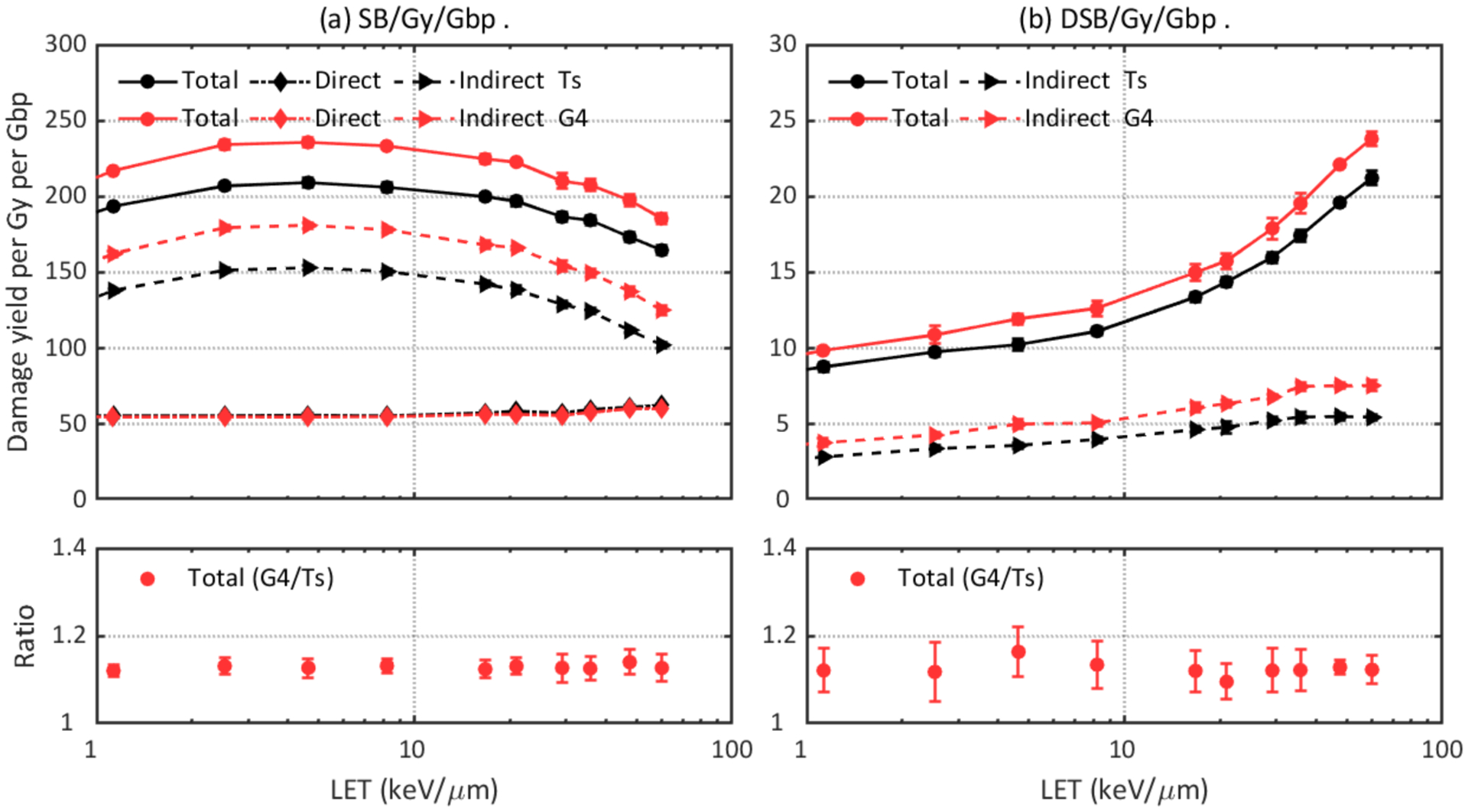
Comparison of DNA damage yields simulated with different chemistry models: Ts (TsEmDNAChemistry) and G4 (G4EmDNAChemistry). Upper left: total, direct, and indirect SB yields. Lower left: ratio of total SB yields. Upper right: total and indirect DSB yields. Lower right: ratio of total DSB yields.
The impact of the time cut of the chemical stage and the probability of hydroxyl radicals to induce DNA damage are shown in Figure 9 and Figure 10, respectively. One expects a higher damage yield with a longer chemical stage length because with a longer time chemical species will travel further before they were terminated, leading to a higher probability to interact with the DNA volume and a higher indirect damage yield. As shown in Figure 9, the 2.5 ns long chemical stage results in up to 36% more SBs and 23% more DSBs than the 1 ns long chemical stage. The 10 ns long chemical stage produces up to 104% more SBs and 51% more DSBs than the 1 ns long chemical stage. As DNA volumes were the only scavengers in the simulation and the typical time scale for scavenging hydroxyl in cellular media is on the order of 1 ns (Lampe et al., 2018a), a 1 ns long chemical stage length may be preferable for DNA damage simulations. As for the hydroxyl damage probability, it is shown that up to 44% more SBs and 71% more DSBs are predicted when a higher damage probability was adopted.
Figure 9.
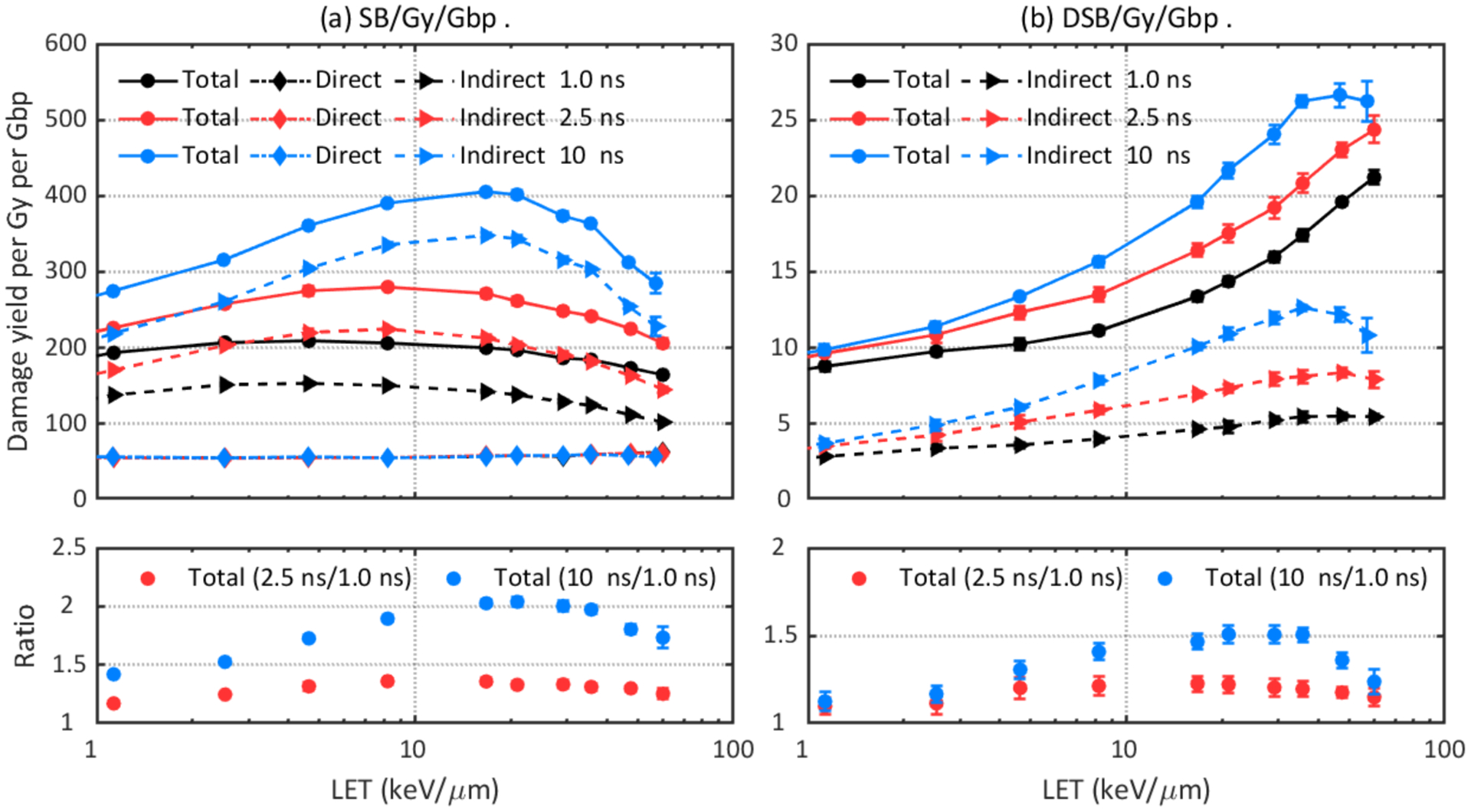
Comparison of DNA damage yields simulated with different chemical stage lengths: 1 ns, 2.5 ns, and 10 ns. Upper left: total, direct, and indirect SB yields. Lower left: ratio of total SB yields. Upper right: total and indirect DSB yields. Lower right: ratio of total DSB yields.
Figure 10.
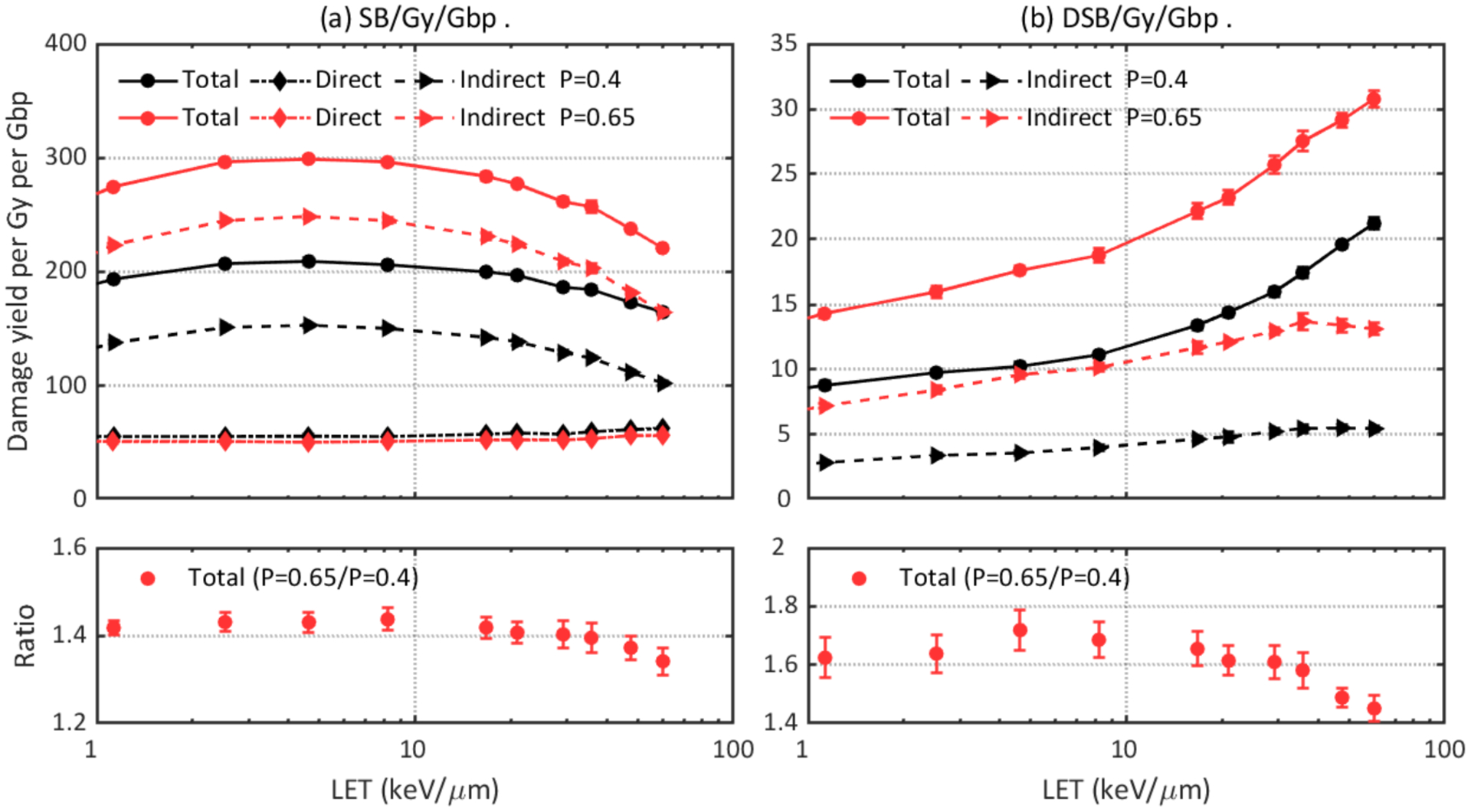
Comparison of DNA damage yields simulated with different hydroxyl damage probabilities: 0.4 and 0.65. Upper left: total, direct, and indirect SB yields. Lower left: ratio of total SB yields. Upper right: total and indirect DSB yields. Lower right: ratio of total DSB yields.
Summary
In this work, we studied the impact of physics and chemistry models as well as three parameters on the DNA damage yields after proton irradiation with a whole nucleus model implemented in TOPAS-nBio. This study aims at providing an estimation for the relative change in DNA damage yields, rather than absolute values, when different models and parameter values are adopted. However, the differences caused by varying DNA geometry configurations and simulation tool on damage results were not evaluated in this work.
The difference in physics constructors intrinsically affect the physical stage interactions, which will sequentially affect the production and interaction of radiolysis in the subsequent pre-chemical and chemical stages. Our simulations show that using G4EmDNAPhysics_option4 results in the highest DNA damage yields and caused up to 34% more DSBs than the G4EmDNAPhysics_option2 model. The G4EmDNAChemistry model predicted ~16% higher DSB yields than the TsEmDNAChemistry model due to different diffusion coefficients and reaction rates. Differences in direct damage threshold, chemical stage length, and hydroxy damage probability can cause differences of up to 26%, 51%, and 71% in predicted DSB yields, respectively, for the DNA geometry and irradiation modalities investigated here.
This study highlights the uncertainties in DNA damage simulations caused by differences in simulation parameters and demonstrates the impact when comparing DNA damage yields from different simulation studies. However, the uncertainty due to the geometry is not considered in this work. A recent study shows that different chromatin compaction forms lead to different DNA damage yields and a change in the contributions from direct and indirect reactions (Tang et al., 2019). In addition, simulation performance critically depends on the adopted cross sections which are prone to uncertainties arising from physics models. For example, it was shown that including the exchange and correlation effects in inelastic electron scattering at low energies (below 10 keV) lead to better agreement with experimental cross section data (Emfietzoglou et al., 2017; Emfietzoglou et al., 2013). The underlying approximations made in cross section calculations may have a sizeable effect on DNA damage calculation. As a result, comparisons of relative effect sizes may be much more stable than absolute predictions as many of the above mentioned uncertainties cancel out.
Acknowledgement
Hongyu Zhu was sponsored by the China Scholarship Council for a year-long study abroad program at Massachusetts General Hospital. This work was supported by the National Cancer Institute under R01 CA187003 (TOPAS – nBio, a Monte Carlo tool for radiation biology research). Simulations were performed on the O2 High Performance Compute Cluster, supported by the Research Computing Group at Harvard Medical School, and the ERISOne cluster, supported by Partners Information Services at Partners Healthcare Systems.
Reference
- Agostinelli S, Allison J, Amako K a, Apostolakis J, Araujo H, Arce P, Asai M, Axen D, Banerjee S and Barrand G 2003. GEANT4—a simulation toolkit Nuclear instruments and methods in physics research section A: Accelerators, Spectrometers, Detectors and Associated Equipment 506 250–303 [Google Scholar]
- Allison J, Amako K, Apostolakis J, Araujo H, Dubois PA, Asai M, Barrand G, Capra R, Chauvie S and Chytracek R J I T o n s 2006. Geant4 developments and applications 53 270–8 [Google Scholar]
- Allison J, Amako K, Apostolakis J, Arce P, Asai M, Aso T, Bagli E, Bagulya A, Banerjee S, Barrand GJNI, Methods in Physics Research Section A: Accelerators S, Detectors and Equipment A 2016 Recent developments in Geant4 835 186–225 [Google Scholar]
- Bernal MA, Bordage MC, Brown JMC, Davidkova M, Delage E, El Bitar Z, Enger SA, Francis Z, Guatelli S, Ivanchenko VN, Karamitros M, Kyriakou I, Maigne L, Meylan S, Murakami K, Okada S, Payno H, Perrot Y, Petrovic I, Pham QT, Ristic-Fira A, Sasaki T, Stepan V, Tran HN, Villagrasa C and Incerti S 2015. Track structure modeling in liquid water: A review of the Geant4-DNA very low energy extension of the Geant4 Monte Carlo simulation toolkit Phys Med 31 861–74 [DOI] [PubMed] [Google Scholar]
- Bordage M, Bordes J, Edel S, Terrissol M, Franceries X, Bardies M, Lampe N and Incerti SJPM 2016. Implementation of new physics models for low energy electrons in liquid water in Geant4-DNA 32 1833–40 [DOI] [PubMed] [Google Scholar]
- Bordes J, Incerti S, Lampe N, Bardiès M and Bordage M-C 2017. Low-energy electron dose-point kernel simulations using new physics models implemented in Geant4-DNA Nuclear Instruments Methods in Physics Research Section B: Beam Interactions with Materials Atoms 398 13–20 [Google Scholar]
- Campa A, Ballarini F, Belli M, Cherubini R, Dini V, Esposito G, Friedland W, Gerardi S, Molinelli S, Ottolenghi A, Paretzke H, Simone G and Tabocchini MA 2005. DNA DSB induced in human cells by charged particles and gamma rays: experimental results and theoretical approaches Int J Radiat Biol 81 841–54 [DOI] [PubMed] [Google Scholar]
- Emfietzoglou D, Kyriakou I, Garcia-Molina R, Abril I and Nikjoo H J R r 2013. Inelastic cross sections for low-energy electrons in liquid water: exchange and correlation effects 180 499–513 [DOI] [PubMed] [Google Scholar]
- Emfietzoglou D, Papamichael G and Nikjoo H 2017. Monte Carlo electron track structure calculations in liquid water using a new model dielectric response function Radiation research 188 355–68 [DOI] [PubMed] [Google Scholar]
- Friedland W, Dingfelder M, Kundrat P and Jacob P 2011. Track structures, DNA targets and radiation effects in the biophysical Monte Carlo simulation code PARTRAC Mutat Res 711 28–40 [DOI] [PubMed] [Google Scholar]
- Friedland W, Jacob P, Bernhardt P, Paretzke HG and Dingfelder M 2003. Simulation of DNA damage after proton irradiation Radiation Research 159 401–10 [DOI] [PubMed] [Google Scholar]
- Friedland W, Schmitt E, Kundrát P, Dingfelder M, Baiocco G, Barbieri S and Ottolenghi A 2017. Comprehensive track-structure based evaluation of DNA damage by light ions from radiotherapy-relevant energies down to stopping Scientific Reports [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SK and Jost D 2018. How epigenome drives chromatin folding and dynamics, insights from efficient coarse-grained models of chromosomes PLoS computational biology 14 e1006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall EJ and Giaccia AJ 2018. Radiobiology for the radiologist (Philadelphia, PA: Lippincottt Williams & Wilkins; ) [Google Scholar]
- Henthorn NT, Warmenhoven JW, Sotiropoulos M, Mackay RI, Kirkby KJ and Merchant MJ 2017. Nanodosimetric Simulation of Direct Ion-Induced DNA Damage Using Different Chromatin Geometry Models Radiat Res 188 690–703 [DOI] [PubMed] [Google Scholar]
- Incerti S, Baldacchino G, Bernal M, Capra R, Champion C, Francis Z, GuÈYe P, Mantero A, Mascialino B, Moretto P, Nieminen P, Villagrasa C and Zacharatou C 2010a. The Geant4-DNA Project International Journal of Modeling, Simulation, and Scientific Computing 01 157–78 [Google Scholar]
- Incerti S, Ivanchenko A, Karamitros M, Mantero A, Moretto P, Tran H, Mascialino B, Champion C, Ivanchenko V and Bernal MJM p 2010b Comparison of GEANT4 very low energy cross section models with experimental data in water 37 4692–708 [DOI] [PubMed] [Google Scholar]
- Incerti S, Kyriakou I, Bernal MA, Bordage MC, Francis Z, Guatelli S, Ivanchenko V, Karamitros M, Lampe N, Lee SB, Meylan S, Min CH, Shin WG, Nieminen P, Sakata D, Tang N, Villagrasa C, Tran HN and Brown JMC 2018. Geant4-DNA example applications for track structure simulations in liquid water: A report from the Geant4-DNA Project Med Phys [DOI] [PubMed] [Google Scholar]
- Karamitros M, Luan S, Bernal MA, Allison J, Baldacchino G, Davidkova M, Francis Z, Friedland W, Ivantchenko V, Ivantchenko A, Mantero A, Nieminem P, Santin G, Tran HN, Stepan V and Incerti S 2014. Diffusion-controlled reactions modeling in Geant4-DNA Journal of Computational Physics 274 841–82 [Google Scholar]
- Kyriakou I, Šefl M, Nourry V and Incerti S J J o A P 2016. The impact of new Geant4-DNA cross section models on electron track structure simulations in liquid water 119 194902 [Google Scholar]
- Lampe N, Karamitros M, Breton V, Brown JM, Kyriakou I, Sakata D, Sarramia D and Incerti S 2018a. Mechanistic DNA damage simulations in Geant4-DNA part 1: A parameter study in a simplified geometry Physica Medica 48 135–45 [DOI] [PubMed] [Google Scholar]
- Lampe N, Karamitros M, Breton V, Brown JMC, Sakata D, Sarramia D and Incerti S 2018b. Mechanistic DNA damage simulations in Geant4-DNA Part 2: Electron and proton damage in a bacterial cell Phys Med 48 146–55 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ and Dorschner M O J s 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome 326 289–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara AL, Ramos-Méndez J, Perl J, Held K, Dominguez N, Moreno E, Henthorn NT, Kirkby KJ, Meylan S and Villagrasa C 2018. Geometrical structures for radiation biology research as implemented in the TOPAS-nBio toolkit Physics in Medicine & Biology 63 175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan S, Incerti S, Karamitros M, Tang N, Bueno M, Clairand I and Villagrasa C 2017. Simulation of early DNA damage after the irradiation of a fibroblast cell nucleus using Geant4-DNA Sci Rep 7 11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokari M, Alamatsaz MH, Moeini H and Taleei R 2018. A simulation approach for determining the spectrum of DNA damage induced by protons Phys Med Biol 63 175003. [DOI] [PubMed] [Google Scholar]
- Nikjoo H, Emfietzoglou D, Liamsuwan T, Taleei R, Liljequist D and Uehara S 2016. Radiation track, DNA damage and response-a review Rep Prog Phys 79 116601. [DOI] [PubMed] [Google Scholar]
- Nikjoo H and Girard P 2012. A model of the cell nucleus for DNA damage calculations Int J Radiat Biol 88 87–97 [DOI] [PubMed] [Google Scholar]
- Nikjoo H, O’Neill P, Wilson W and Goodhead D 2001. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation Radiation research 156 577–83 [DOI] [PubMed] [Google Scholar]
- Nikjoo H, Uehara S, Emfietzoglou D and Cucinotta FA 2006. Track-structure codes in radiation research Radiation Measurements 41 1052–74 [Google Scholar]
- Perl J, Shin J, Schumann J, Faddegon B and Paganetti H 2012. TOPAS: an innovative proton Monte Carlo platform for research and clinical applications Med Phys 39 6818–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante I and Devroye L 2017. Considerations for the independent reaction times and step-by-step methods for radiation chemistry simulations Radiation Physics and Chemistry 139 157–72 [Google Scholar]
- Ramos-Méndez J, Perl J, Schuemann J, McNamara A, Paganetti H and Faddegon B 2018. Monte Carlo simulation of chemistry following radiolysis with TOPAS-nBio Physics in Medicine & Biology 63 105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata D, Lampe N, Karamitros M, Kyriakou I, Belov O, Bernal MA, Bolst D, Bordage MC, Breton V, Brown JMC, Francis Z, Ivanchenko V, Meylan S, Murakami K, Okada S, Petrovic I, Ristic-Fira A, Santin G, Sarramia D, Sasaki T, Shin WG, Tang N, Tran HN, Villagrasa C, Emfietzoglou D, Nieminen P, Guatelli S and Incerti S 2019. Evaluation of early radiation DNA damage in a fractal cell nucleus model using Geant4-DNA Phys Med 62 152–7 [DOI] [PubMed] [Google Scholar]
- Schuemann J, McNamara AL, Ramos-Mendez J, Perl J, Held KD, Paganetti H, Incerti S and Faddegon B 2019a. TOPAS-nBio: An Extension to the TOPAS Simulation Toolkit for Cellular and Sub-cellular Radiobiology Radiat Res 191 125–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuemann J, McNamara AL, Warmenhoven JW, Henthorn NT, Kirkby KJ, Merchant MJ, Ingram S, Paganetti H, Held KD, Ramos-Mendez J, Faddegon B, Perl J, Goodhead DT, Plante I, Rabus H, Nettelbeck H, Friedland W, Kundrat P, Ottolenghi A, Baiocco G, Barbieri S, Dingfelder M, Incerti S, Villagrasa C, Bueno M, Bernal MA, Guatelli S, Sakata D, Brown JMC, Francis Z, Kyriakou I, Lampe N, Ballarini F, Carante MP, Davidkova M, Stepan V, Jia X, Cucinotta FA, Schulte R, Stewart RD, Carlson DJ, Galer S, Kuncic Z, Lacombe S, Milligan J, Cho SH, Sawakuchi G, Inaniwa T, Sato T, Li W, Solov’yov AV, Surdutovich E, Durante M, Prise KM and McMahon SJ 2019b. A New Standard DNA Damage (SDD) Data Format Radiat Res 191 76–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin W-G, Bordage M-C, Emfietzoglou D, Kyriakou I, Sakata D, Min C, Lee SB, Guatelli S and Incerti S 2018. Development of a new Geant4-DNA electron elastic scattering model for liquid-phase water using the ELSEPA code Journal of Applied Physics 124 224901 [Google Scholar]
- Shin W-G, Ramos-Mendez J, Faddegon B, Tran H, Villagrasa C, Perrot Y, Okada S, Karamitros M, Emfietzoglou D and Kyriakou I 2019. Evaluation of the influence of physical and chemical parameters on water radiolysis simulations under MeV electron irradiation using Geant4-DNA Journal of Applied Physics 126 114301 [Google Scholar]
- Tang N, Bueno M, Meylan S, Incerti S, Tran HN, Vaurijoux A, Gruel G and Villagrasa C 2019. Influence of chromatin compaction on simulated early radiation-induced DNA damage using Geant4-DNA Medical Physics 46 1501–11 [DOI] [PubMed] [Google Scholar]
- Tran H, El Bitar Z, Champion C, Karamitros M, Bernal M, Francis Z, Ivantchenko V, Lee S, Shin J, Incerti SJNI, Materials M i P R S B B I w and Atoms 2015. Modeling proton and alpha elastic scattering in liquid water in Geant4-DNA 343 132–7 [Google Scholar]
- Zhu H, McNamara A, McMahon SJ, Ramos-Mendez J, Henthorn NT, Faddegon B, Held K, Perl J, Li J, Paganetti H and Schuemann J 2019. Cellular response to proton irradiation: a simulation study with TOPAS-nBio (under review) [DOI] [PMC free article] [PubMed] [Google Scholar]


