Abstract
N-terminal acetylation (NTA) (see Glossary) is one of the most widespread protein modifications, which occurs on most eukaryotic proteins, but is significantly less common on bacterial and archaea proteins. This modification is carried out by a family of enzymes called N-terminal acetyltransferases (NATs) (see Glossary). To date, twelve NATs have been identified, harboring different composition, substrate specificity and, in some cases, modes of regulation. Recent structural and biochemical analysis of NAT proteins now allows for a comparison of their molecular mechanisms and modes of regulation, which are described here. Although sharing an evolutionarily conserved fold and related catalytic mechanism, each catalytic subunit employs unique elements to mediate substrate-specific activity and employ NAT-type specific auxiliary and regulatory subunits for their cellular functions.
Keywords: co-translational modification, post-translational modification enzyme mechanism, N-terminal acetylation, NATs, HYPK, IP6, ribosome
Evolutionarily Conserved NTA has Diverse Biological Functions
The majority of eukaryotic proteins are subject to several N-terminal modifications at the early stage of their biogenesis by the ribosome, including Nt-methionine excision (NME), Nt-acetylation (NTA), Nt-myristoylation (MYR), Nt-methylation and Nt-arginylation [1–3]. The evolutionarily conserved and irreversible NME and NTA modifications affect the functions of many proteins and increase the diversity of the proteome [2, 3]. NTA occurs when the iMet is removed or retained, co-translationally (see Glossary) or post-translationally (see Glossary) [4–6] and on 50–90% of eukaryotic proteins but only 10–29% of bacterial and archaea proteins [7–13], correlating the abundance of NTA with organism complexity.
NTA changes the chemical properties of the protein N-termini: neutralizing the charge, creating a new h-bond acceptor, changing the α-amino nitrogen nucleophilicity and basicity, and increasing its hydrophobicity and size. These changed chemical properties have diverse biological consequences on protein function including protein half-life, folding, complex formation, and localization [5, 14]. Correlating with these diverse functions, knockout of individual eukaryotic N-terminal acetyltransferases (NATs) display different phenotypes, presumably associated with the differential misregulation of their respective client proteins [5]. Alteration in NTA activity is also linked to disease such as various developmental and neurodegenerative disorders and cancers [15–17] in humans and stress response in plants [6, 18]. While several other reviews cover the biology of NATs [4, 5, 17, 19], this review will focus on their structure, mechanism and regulation over evolution.
General Molecular Features of NATs
The GCN5 related N-acetyltransferases (GNAT) (see Glossary) superfamily of proteins is one of the largest protein families, with over 10,000 protein members present in all domains of life. These proteins catalyze a bi-reactant process: transferring an acetyl group from the donor acetyl-CoA cofactor to the primary amine group of a variety of biomolecules including small molecules and protein or peptides lysine sidechains and N-termini. GNATs share a common structurally conserved α/β fold, despite their low degree of sequence identity; and mode of acetyl-CoA binding [20]. GNATs typically contain distinct flanking N and C terminal regions that directly contributing to their distinct functions.
NTA is carried out by the NAT subfamily of GNATs, which typically contain four α-helices and seven β-strands, although addition secondary structural elements are often present at their N or C termini (Figure 1A). β1-β4 and β5-β6 are arranged antiparallelly, while the parallel β4 and β5 strands split to create a splay in between for binding peptide and acetyl-CoA (Figure 1A). The peptide binding site of NATs is typically flanked by the β6-β7 and α1-α2 loops, which directly participate in N-terminal substrate binding and take on a relatively closed configuration relative to GNATs that acetylate internal lysine substrates, thereby disfavoring lysine sidechain binding [21–23]. NATs typically make extensive h-bond interactions with the backbone atoms of the first 2–3 N-terminal residues further contributing to specificity for protein N-termini. These two distinct features distinguish NATs from KATs (lysine acetyltransferases) (see Glossary) and other members of GNATs. NTA by NATs follows an ordered Bi-Bi reaction mechanism, where acetyl-CoA cofactor binding promotes subsequent N-termini binding prior to direct acetyl-group transfer from the cofactor to protein [24]. One or two residues function as a general base(s) (often through an intervening water molecule) to deprotonate the terminal amino group to facilitate its nucleophilic attack of the acetyl group of acetyl-CoA (Figure 1B). This reaction proceeds through a tetrahedral intermediate, thus making CoA-peptide conjugate bi-substrate analogues in which N-terminal peptides are linked to CoA through an acetaldehyde group potent NAT inhibitors (Figure 1C) [25].
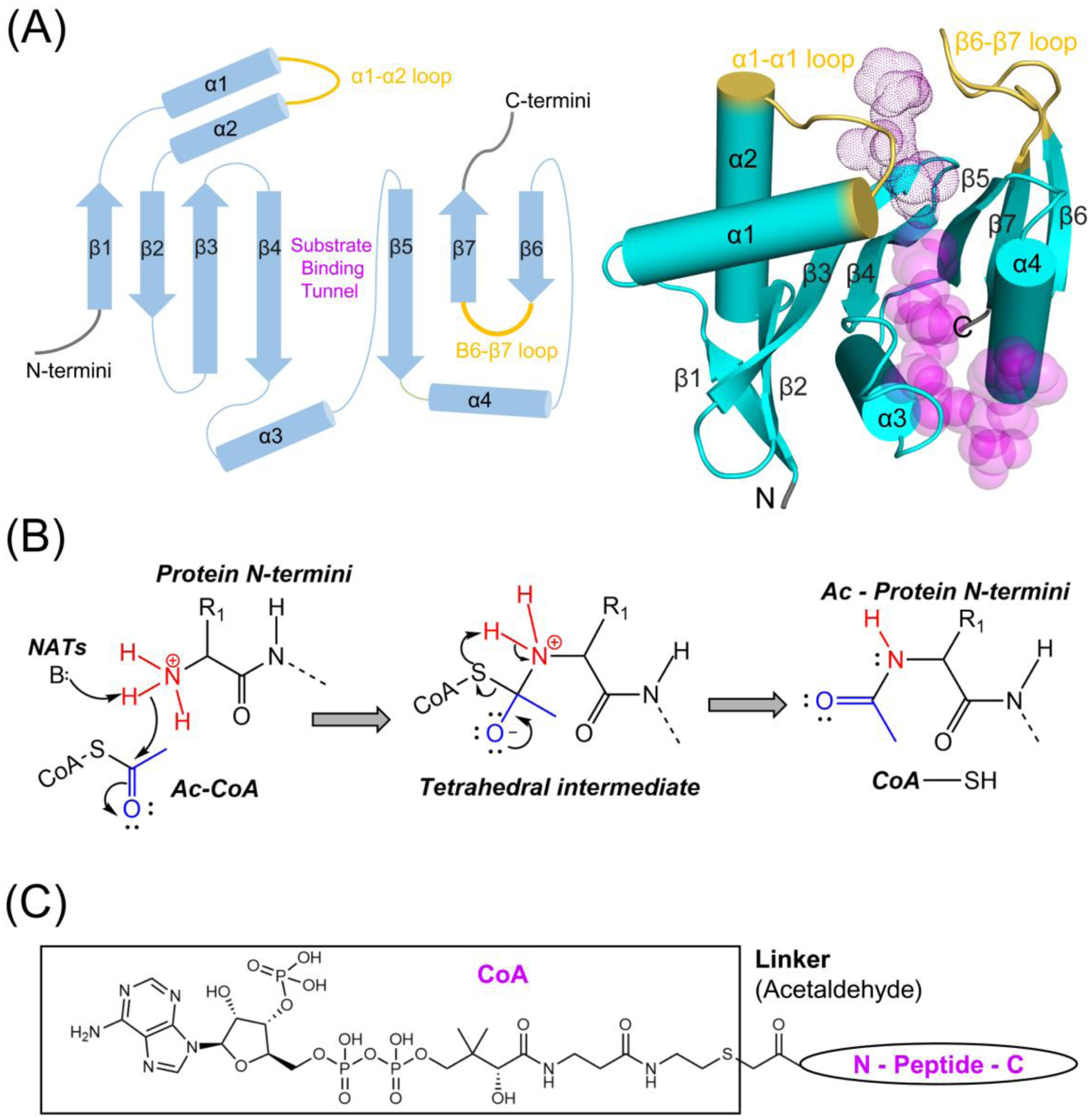
Figure 1.
NAT catalytic domains share common topology and related catalytic mechanism. (A) The general topology of NAT catalytic subunits is depicted as in 2D cartoon on the left and 3D on the right panel with secondary structures shown. NATs usually contain seven β strands and four helices, but additional secondary structural elements within this topology and at their N-, C- termini are sometimes present. The α1- α2 and β6-β7 substrate binding loops are highlighted in yellow. The length of α helices and β strands does not accurately reflect actual scale in the 2D representation. The transparent magenta dots and spheres represent in the 3D representation represent the peptide and acetyl-CoA substrates, respectively. The 3D representation was generated using human NAA50 PDB: 3TFY (B) General catalytic mechanism of NATs, which transfer an acetyl group from acetyl-CoA to protein Nt-amino group is depicted. The acetyl and Nt-amino groups are colored as blue and red, respectively. A general base (or two, sometimes through a coordinated water molecule) is utilized to deprotonate the protein Nt-amino group, which subsequently attacks the acetyl group to form a tetrahedral intermediate. The deprotonated CoA is then deprotonated by a general acid (not shown) and released as the tetrahedral intermediate collapses. (C) The chemical structural of a CoA-peptide conjugate Bi-substrate analogue is shown. A linker (acetaldehyde group) is used to covalently link the CoA and peptide substrate.
A wealth of mechanistic information about the substrate specific activities of NATs has been derived from their structures and associated biochemical studies. The NATs that have been structurally characterized to date are listed in Table 1. As described below, each NAT employs a unique strategy to achieve substrate-specific acetylation and regulation.
Table. 1.
N-terminal acetyltransferases are characterized in all domains of life.
| ╱ | NATs | Subunits | Substrates specificity | PDBb and References | ||
|---|---|---|---|---|---|---|
| Bacteria | RimI | monomer | Ribosomal protein S18, relaxed | S. typhimurium | 2CNMc,e | [29] |
| RimL | homodimer | Ribosomal protein L7/L12 | S. typhimurium | 1S7Nc | [31] | |
| RimJ | monomer | Ribosomal protein S5, relaxed | Mycobacterium | 6C32c | - | |
| Archaea | ssNAT | monomer | relaxed | S. solfataricus | 4LX9c | [36] |
| Eukaryotes | NAA10 | monomer | Acidic substrates | S. pombe | 4KVXc | [46] |
| NatAa | NAA10, NAA15 | A-, S-, T-, V-, G- | S. pombe Human | 4KVMc,e 6C9Mc |
[46] [49] |
|
| NatA/HYPKa | NAA10, NAA15, HYPK | C. thermophilum Human | 5NNRc 6C95d |
[47] [49] |
||
| NatBa | NAA20, NAA25 | M-D/E/N/Q- | C. albicans Human | 5K18c,e 6VP9d,e | [53] [54] |
|
| NatCa | NAA30, NAA35, NAA38 | M-L/I/F/Y/K- | [57] | |||
| NatDa | NAA40, monomer | Histones H2A/H4 (S-G-R-G-) | S. pombe Human | 4UA3c 4U9Wc,e |
[60] [60] |
|
| NAA50 | monomer | M-S/T/A/V/L/I/F/Y/K- | Human | 3TFYc,e | [61] | |
| NatEa | NAA50, NAA10, NAA15 | NatA- and NAA50-type | S. cerevisiae Human | 6O07c 6PPLd |
[48] [50] |
|
| NatE/HYPKa | NAA50, NAA10, NAA15, HYPK | Human | 6PW9d | [50] | ||
| NatF | Homodimer/monomer | M-L/I/F/Y/K- | Human A. thaliana | 5ICWc,5ICVc,e 6TGXc,e |
[70] [71] |
|
| NatG | NAA70 | M-, A-, S-, T- | [72] | |||
| NatH | NAA80, monomer | Actins (D/E-D/E-D/E-) | Drosophila | 5WJEc,e | [45] | |
| NatH/profilin/actin | NAA80, profilin, actin | Human | 6NBEc | [76] | ||
These NATs act co-translationally.
Representative NAT member for which a 3D structures is determined.
Structures solved by X-ray crystallography.
Structures solved by Cryo-EM.
Structures include a peptide substrate fragment as a ligand or as part of a ligand.
Bacterial NATs
The three E. coli homologs, RimI, RimJ and RimL were among the first characterized NATs, showing post-translational NTA activity towards the ribosomal proteins S18, S5, and L7/L12, respectively [26]. Some recombinant proteins overexpressed in E coli were later found to undergo NTA [27], and more recent proteome-wide analysis found that ~30–100 E. coli N-termini are at least partially acetylated, predominantly at N-terminal serine, alanine, methionine and threonine residues [10, 11]. The abundance of NTA in Pseudomonas aeruginosa PA14 and Mycobacterium tuberculosis appears to be in the range of 10% ~ 29% [9, 28], although their biological importance have not been rigorously evaluated.
Bacterial orthologs of each of the Rim proteins have been structurally characterized. The Salmonella typhimurium RimI (StRimI) structure was determined in complex with CoA, acetyl-CoA or a CoA-peptide conjugate showing a typical GNAT fold (Figure 2A) [29]. Several StRimI residues change conformation when in complex with the peptide portion of the conjugate with several residues making backbone h-bonds to carbonyl and nitrogen atoms of the first three residues of the peptide portion, while several RimI residues make van der Waals and h-bonds to side chains 2–4 (Figure 3). RimI Glu103 is proposed to function as a general base through an intervening water molecule, while Tyr115 is proposed to act as the general acid (Figure 3). A more recent biochemical characterization of Mycobacterium tuberculosis RimI demonstrated a conventional eukaryotic NatA/NatC/NatE substrate preference, consistent with a relaxed substrate specificity for RimI [30].
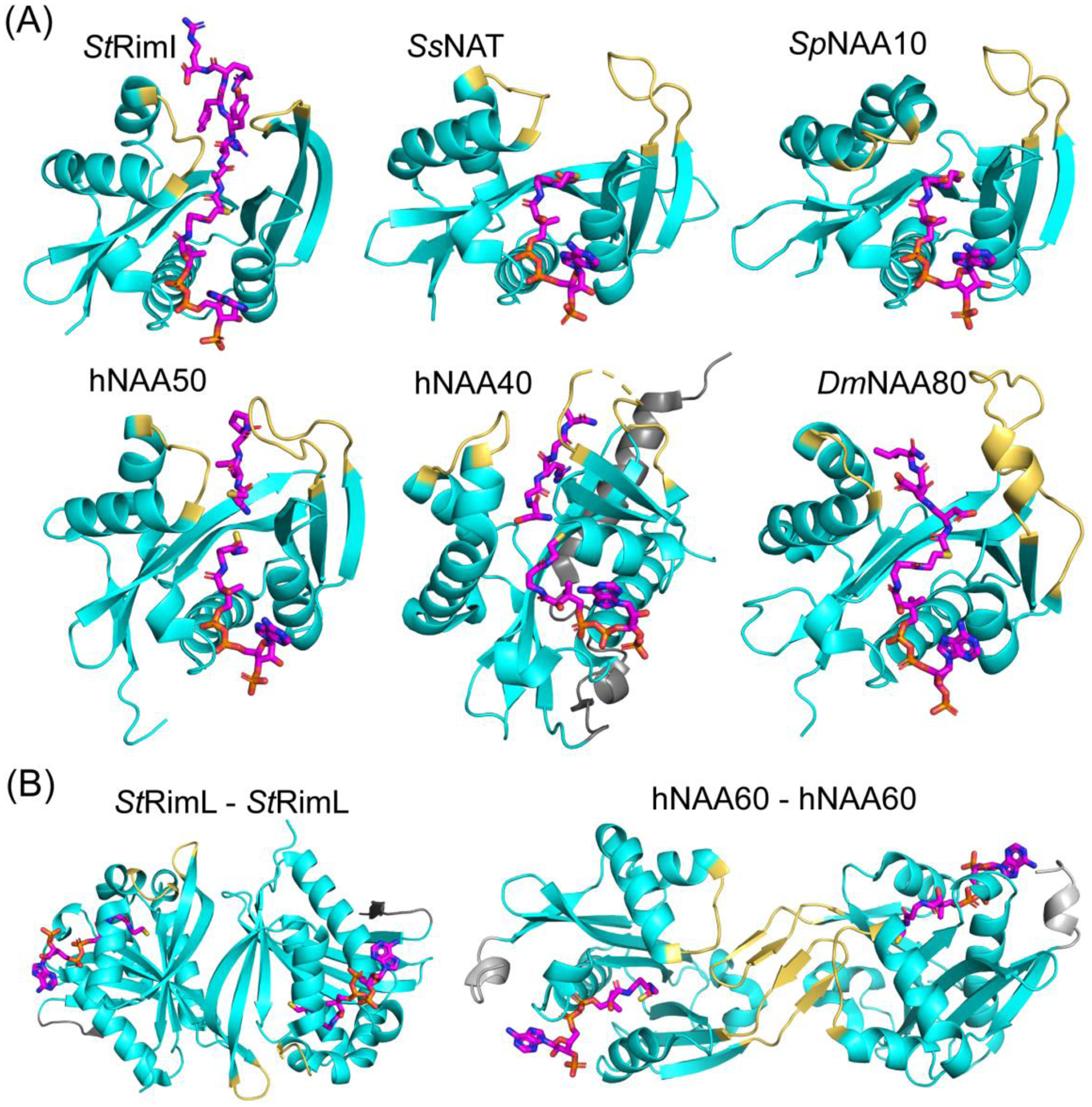
Figure 2.
Some NATs function independently, either in monomeric or homodimer form. (A) Structures of monomeric StRimI (PDB: 2CNM), SsNAT (PDB: 4LX9), uncomplexed SpNAA10 (PDB: 4KVX), hNAA50 (PDB: 3TFY), hNAA40 (PDB: 4U9W), and DmNAA80 (PDB: 5WJE) are shown in cartoon and color in cyan. The α1- α2 and β6-β7 substrate binding loops are highlighted in yellow. Substrate in the structures are shown in stick and colored in magenta. The hNAA40-specific N terminal domain is highlighted in grey. (B) Dimeric RimL (PDB:1S7N) and hNAA60 (PDB: 5ICW) are shown. The α1- α2 and β6-β7 substrate binding loops are highlighted in yellow. Substrate in the structures are shown in stick and colored in magenta. To form a dimer, StRimL utilizes the two β6 strands from each subunit, while hNAA60 uses the extended β6- β7 loops. The NAA60-specific N terminal domain is highlighted in grey. NAT catalytic subunits that have not been shown to function independently are not shown.
Figure 3.
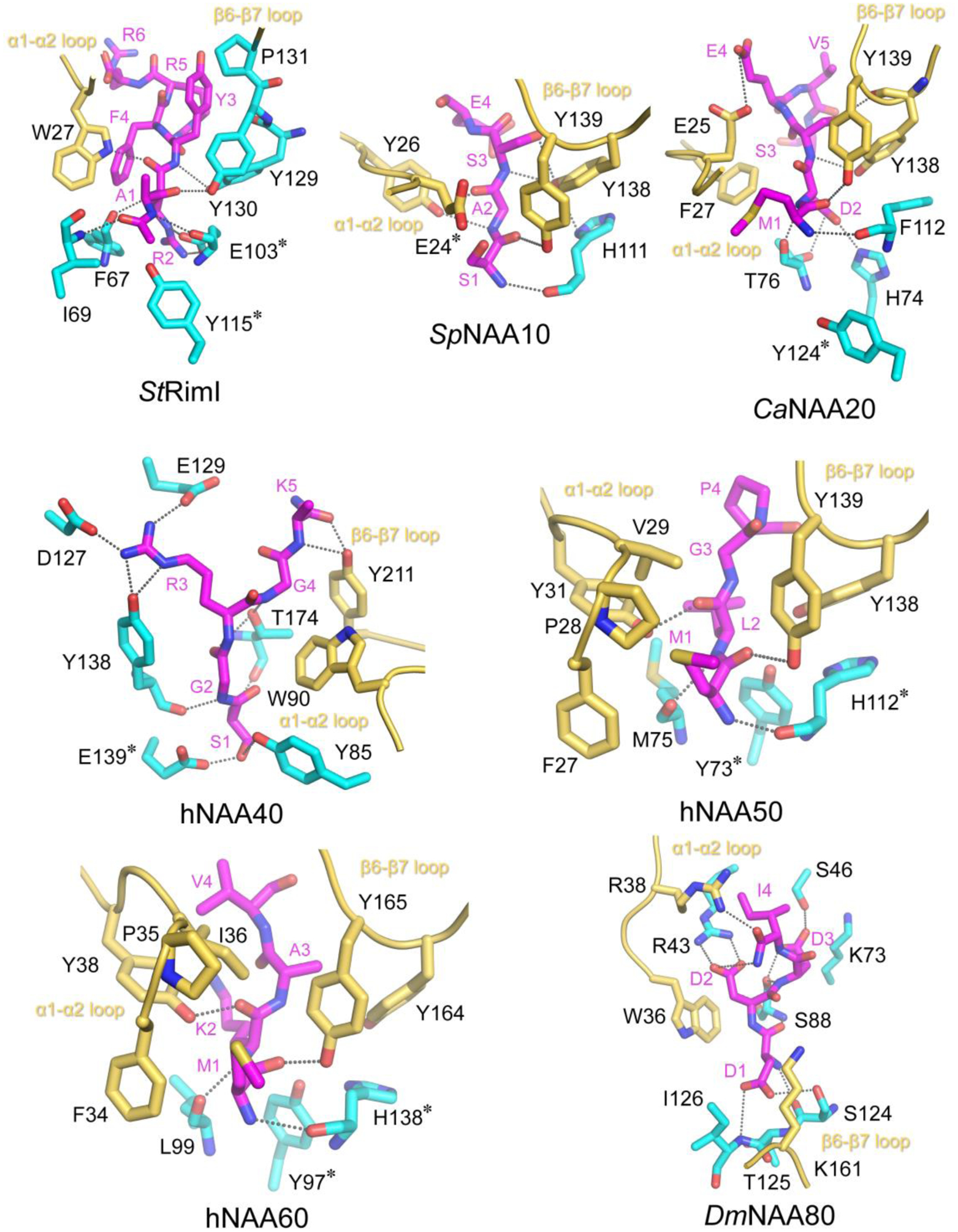
NATs use related but distinct mechanism to recognize their peptide substrate N-termini. Peptide binding sites of StRimI (PDB: 2CNM), complexed SpNAA10 (PDB: 4KVM, with SpNAA15 hidden), CaNAA20(PDB: 5K04, with CaNAA25 hidden), hNAA40 (PDB: 4U9W), hNAA60 (PDB: 5ICV), and DmNAA80 (PDB: 5WJE) are shown in cartoon. Peptide substrates are shown in magenta sticks. The residues labeled with a * symbol are proposed catalytic residues. Dashed lines indicate h-bonds formed between atoms. The α1- α2 and β6-β7 substrate binding loops are highlighted in yellow. Water-mediate interactions in the PDB structures are not shown.
Crystal structures of StRimL in apo form and in complex with CoA reveals a similar overall fold to bacterial StRimI, however StRimL forms a homodimer through antiparallel β-strands (β6) from opposing subunits to form a contiguous β-sheet that spans the entire dimer (Figure 2B) [31]. This 2-fold symmetry appears to be precisely configured to acetylate its cognate dimeric L7 N-termini [32, 33]. Compared to the apo StRimL structure, the bound of CoA appears to stabilize the α1-α2 loop, which could also facilitate the cooperative binding of the dimeric L7/L12. Like StRimI, StRimL is proposed to use a ternary complex mechanism involving a glutamate general base residue (Glu160), while a tyrosine (Tyr98) is proposed to stabilize the tetrahedral intermediate, with a serine residue (Ser141) functioning as a general acid.
Mycobacterium smegmatis RimJ structures alone and in complex with cofactor or cofactor analogs has recently been deposited to the PDB (PDB: 6C32, 6C30, 6C37. Taken together, the bacterial NAT proteins reveal a general GNAT fold in either monomeric or homodimeric form with substrate protein recognition occurring through backbone and sidechain interaction to the first few N-terminal residues, with acid-base catalysis proceeding through dedicated residues.
Archaea NATs
Like in bacteria, while earlier studies indicated that NTA occurs on only ribosomal proteins in archaea, more recent N-terminomics studies reveal that NTA occurs on 14–29% of archaeal proteins [12, 13, 34]. Archaea contains only one conserved NAT, which exhibits a very relaxed substrate spectrum including substrates by eukaryotic NatA/B/C/E [35, 36]. The Sulfolobus solfataricus NAT (SsNAT) shows the greatest sequence conservation with the NAA10 catalytic subunit of the eukaryotic NatA complex, with 33% sequence identity [36], as confirmed by its structure bound to acetyl-CoA (PDB: 4LX9) (Figure 2A) [36]. Interestingly, the monomeric SsNAT is more structurally similar to the active form of Naa10 within the binary NatA complex, rather than NAA10 alone, which is not active towards NatA substrates. A notable place where SsNAT and NAA10 diverge is in the C-terminal end of the α1 helix that is part of the α1-α2 loop substrate binding region of NATs. This region of SsNAT is more similar to the eukaryotic NAA50, which is also active towards NatE substrates as a monomer. This suggests that SsNAT is a hybrid of the NAA10 and NAA50 eukaryotic NATs. This is consistent with biochemical studies on SsNAT demonstrating that different catalytic residues are used to acetylate NatA (SsNAT-Glu84) and NAA50/NatE (SsNAT-His137/E176 -like substrates, analogous to corresponding residues in these eukaryotic NATs [36]. It is possible that the archaea NAT serves as one of the ancestors of eukaryotic NATs, which subsequently evolved to be substrate-specific. An unusually long β3-β4 loop in SsNAT, was also shown to play a more selective role in acetylating NAA50-type substrates, although the mechanism for this is still unclear. A more recent SsNAT structure (called Ard1) [37] and Thermoplasma volcanium Ard1 [38], were consistent with the earlier study, reinforcing the substrate flexibility of the archaeal NATs. Interestingly, it was demonstrated that mutations of the catalytic Glu residue of ssNAT could shift its substrate preference [37].
Eukaryotic NATs
NatA:
NatA acetylates ~ 40% of human proteins, the most of any NAT [7]. NatA is a heterodimer of catalytic NAA10 (also named ARD1) and auxiliary NAA15 subunits [39, 40]. Notably, NAA10 can also exist independently [41, 42]. When the second residue of a protein is relatively small and uncharged (Table 1), it will be acetylated by NatA after the initiator methionine is cleaved [7, 43]. In contrast, the NAA10 monomer is not able to acetylate canonical NatA substrate, but instead exhibits specificity for acidic N-terminal sequences in vitro [42, 44, 45].
The structure of Schizosaccharomyces pombe NatA (SpNatA) bound to a CoA-peptide conjugate reveals that the SpNAA15 auxiliary subunit adopt a ring-like structure of tetratricopeptide repeat (TPR) motifs, which completely wraps around the SpNAA10 catalytic subunit, with the two proteins making extensive hydrogen bonding and van der Waals interactions (Figure 4A) [46]. The Chaetomium thermophilum [47] and Saccharomyces cerevisiae [48] NatA display similar overall folds. A structural comparison of the SpNatA complex with SpNAA10 alone, reveals that SpNAA15 binding to SpNAA10 induces a conformational change of the α1-α2 loop region of SpNat10 to position key catalytic residues for substrate-specific recognition and catalysis [46]. The SpNAA10 active site reveals that h-bonds to the backbone carbonyl groups of the first two residues and van der Waals contacts to Ser1 anchor the peptide portion of the CoA-peptide conjugate (Figure 3). Glu24 was also proposed to play role as a general base for catalysis. Each of the residues revealed to play important roles in substrate binding and catalysis are highly conserved within NatA orthologs and shown to result in decreased NatA activity when mutated [46]. Importantly, modeling of amino acid side chains in the Ser1 position, reveals that only small uncharged amino acids could be accommodated, consistent with the known substrate specificity of NatA [46]. Interestingly, this same study showed that a non-cognate EEE peptide could be acetylated by the SpNAA10-E24A mutant but not by WT SpNatA, suggesting that free NAA10 uses a different catalytic strategy to acetylate NatH-type substrates.
Figure 4.
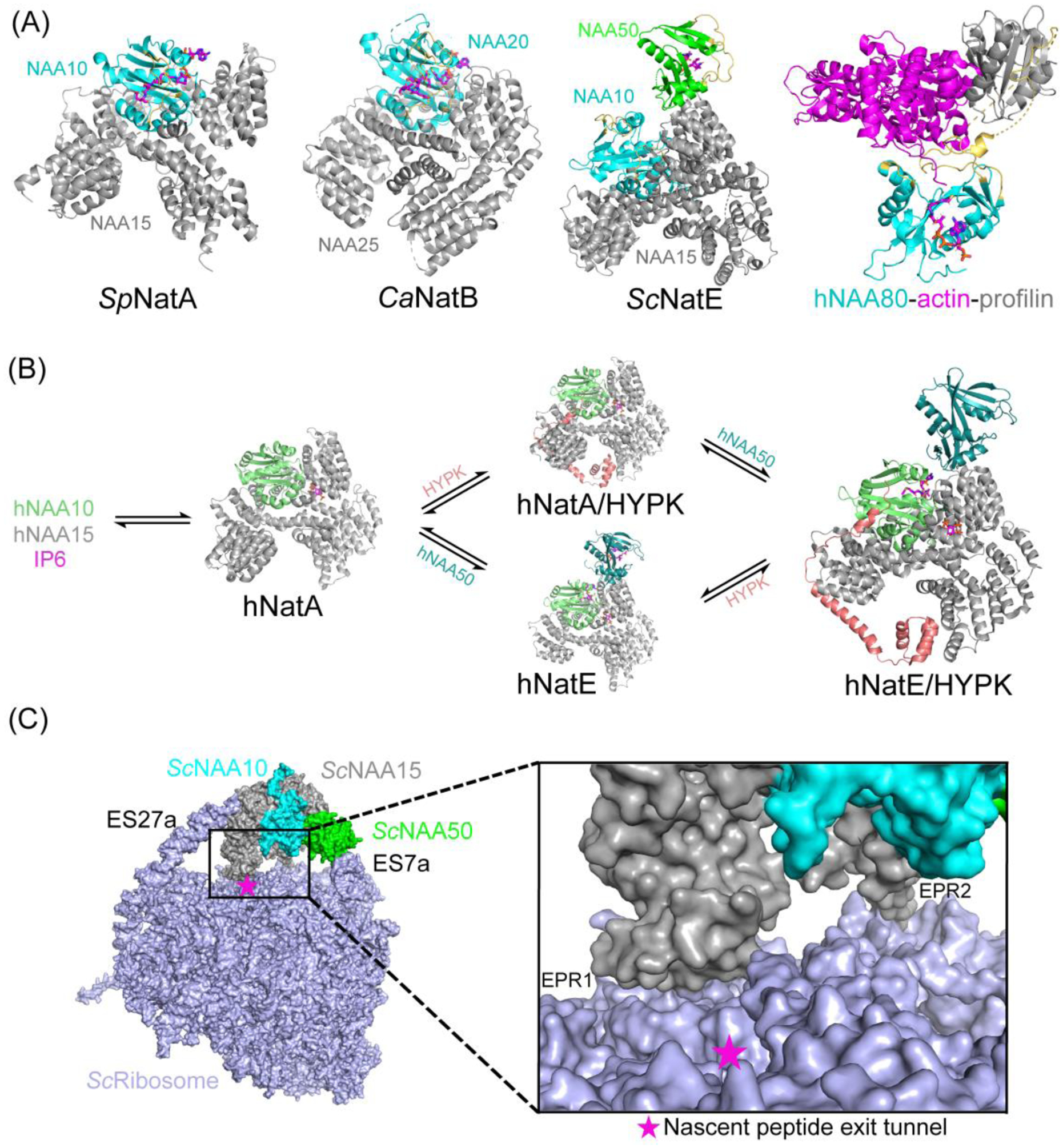
Some NATs function by forming complexes with auxiliary subunits or regulatory proteins. (A) Structures of SpNatA (PDB:4KVM), CaNatB (PDB:5K04), ScNatE (PDB: 6O07) and hNAA80-actin-profilin complexes (PDB:6NBE) are shown in cartoon. (B) In humans, the dynamics and interplay between hNatA, hNAA50 and HYPK are shown. hNAA10 can exist independently. Two subunits and Inositol hexaphosphate (IP6) form hNatA (PDB: 6C9M) complex. HYPK and hNAA50 each can associate with hNatA to form competing complexes (hNatA/HYPK PDB: 6C95, hNatE PDB:6PPL). Tetrameric complex hNatE/HYPK (PDB: 6PW9) can be formed when both HYPK and hNAA50 are bound to hNatA. hNAA10, HYPK, hNAA50, hNAA15 are shown in chartreuse, salmon, teal and grey, respectively. (C) Structure of ScNatE bound to ribosome (PDB: 6HD7) is shown. Two rRNA expansion segments ES27a and ES7a are contacting NAA15 and NAA50, respectively. Two electropositive regions ERP1(N terminus of NAA15) and EPR2 (internal basic helix) on NAA15 which directly contact ribosome are shown in the zoom-in view.
The structure of the human NatA (hNatA) shows a high degree structural conservation with SpNatA [49], except for the presence of an extended metazoan-specific Sel1-like repeat region at the C-terminal end of the hNAA15 auxiliary subunit, which was shown to play a role in overall hNatA stability [49]. In addition, the hNatA structure reveals the presence of an endogenously bound inositol hexaphosphate (IP6) (see Glossary) molecule at an interface region between hNAA10 and hNAA15 via electrostatic interactions [49, 50]. A recently identified hNatA mutant K450E displays defects in IP6 binding and leads to NatA activity loss which can be rescued by the addition of IP6 [51], consistent with a role of IP6 in hNatA stability and activity.
NatB:
NatB, conserved from yeast to man, acetylates ~21% of human proteins, such as actin, tropomyosin, CDK2, and α-synuclein. NatB is a binary complex containing catalytic NAA20 and auxiliary NAA25 subunits and has specificity for N termini containing MD-, ME-, MN- and MQ- sequences [52]. The structure of Candida albicans NatB (CaNatB) was determined alone and in complex with a CoA-peptide conjugate [53]. The catalytic CaNaa20 subunit shows a typical GNAT fold, and the CaNAA25 auxiliary subunit also displays high structural similarity to SpNAA15, particularly in the way it wraps around the catalytic subunit (Figure 4A). Unlike NAA10, NAA20 is unstable in the absence of NAA25. Extensive interactions are observed between CaNAA25 and CaNAA20, with the major contact interface mediated by the α1- α2 loop of CaNAA20 [53], similar to the NatA complex. The interaction between NAA20 and the peptide substrate mainly involves h-bond interactions to the backbone amides of the first three residues of the peptide portion of the CoA-peptide conjugate, with side chain contacts to residues Met1 and Asp2 through van der Walls and h-bond interactions (Figure 3). Mutation of contact residues generally reduce NatB acetylation activity, consistent with the structural observations[53]. A more recent cryo-EM structure of human NatB bound to a CoA-α-synulein peptide conjugate shows a high degree of structural similarity to CaNatB, although provides structural and biochemical data consistent with a role for Tyr124 (hNAA20-Tyr123) acting as a general base for catalysis [54].
NatC:
NatC is a heterotrimer of a catalytic subunit NAA30 and two auxiliary subunits NAA35 and NAA38 [55]. NatC acts co-translationally on the peptide substrate staring with ML-, MF, MI, MW-, playing roles in viral particle assembly, maintaining mitochondrial integrity and apoptosis [56, 57]. As of the writing of this review, a a detailed structure/function analysis of NatC had not yet been reported.
NatD:
NatD (NAA40), which functions as a monomer, is conserved from yeast to human, and is the most selective NAT, carrying out NTA of only histones H4 and H2A [58, 59], which most commonly contain the sequence SGRGK. The structure of hNatD bound to acetyl-CoA and CoA and an N-terminal histone H4/H2A peptide, reveals that while NatD adopts a GNAT fold, it also contains a unique N terminal helix-loop-strand segment, which wraps around the GNAT domain and plays an important role in hNatD stability, partially mimicking similar roles of the auxiliary subunits of NatA and NatB [60] (Figure 2A). Another unique feature of NatD is found in its substrate binding loops, where the α1- α2 loop is extended and flipped towards the peptide substrate, such that the opposing β6-β7 loop is flipped away [60]. This alters the peptide binding site to make it uniquely suited for its cognate N-termini. Indeed, the structure of hNatD bound to CoA and peptide reveals that nearly every hydrogen bond donor and acceptor atom within the first four residues are engaged in hydrogen bounding interactions, with Arg3 playing a particularly important role, and the small sizes of Gly2 and Gly4 also being critical (Figure 3). Activity assays also revealed that Glu139 is essential for catalyst, possibly acting as a general base (Figure 3).
NatE:
NatE refers to the complex between the NAA50 catalytic subunit and the NatA complex (also containing the NAA10 catalytic subunit). Monomeric NAA50 is also stable. However, yeast NAA50 is inactive, while in higher eukaryotes, such as human and Drosophila, NAA50 is active in the absence or presence of NatA [22, 48, 61–63]. NAA50 has specificity for N-terminal Met residues followed by a less restricted set of amino acids at the second position (Table 1) [22]. The structure of hNAA50 bound to CoA and an N-terminal peptide reveals that the peptide is recognized almost exclusively through interactions to residues one and two [61] (Figures 2A and 3). While the carbonyl and amino groups of residues Met1 and Leu2 engage in h-bond interactions, the side chains make extensive van der Walls interactions. Consistent with the known specificity of NAA50, the active site appears exquisitely suited for Met1, while the Val2 binding site appears to have greater flexibility to accommodate several amino acid substitutions (Figure 3). A combination of mutagenesis, kinetics and structural observations suggest that hNAA50 Tyr73 and His112 are essential for catalysis via an ordered water molecule, likely serving as general acid or base residues [61] (Figure 3). Interestingly, these two catalytic residues of hNAA50 are absent in the corresponding positions of yeast homologous, which likely explains the inactivity of the yeast NAA50 proteins. A recent structure of ScNatE and related biochemical studies also shows that ScNAA50 has relatively poor acetyl-CoA binding capacity, consistent with its catalytic inactivity [48].
The binding affinity between NAA50 and NatA is in the nanomolar range[48], consistent with the fact that a significant portion of NAA50 in the cell is bound to NatA [64]. The molecular basis for association between NAA50 and NatA from yeast and human were recently revealed [48, 50], demonstrating that NAA50 docks onto a unique surface of the NAA15 auxiliary subunit primarily through hydrophobic interactions, while making significantly more modest electrostatic interactions with NAA10 (Figure 4). Nonetheless biochemical experiments reveal that NAA10 and NAA50 can influence each other’s activity within the NatE complex [48, 50].
NatF:
NatF (NAA60), found only in higher eukaryotes [65, 66], is located on the cytosolic side of Golgi membranes to acetylate transmembrane protein [67, 68]. It has an unique membrane binding domain at its C-termini [67–70], which mediates Golgi membrane binding [68]. Recently, Arabidopsis thaliana NatF was demonstrated to associate with the plasma membrane also via its C-termini [71]. NatF has specificity for N-terminal Met residues followed by Leu, Ile, Phe, Tyr and Lys residues (Table 1). Unlike other NATs, NatF is homodimeric in solution but shifts to a monomer state when peptide substrate binds (Figure 2B) [70]. The structure of hNatF in complex with CoA or a CoA-peptide conjugate reveals that it has a unique extended β -β7 loop, which mediates dimerization and sterically occludes peptide binding, but participates in peptide recognition of the NatF monomer when cofactor and peptide are both bound [70]. hNatF peptide recognition in hNatF is mainly to the first two residues, through backbone h-bonds and a hydrophobic pocket (similar to NAA50) that recognizes Met1 with less specificity for residue Lys2 (Figure 3) [70]. Tyr97 and His138 are proposed to function as general base residues in hNatF, similar to roles played by residues of Tyr73 and His112 from hNAA50 [70], which is consistent with mutagenesis studies (Figure 3) [69].
NatG:
NatG is localized within the chloroplast of plant, processing M-, A-, S-, T-staring-termini as substrates [72]. As of the writing of this review, a detailed structure/function analysis of NatG had not yet been reported.
NatH:
NatH (NAA80) is widespread only in animals [44], and post-translationally active toward processed cytoplasmic β- and γ-actin in vivo with acidic N termini DDDI and EEEI, respectively [44, 73, 74]. The structure of Drosophila melanogaster Naa80 (DmNaa80) bound to a CoA-peptide conjugate reveals a more open and highly basic substrate binding site than other NATs, which is specifically configured to bind its acidic substrates [45] (Figure 2A). H-bonds extend through the first three backbone and side-chain residues (Figure 3), explaining the specificity for highly acidic substrates. An unusual intramolecular hydrogen bond is also observed between the backbone amide nitrogen of I4 and the sidechain of D2 (Figure 3). Mutational and activity assays is consistent with substrate preference deriving mostly from the acidic residues at positions two and three [45], which is unusual for NAT proteins that typically specify residues one and two.
Regulation of NATs.
HYPK regulates both NatA and NatE activity.
The Huntingtin-Interacting Protein K (HYPK) (see Glossary), was found to associate with the human NatA complex [75]. It is absent in most yeast, but found in Chaetomium thermophilum [47] and human [49, 50] where it was structurally characterized bound to NatA. In both cases, HYPK was shown to have intrinsic NatA inhibitory activity, and in the study of the human complex proposed to contribute to cognate substrate specificity as HYPK was shown to be partially uncompetitive and noncompetitive with respect to acetyl-CoA and peptide substrate, respectively [49]. When bound to NatA, HYPK forms a bipartite structure: a C-terminal ubiquitin-associated domain (α3-α5) binds extensively to the NAA15 auxiliary subunit, a N-terminal loop-α1-helix region binds across the catalytic NAA10 subunit to distort its active site, and a long α2 helix connects these two ends of HYPK. Biochemical data is consistent with the structural findings that the ubiquitin-associated domain forms the high affinity interaction, while the loop-α1-helix region harbors the catalytic inhibitory activity. More recently, structural and biochemical studies reveal that HYPK can also form a stable complex with the NatE complex (Figure 4B), although HYPK and NAA50 where shown to allosterically reduce the binding affinity of each other [50].
Naa80 activity is regulated by the actin chaperone profilin.
Naa80 is preferentially active towards monomeric actin and the presence of profilin has been shown to increase its catalytic efficiency [76]. The crystal structure of hNaa80 in complex with monomeric actin and profilin with cofactor and cofactor analogs reveals the first structure of a NAT bound to its intact protein substrate [76]. The structure shows that a Naa80-specific extended β6-β7 proline-rich loop and α2-helix of hNAA80 is utilized to meditate interaction with both actin and profilin [76]. Thus formation of hNAA80-actin-profilin complex orients the N termini of actin perfectly into the substrate binding groove of hNAA80 (Figure 4A) [76]. It is proposed that, with the aid of profilin, hNAA80 can act more efficiently to compensate for the low abundance of hNAA80 relative to actin in the cell.
Ribosome association by NATs
In eukaryotes, NatA/B/C/E associate with the ribosome for co-translational activity, utilizing their auxiliary subunits as the predominant anchoring point [77], while NatD could potentially use its extended N terminal region [59, 60]. This proximity of the NATs to the ribosome likely facilitates the relatively high stoichiometry of NTA that is observed in vivo.
The current molecular understanding of the interactions between NATs and the ribosome is limited to NatA or NatE. Early studies suggested that NatA contacts ribosomal protein uL23 and uL29 around the peptide exit tunnel [77] to interact with the nascent chain [78]. The ratio of ribosome to NatA is around 40:1 in yeast, leading to questions about the dynamics of NatA/ribosome interactions [79]. Purified non-translating ribosome can also interact with NatA in vitro via two conserved electropositive regions (EPR) on NAA15 [80], which is confirmed through mutational studies [80] and in vivo [81]. A recent structure of a ScNatE/ribosome complex reveals that ScNatE docks at the nascent peptide exit tunnel near uL31 and uL22, with rRNA expansion segments making key contacts to ScNAA15 and ScNaa50 (Figure 4C) [82].
Concluding Remarks
NTA occurs on countless proteins in three domains of life both co-translationally and post-translationally, impacting diverse cellular functions of their client protein. Although sharing an evolutionarily conserved GNAT fold and related catalytic mechanism, each NAT employs unique elements to mediate substrate-specific activity to carry out their distinct cellular functions. NAT auxiliary and regulatory subunits also play NAT-type specific roles. While some NATs have dedicated co-translational and post-translational roles, the mechanisms that may toggle some NATs such as NAA10 and NAA50 between the two roles requires further investigation. Still missing from the PDB is NatC, which is the only NAT that used two auxiliary subunits, and the chloroplast localized NATs, which may have unique functions. Do other NAT agonists or antagonists exist and how might they work (see Outstanding Questions)? Finally, a greater molecular understanding of how NATs carry out and navigate between each other co-translational NTA on the ribosome warrants further study.
Structural Basis, Mechanism, and Regulation of Protein N-Terminal Acetylation: Balancing Evolutionary Conservation and Versatility.
Outstanding Questions
How do NATs toggle between post- and co-translational N-terminal acetylation?
To date, the structure and mechanism of the NatC complex is not known. Why does NatC need two auxiliary subunits for function?
To date, the structure and mechanism of the chloroplast localized NATs are unknown. What are the unique features of these NATs?
Do other NAT agonists or antagonists exist?
What is the molecular mechanism of co-translational acetylation on the ribosome of NatA/E and other NATs that function co-translationally such as NatC/B/D?
How do NATs with co-translational activities navigate the same or different ribosomes?
Highlights
To date, in total of 12 different NATs have been identified to collectively N-terminally acetylate countless proteins from all domains of life to mediate many biological processes
NATs uniquely mediate both post- and co-translational N-terminal acetylation
The currently availability of structures of many NATs bound to their cognate substrates now allows for a detailed molecular comparison to derive conserved and unique features underlying NAT activity and substrate specificity
NATs are subject to regulation by inhibitor and stimulatory proteins and the molecular basis for this regulation has recently come to light
Acknowledgements
This work was supported by NIH grant R35 GM118090 awarded to R.M.
Glossary
- Co-translationally
a process that occurs when the protein nascent chain is still bound to the ribosome
- GCN5-related N-acetyltransferases (GNATs)
an enzyme superfamily that transfers an acetyl group from the donor acetyl-CoA to the primary amine group of varied acceptors including small molecules, peptides, protein N-termini, and sidechains of protein internal lysine residues
- Huntingtin-interacting protein K (HYPK)
initially found as a huntingtin interacting partner, this protein can physically associate with NatA and harbors intrinsic NatA inhibitory activity
- Inositol hexaphosphate (IP6)
A small molecule that is the primary storage form of phosphorus in plant seeds. Present at the interface between NAA15 and NAA10, IP6 is essential for NatA complex formation and normal function
- KATs (lysine acetyltransferases
enzymes that catalyze the transfer of an acetyl group from acetyl-CoA to the primary amine in the ε-position of the lysine side chain on proteins including histones, formerly knowns as histone acetyltransferases or HATs
- N-terminal acetylation (NTA)
an enzymatic modification that covalently attaches an acetyl group from acetyl-CoA to the N-terminal amino (α-amino) group of the first residue of a protein, also known as N-α acetylation and Nt-acetylation
- N-terminal acetyltransferase (NAT)
enzyme or enzyme complex that catalyzes protein NTA. In eukaryotes, the catalytic subunits of NatA-NatH are named as NAA10/20/30/40/50/60/70/80, while the auxiliary subunits of NatA/B/C are named as NAA15, NAA25, NAA35/NAA38, respectively
- Post-translationally
a process that occurs after the synthesized protein is released from the ribosome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mohamed AE and Mohamed AR (2018) Post-translational N-terminal Arginylation of Protein Fragments: A Pivotal Portal to Proteolysis. Current Protein & Peptide Science 19, 1214–1223 [DOI] [PubMed] [Google Scholar]
- 2.Varland S, et al. (2015) N‐terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics 15, 2385–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giglione C, et al. (2015) N-terminal protein modifications: Bringing back into play the ribosome. Biochimie 114, 134–146 [DOI] [PubMed] [Google Scholar]
- 4.Aksnes H, et al. (2019) Co-translational, Post-translational, and Non-catalytic Roles of N-Terminal Acetyltransferases. Mol Cell 73, 1097–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aksnes H, et al. (2016) First Things First: Vital Protein Marks by N-Terminal Acetyltransferases. Trends Biochem Sci 41, 746–760 [DOI] [PubMed] [Google Scholar]
- 6.Linster E and Wirtz M (2018) N-terminal acetylation: an essential protein modification emerges as an important regulator of stress responses. Journal of Experimental Botany 69, 4555–4568 [DOI] [PubMed] [Google Scholar]
- 7.Arnesen T, et al. (2009) Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci U S A 106, 8157–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bienvenut WV, et al. (2012) Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-alpha-acetylation features. Mol Cell Proteomics 11, M111.015131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelkar DS, et al. (2011) Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol Cell Proteomics 10, M111.011627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bienvenut WV, et al. (2015) Proteome-wide analysis of the amino terminal status of Escherichia coli proteins at the steady-state and upon deformylation inhibition. Proteomics 15, 2503–2518 [DOI] [PubMed] [Google Scholar]
- 11.Schmidt A, et al. (2016) The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol 34, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkland PA, et al. (2008) Shotgun proteomics of the haloarchaeon Haloferax volcanii. Journal of proteome research 7, 5033–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aivaliotis M, et al. (2007) Large-Scale Identification of N-Terminal Peptides in the Halophilic Archaea Halobacterium salinarum and Natronomonas pharaonis. Journal of Proteome Research 6, 2195–2204 [DOI] [PubMed] [Google Scholar]
- 14.Oh JH, et al. (2017) Control of Hsp90 chaperone and its clients by N-terminal acetylation and the N-end rule pathway. Proc Natl Acad Sci U S A 114, E4370–E4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myklebust LM, et al. (2015) Naa10 in development and disease. Oncotarget 6, 34041–34042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalvik TV and Arnesen T (2013) Protein N-terminal acetyltransferases in cancer. Oncogene 32, 269–276 [DOI] [PubMed] [Google Scholar]
- 17.Dorfel MJ and Lyon GJ (2015) The biological functions of Naa10 - From amino-terminal acetylation to human disease. Gene 567, 103–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber M, et al. (2020) NatB-Mediated N-Terminal Acetylation Affects Growth and Biotic Stress Responses. Plant Physiology 182, 792–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ree R, et al. (2018) Spotlight on protein N-terminal acetylation. Exp Mol Med 50, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salah Ud-Din AI, et al. (2016) Structure and Functional Diversity of GCN5-Related N-Acetyltransferases (GNAT). International journal of molecular sciences 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magin RS, et al. (2016) The N-terminal Acetyltransferase Naa10/ARD1 Does Not Acetylate Lysine Residues. The Journal of biological chemistry 291, 5270–5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evjenth R, et al. (2009) Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. The Journal of biological chemistry 284, 31122–31129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abboud A, et al. (2020) Dynamics-function relationship in the catalytic domains of N-terminal acetyltransferases. Computational and Structural Biotechnology Journal 18, 532–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evjenth RH, et al. (2012) Human protein N-terminal acetyltransferase hNaa50p (hNAT5/hSAN) follows ordered sequential catalytic mechanism: combined kinetic and NMR study. The Journal of biological chemistry 287, 10081–10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foyn H, et al. (2013) Design, synthesis, and kinetic characterization of protein N-terminal acetyltransferase inhibitors. ACS chemical biology 8, 1121–1127 [DOI] [PubMed] [Google Scholar]
- 26.Favrot L, et al. (2016) Bacterial GCN5-Related N-Acetyltransferases: From Resistance to Regulation. Biochemistry 55, 989–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernal-Perez LF, et al. (2012) RimJ-mediated context-dependent N-terminal acetylation of the recombinant Z-domain protein in Escherichia coli. Molecular bioSystems 8, 1128–1130 [DOI] [PubMed] [Google Scholar]
- 28.Ouidir T, et al. (2015) Characterization of N-terminal protein modifications in Pseudomonas aeruginosa PA14. J Proteomics 114, 214–225 [DOI] [PubMed] [Google Scholar]
- 29.Vetting MW, et al. (2008) Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for Nα-acetylation of ribosomal protein S18. Protein Science 17, 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathak D, et al. (2016) Biochemical evidence for relaxed substrate specificity of Nα-acetyltransferase (Rv3420c/rimI) of Mycobacterium tuberculosis. Scientific Reports 6, 28892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vetting MW, et al. (2005) A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium. The Journal of biological chemistry 280, 22108–22114 [DOI] [PubMed] [Google Scholar]
- 32.Wahl MC, et al. (2000) Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12. The EMBO Journal 19, 174–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bocharov EV, et al. (2004) From Structure and Dynamics of Protein L7/L12 to Molecular Switching in Ribosome. J. Biol. Chem 279, 17697–17706 [DOI] [PubMed] [Google Scholar]
- 34.Falb M, et al. (2006) Archaeal N-terminal protein maturation commonly involves N-terminal acetylation: a large-scale proteomics survey. J Mol Biol 362, 915–924 [DOI] [PubMed] [Google Scholar]
- 35.Mackay DT, et al. (2007) An acetylase with relaxed specificity catalyses protein N-terminal acetylation in Sulfolobus solfataricus. Mol Microbiol 64, 1540–1548 [DOI] [PubMed] [Google Scholar]
- 36.Liszczak G and Marmorstein R (2013) Implications for the evolution of eukaryotic amino-terminal acetyltransferase (NAT) enzymes from the structure of an archaeal ortholog. Proc Natl Acad Sci U S A 110, 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y-Y and Hsu C-H (2015) Structural Basis for Substrate-specific Acetylation of Nα-acetyltransferase Ard1 from Sulfolobus solfataricus. Scientific Reports 5, 8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma C, et al. (2014) Structure of Thermoplasma volcanium Ard1 belongs to N-acetyltransferase family member suggesting multiple ligand binding modes with acetyl coenzyme A and coenzyme A. Biochimica et biophysica acta 1844, 1790–1797 [DOI] [PubMed] [Google Scholar]
- 39.Mullen JR, et al. (1989) Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. Embo j 8, 2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park EC and Szostak JW (1992) ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. Embo j 11, 2087–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnesen T, et al. (2005) Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem J 386, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Damme P, et al. (2011) Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of N(alpha)-acetyltransferases and point to hNaa10p as the post-translational actin N(alpha)-acetyltransferase. Mol Cell Proteomics 10, M110.004580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polevoda B, et al. (1999) Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. Embo j 18, 6155–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drazic A, et al. (2018) NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc Natl Acad Sci U S A 115, 4399–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goris M, et al. (2018) Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80. Proc Natl Acad Sci U S A 115, 4405–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liszczak G, et al. (2013) Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat Struct Mol Biol 20, 1098–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weyer FA, et al. (2017) Structural basis of HypK regulating N-terminal acetylation by the NatA complex. Nat Commun 8, 15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng S, et al. (2019) Structure and Mechanism of Acetylation by the N-Terminal Dual Enzyme NatA/Naa50 Complex. Structure 27, 1057–1070 e1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gottlieb L and Marmorstein R (2018) Structure of Human NatA and Its Regulation by the Huntingtin Interacting Protein HYPK. Structure 26, 925–935 e928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng S, et al. (2020) Molecular basis for N-terminal acetylation by human NatE and its modulation by HYPK. Nature Communications 11, 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng H, et al. (2019) Phenotypic and biochemical analysis of an international cohort of individuals with variants in NAA10 and NAA15. Hum Mol Genet 28, 2900–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Damme P, et al. (2012) N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc Natl Acad Sci U S A 109, 12449–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong H, et al. (2017) Molecular Basis of Substrate Specific Acetylation by N-Terminal Acetyltransferase NatB. Structure 25, 641–649 e643 [DOI] [PubMed] [Google Scholar]
- 54.Marmorstein R, et al. (2020) Molecular Basis for N-terminal Alpha-Synuclein Acetylation by Human NatB. bioRxiv, 2020.2005.2011.089318 [DOI] [PMC free article] [PubMed]
- 55.Polevoda B and Sherman F (2001) NatC Nalpha-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p, and Mak31p. The Journal of biological chemistry 276, 20154–20159 [DOI] [PubMed] [Google Scholar]
- 56.Van Damme P, et al. (2016) A Role for Human N-alpha Acetyltransferase 30 (Naa30) in Maintaining Mitochondrial Integrity. Mol Cell Proteomics 15, 3361–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Starheim KK, et al. (2009) Knockdown of human N alpha-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol Cell Biol 29, 3569–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song OK, et al. (2003) An Nalpha-acetyltransferase responsible for acetylation of the N-terminal residues of histones H4 and H2A. The Journal of biological chemistry 278, 38109–38112 [DOI] [PubMed] [Google Scholar]
- 59.Hole K, et al. (2011) The human N-alpha-acetyltransferase 40 (hNaa40p/hNatD) is conserved from yeast and N-terminally acetylates histones H2A and H4. PLoS One 6, e24713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Magin RS, et al. (2015) The molecular basis for histone H4- and H2A-specific amino-terminal acetylation by NatD. Structure 23, 332–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liszczak G, et al. (2011) Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. The Journal of biological chemistry 286, 37002–37010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Damme P, et al. (2015) N-terminal acetylome analysis reveals the specificity of Naa50 (Nat5) and suggests a kinetic competition between N-terminal acetyltransferases and methionine aminopeptidases. Proteomics 15, 2436–2446 [DOI] [PubMed] [Google Scholar]
- 63.Rong Z, et al. (2016) Opposing Functions of the N-terminal Acetyltransferases Naa50 and NatA in Sister-chromatid Cohesion. The Journal of biological chemistry 291, 19079–19091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hou F, et al. (2007) The acetyltransferase activity of San stabilizes the mitotic cohesin at the centromeres in a shugoshin-independent manner. J Cell Biol 177, 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rathore OS, et al. (2016) Absence of N-terminal acetyltransferase diversification during evolution of eukaryotic organisms. Sci Rep 6, 21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Damme P, et al. (2011) NatF Contributes to an Evolutionary Shift in Protein N-Terminal Acetylation and Is Important for Normal Chromosome Segregation. PLoS Genet. 7, e1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aksnes H, et al. (2015) An organellar nalpha-acetyltransferase, naa60, acetylates cytosolic N termini of transmembrane proteins and maintains Golgi integrity. Cell Rep 10, 1362–1374 [DOI] [PubMed] [Google Scholar]
- 68.Aksnes H, et al. (2017) Molecular determinants of the N-terminal acetyltransferase Naa60 anchoring to the Golgi membrane. The Journal of biological chemistry 292, 6821–6837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen JY, et al. (2016) Structure and function of human Naa60 (NatF), a Golgi-localized bi-functional acetyltransferase. Sci Rep 6, 31425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stove SI, et al. (2016) Crystal Structure of the Golgi-Associated Human Nalpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation. Structure 24, 1044–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Linster E, et al. The Arabidopsis Nα-acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response. New Phytologist n/a [DOI] [PubMed] [Google Scholar]
- 72.Dinh TV, et al. (2015) Molecular identification and functional characterization of the first Nalpha-acetyltransferase in plastids by global acetylome profiling. Proteomics 15, 2426–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arnesen T, et al. (2018) Actin’s N-terminal acetyltransferase uncovered. Cytoskeleton 75, 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wiame E, et al. (2018) NAT6 acetylates the N-terminus of different forms of actin. The FEBS journal 285, 3299–3316 [DOI] [PubMed] [Google Scholar]
- 75.Arnesen T, et al. (2010) The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of Huntingtin aggregation. Mol Cell Biol 30, 1898–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rebowski G, et al. (2020) Mechanism of actin N-terminal acetylation. Science Advances 6, eaay8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Polevoda B, et al. (2008) Yeast N(alpha)-terminal acetyltransferases are associated with ribosomes. J Cell Biochem 103, 492–508 [DOI] [PubMed] [Google Scholar]
- 78.Gautschi M, et al. (2003) The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol Cell Biol 23, 7403–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raue U, et al. (2007) Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. The Journal of biological chemistry 282, 7809–7816 [DOI] [PubMed] [Google Scholar]
- 80.Magin RS, et al. (2017) Probing the interaction between NatA and the ribosome for co-translational protein acetylation. PLoS One 12, e0186278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Varland S and Arnesen T (2018) Investigating the functionality of a ribosome-binding mutant of NAA15 using Saccharomyces cerevisiae. BMC Research Notes 11, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Knorr AG, et al. (2019) Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation. Nat Struct Mol Biol 26, 35–39 [DOI] [PubMed] [Google Scholar]


