Abstract
Arginylation is a protein post-translational modification catalyzed by arginyl-tRNA transferases (ATE1s), which are critical enzymes conserved across all eukaryotes. Arginylation is a key step in the Arg N-degron pathway, a hierarchical cellular signaling pathway that links the ubiquitin-dependent degradation of a protein to the identity of its N-terminal amino acid side chain. The fidelity of ATE1-catalyzed arginylation is imperative, as this post-translational modification regulates several essential biological processes such as cardiovascular maturation, chromosomal segregation, and even the stress response. While the process of ATE1-catalyzed arginylation has been studied in detail at the cellular level, much remains unknown about the structure of this important enzyme, its mechanism of action, and its regulation. In this work, we detail the current state of knowledge on ATE1-catalyzed arginylation, and we discuss both ongoing and future directions that will reveal the structural and mechanistic details of this essential eukaryotic cellular regulator.
Keywords: ATE1: an abbreviation for arginyl-tRNA transferase (also known as R-transferase), a eukaryotic enzyme that catalyzes post-translational arginylation in a tRNA-dependent manner; arginylation: the post-translational covalent addition of the amino acid arginine (Arg) to a peptide or a protein arginylome: a compendium of all arginylation targets within an organism; GNAT fold: an abbreviation for the Gcn5-related N-acetyl transferase fold, a protein fold that typically comprises a six-stranded β-sheet of mixed polarity surrounded by four α-helices of mixed orientations and that catalyzes the acetylation of various substrates; isoform: a functionally similar protein of nonidentical amino acid sequence encoded by either the same gene from which exons have been removed or by a completely separate gene; L/F transferase: an abbreviation for leucyl/phenylalanyl-tRNA transferase, a bacterial enzyme that catalyzes posttranslational leucylation/phenylalanylation in a tRNA-dependent manner; N-degron: an N-terminal chemical moiety that signals the degradation rate of a peptide or protein; N-degron pathway: formerly the N-end rule pathway, a hierarchical cellular signaling pathway that links the ubiquitin-dependent degradation of a protein to the identity of its N-terminal amino acid side chain; post-translational modification (PTM): covalent additions of, or chemical modifications to, protein functional groups that occur after protein synthesis and are essential for normal cellular function; tRNA: an abbreviation for transfer ribonucleic acid, a type of RNA that commonly decodes mRNA, but may also be used for purposes other than protein translation; ubiquitin proteasome system (UPS): one of the major eukaryotic mechanisms of intracellular protein degradation and protein turnover
Graphical Abstract
POST-TRANSLATIONAL MODIFICATIONS
Post-translational modifications (PTMs) are covalent additions of, or chemical modifications to, protein functional groups that occur after protein synthesis and are essential for normal cellular function.1,2 Proteins generally function as the work horses of biological macromolecules; however, not all necessary cellular functions can be easily achieved by the combination of the 20 canonical amino acids that make up most polypeptides. Despite the relatively low number of genes in a eukaryotic cell (ca. 105), the breadth of the proteome is estimated to be significantly higher (ca. 107).3,4 To expand the repertoire of proteins, nature relies on PTMs to diversify function and to provide regulation. These post-translational chemical modifications can produce a variety of changes to protein function by affecting diverse properties of the polypeptide, including stability, conformation, dynamics, binding propensity, and even activity.5 Unsurprisingly, the fidelity of these processes is crucial for normal cellular function, and errors in PTM pathways are linked to diseases in humans.6–8
This biological importance has spurred decades of research aimed at understanding the nature of PTMs, including the types of chemical modifications that occur, the enzymes that accomplish such modifications, and the biochemical ramifications of these modifications.2 Consequently, some of the best-known PTMs, such as acetylation,9,10 methylation,11 and phosphorylation12 have been well characterized, and many of their biological functions are known. For example, acetylation and methylation both play a role in epigenetics through histone modifications.13,14 Acetylated histones are associated with euchromatin, or relaxed chromatin, and expression of genes,14 while methylated histones are associated with gene silencing and condensed heterochromatin.13 Furthermore, both acetylation and methylation can affect the stability, localization, and even oligomerization of target polypeptides, dramatically changing function.2 As another example, phosphorylation is especially common in signaling cascades, where stimuli can trigger a series of modifications of various proteins, often leading to the activation of a group of responsive genes or affecting metabolic pathways through allosteric regulatory mechanisms.12 However, despite their importance and ubiquity, these well-characterized PTMs represent only a fraction of protein modifications known to occur in eukaryotic cells.1
Several PTMs remain understudied and enigmatic, and this work will focus on one such PTM known as arginylation. Catalyzed by an enzyme known as arginyl-tRNA transferase (ATE1), arginylation is beginning to emerge as a global controller of eukaryotic cellular function. However, much is unknown about the structure, function, and regulation of ATE1s, prohibiting the rational development of chemical therapeutics to target this pathway. This review summarizes what is known about this underappreciated PTM, with a focus from the perspective of structure and function. This work then frames future approaches that may be taken to break open our mechanistic understanding of this essential eukaryotic protein modification.
DISCOVERY OF ATE1
In the early 1960s, a new soluble enzyme system in Escherichia coli was discovered that incorporated amino acids into proteins in a manner distinct from protein synthesis, as it did not require ribosomal machinery.15 The authors of this discovery attributed the amino acid incorporation to an “amino acyl soluble ribonucleic acid transfer factor,” which was later termed aminoacyl-tRNA-protein transferase (aa-tRNA transferase). Further, it was shown that this prokaryotic, nonribosomal aatRNA transferase incorporated Leu and Phe nearly 10 times greater than other amino acids, with the majority of these amino acids being covalently attached to the amino terminus. We now know these proteins as L/F-transferases, bacterial enzymes that catalyze the nonribosomal peptide bond formation between the N-termini of target peptides and the amino acids Leu and Phe. While these initial transferases were isolated from prokaryotes,15 aminoacyl-tRNA-protein transferases from eukaryotes were discovered around the same time.16,17
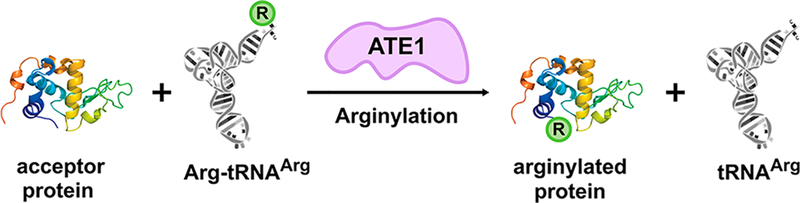
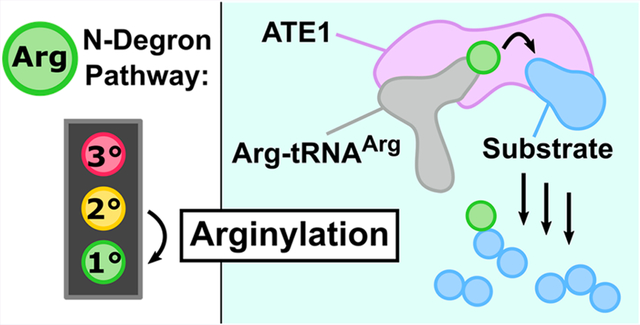
The first reports of a eukaryotic aminoacyl-tRNA-protein transferase were also described in the early 1960s. These aminoacyl-tRNA transferases were initially isolated from rat liver homogenates and sheep thyroid and were shown to catalyze the transfer of Arg from a charged tRNA to preassembled proteins.16–19 At the time, the function of the arginylation reaction was unknown but was thought to be linked to cellular stress and regeneration.20 Similar systems were subsequently identified in plants21 and other mammals.22 In the early 1990s, the gene encoding for this Arg-specific aminoacyl-tRNA transferase was identified in Saccharomyces cerevisiae, cloned, and characterized.23 It is now well established that the family of enzymes present in all eukaryotes24 that catalyze the energy-independent transfer of the amino acid Arg to a target polypeptide (Figure 1) are known as arginyl-tRNA transferases (also referred to as R-transferases or ATE1s). The importance of ATE1-mediated arginylation lies in its connection to the N-degron pathway (formally termed the N-end rule pathway), which links the ubiquitin-dependent degradation of a protein to the identity of its N-terminal amino acid residue, subsequently regulating a number of crucial biological pathways.25
Figure 1.
Cartoon depiction of ATE1-mediated post-translational arginylation. Arginyl-tRNA transferases (ATE1; lavender) catalyze the energy-independent transfer of the amino acid Arg (R, green circle) from an aminoacylated tRNA (gray) to an acceptor protein (rainbow colored from blue to red, N-terminus to C-terminus, respectively). The process of arginylation occurs most frequently at the N-terminus of a polypeptide, although more recent studies have shown that internal (midchain) arginylation is also possible.
THE ARG N-DEGRON PATHWAY AND ATE1
The proteasomal degradation of a cellular protein is frequently dependent on the identity of the terminal residues of the polypeptide.26,27 These signals of degradation rates are known as degrons, which often (but not always) exist at either the N- or C-termini and function as molecular flags for ubiquitin-dependent proteolysis.28 N-terminal degrons (N-degrons; the largest class of such degrons discovered so far) consist of three components: (1) an N-terminal (Nt-) destabilizing residue, which is an amino acid that, if present at the N-terminus of a protein, will eventually lead to degradation of said protein; (2) an internal lysine where polyubiquitination occurs; and (3) a sterically unstructured region to serve as the site of initiation for unfolding of the substrate.28 Currently, there are two major branches of the N-degron pathway (while other minor branches exist)29–31: the acetylation (Ac) N-degron pathway, which involves acetylation of N-terminal destabilizing residues as the determinant for degradation,32 and the arginylation (Arg) N-degron pathway, which involves arginylation as the degradation signal.33 N-terminal acetylation as a general protein modifier has been an historic focus by many research groups, whereas N-terminal arginylation and its related N-degron pathway is more poorly understood at the molecular level.
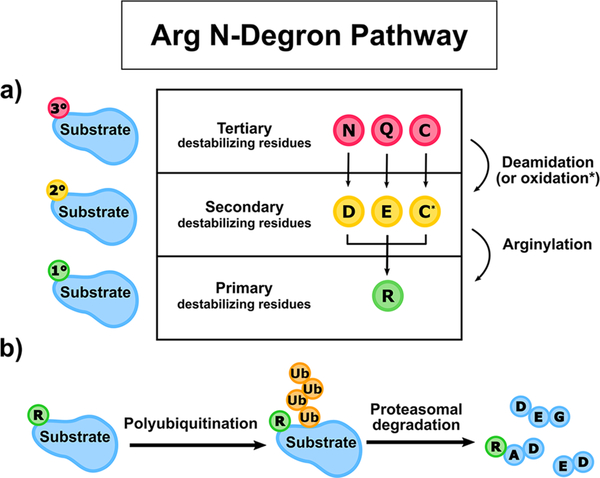
The Arg N-degron pathway is hierarchical and describes three strata of amino acids that, if present at the N-terminus of a protein, may be destabilizing residues (Figure 2).28,33 In this sense, destabilization does not mean creating thermodynamic instability of the polypeptide; rather, destabilizing describes a cellular context in which the polypeptide becomes shorter lived due to signals received by proteolytic machinery. These destabilizing residues may be exposed via a number of mechanisms, such as cleavage of nascent Nt-Met residues by Met-aminopeptidases,33 or by proteolysis catalyzed by calpains and caspases, both of which are cysteine proteases involved in the apoptosis signaling cascade.33,34 If the exposed residue is Asn, Gln, or Cys, it is known as a tertiary (3°) destabilizing residue and requires further processing before the target protein is ubiquitinated (Figure 2A). Asn and Gln may be enzymatically deamidated by N-terminal Asn amidohydrolase (NTAN)35 and N-terminal Gln amidohydrolase (NTAQ)36 to Asp and Glu, respectively, to become secondary (2°) destabilizing residues (Figure 2A). Cys, however, is not enzymatically processed but can be oxidized by nitric oxide (NO) or other oxidizing molecules to form either Cys-sulfinic acid (CysO2H) or Cys-sulfonic acid (CysO3H; Figure 2A).37 These oxidized Cys residues (C*) are also secondary (2°) destabilizing amino acids in most eukaryotes, although notably not in yeast. ATE1s recognize secondary destabilizing residues and condense Arg to the target polypeptide, resulting in the exposure of a primary (1°) destabilizing residue (Figure 2A).
Figure 2.
Cartoon schematic of the Arg N-degron pathway. (a) The hierarchical nature of the Arg N-degron pathway. Tertiary destabilizing residues (red circles) are first either enzymatically deamidated (N or Q) or oxidized (C) to become secondary destabilizing residues (yellow circles). Secondary destabilizing residues (D, E, and oxidized C, denoted C*) are directly recognized by ATE1s, which catalyze the process of arginylation, the nonribosomal conjugation of Arg to a target polypeptide, resulting in the transfer of a primary destabilizing residue (green circles). (b) Arginylation primes a protein substrate for proteolysis. After arginylation, the primary destabilizing residue R (green circle) is recognized by cellular N-recognins, E3 ligases of the ubiquitin-proteasome pathway that ubiquitinate (orange circles) proteins for subsequent proteasomal degradation.
Primary destabilizing residues are bound by N-recognins, which are individual enzymes or complexes that recognize N-degrons.38 N-recognins of the Arg N-degron pathway involve a number E3 ligases of the ubiquitin proteasome system (UPS). The general process of ubiquitination involves four enzymes, numbered E1–E4. The first step of ubiquitination is the ATP-dependent conjugation of ubiquitin to the activating enzyme E1, forming a reactive thioester bond, followed by the transfer of the ubiquitin to the conjugating enzyme, E2.39 Finally, the ligating enzyme E3 catalyzes ubiquitin transfer to the substrate at an internal Lys.39 Where polyubiquitination is required, E4 enzymes will aid in elongating ubiquitin chains. E3 enzymes are the substrate-determination part of the UPS and, as such, display the most diversity among the four classes of UPS enzymes.40
The E3 ligases that participate in the Arg N-degron pathway vary from organism to organism. For example, S. cerevisiae has two E3 ligases with roles in the Arg N-degron targeting complex: Ubr1, a Really Interesting New Gene (RING)-type E3 ligase, and Ufd4, a homologous to the E6AP carboxy-terminus (HECT)-type ligase.41 Ubr1 can recognize N-degrons on its own as well as in complex with Ufd4, but it is not understood how recognition differs between the two cases.28,41 There are at least four different E3 ligases in mammals that recognize Arg N-degrons: Ubr1, Ubr2, Ubr4, and Ubr5.28,42 The Ubr proteins are large, ranging in size from ≈200 kDa to ≈500 kDa, and there are currently no structures of any full-length Ubr enzymes.28,42 However, several of the key features of the Ubrs have been structurally characterized: the RING domain, which coordinates two Zn2+ ions to form a platform for binding to E2, and the UBR box, an ≈80 residue zinc-finger like domain that binds the primary destabilizing residue Arg (with Lys and His being bound to a much lesser extent).33,42,43 There are several structures of the UBR box deposited in the Protein Data Bank (PDB), many of them in complex with various N-degrons. Yet, while some of the Ubrs that participate in the Arg N-degron pathway have been structurally and mechanistically characterized, much less is known about the structure and mechanism of ATE1, which is the essential upstream enzyme that catalyzes arginylation, initiating the protein degradation cascade.
NONDEGRADATIVE ARGINYLATION
Due to the initial discovery of ATE1’s involvement in the N-degron pathway, much of the early arginylation work focused on ATE1’s connection to protein degradation. However, more recent work has demonstrated that arginylation is not always degradative; that is, the post- or cotranslational addition of Arg to a protein does not always result in its removal by the proteasome, and in some cases can even increase its metabolic stability.44,45 For example, in the case of β-amyloid, the main protein comprising plaques found in the brains of Alzheimer’s patients, arginylation seems to prevent its aggregation and its subsequent accumulation.46 Similarly, arginylation of the nonmuscle cytoskeletal protein β-actin, normally involved in cell morphology and cell migration, is essential to its polymerization.47,48 ATE1-null mouse fibroblasts display abrogated actin filament formation attributed to aggregated actin.47 The effect of arginylated β-actin has been further explored in the slime mold Dictyostelium discoideum, where ATE1 was shown to be enriched near lateral protrusions of lamellipodia.49 Knockouts of ATE1 in this organism also result in disrupted cell adhesion, emphasizing the importance of arginylation to normal β-actin function.49 Finally, arginylation may also serve to change a protein’s function. An example of this functional modification is the retrotranslocation of the ER-native protein calreticulin to the cytoplasm. In the cytoplasm, calreticulin is exposed to ATE1, and its arginylation induces dimerization, which is a key structural feature associating calreticulin with stress granules.50 It is not known what percentage of arginylation events are degradative versus nondegradative, but the evidence is clear that both play important roles in the essential functioning of eukaryotes. Given the increase in functional diversity provided by other post-translational modifications, arginylation likely has many undiscovered impacts on its protein substrates (vide infra).
ATE1 STRUCTURE
Genes and Isoforms.
ATE1s are encoded along at least one ate1 gene in eukaryotes, although some organisms may have more than a single ate1 gene, and varied splicing mechanisms appear to result in multiple different isoforms that may be translated.51 In some lower-order eukaryotes, such as yeast, only a single (iso)form is thought to exist encoded along a single gene.23 In contrast, while there is believed to be a single (iso)form of Drosophila melanogaster ATE1,52 it was postulated that A. thaliana has two genes encoding for ATE1.53 Initially, it was unknown whether both were functional, but a more recent study has shown that both AtATE1 genes are operative and may be redundant.54 For human ATE1, the gene has been mapped to chromosome 10, and at least two52 to as many as five isoforms are predicted, although unverified at the protein level. Thus, some organisms encode for as few as a single (iso)form of ATE1, while other organisms may encode for several isoforms of ATE1, all of which could have differential activity and be targeted to specific tissues and/or organelles.
Of the studied mammalian ATE1s, previous research has focused chiefly on the Mus musculus (mouse) arginyl-tRNA transferase isoforms. Initially, at least two distinct M. musculus ATE1 mRNAs were detected from a single gene.55 Further work demonstrated that the single M. musculus ate1 gene encodes at least four, and possibly up to six, splicing-derived isoforms under the control of a promoter that is also bidirectional, regulating in the opposite direction the expression of the mouse protein Dfa (divergent from ATE1), a repressor of TATA-box promoter sequences.24,51,56 There are 12 exons in the M. musculus ate1 gene: exons 1 and 7 may each be processed differently resulting in translation into distinct ATE1 isoforms, while exons 2–6 and 8–12 are present in the mRNA encoding for all mouse ATE1 isoforms. The difference in the isoforms is due to the presence of either exon 1A or exon 1B, and exon 7A, exon 7B, or both. Reflecting these differences, the four major isoforms have been designated as ATE1–1 (ATE11B7A), ATE1–2 (ATE11B7B), ATE1–3 (ATE11A7A), and ATE1–4 (ATE11A7B) and minor isoforms ATE11A7A7B and ATE11B7A7B; most isoforms differ by a modest change of ca. 5 kDa once translated.51,57 However, further corroboration is necessary to verify the generality of the six ATE1 isoforms in other mammals.
Isoforms of M. musculus ATE1s have variable expression levels, tissue localization, and relative activities. Differential expression of ATE1s was initially suggested by observing differential mRNA levels (a proxy for its relative expression level) of at least two isoforms in tissues excised from skeletal muscle, spleen, liver, brain, and testis.55 Once all six M. musculus ATE1 isoforms were discovered, RT-PCR was used to show differing levels of each isoform’s mRNA across multiple mouse tissues. Four major isoforms were identified: ATE1–3 (ATE11A7A; prominent in muscle tissue), ATE1–1 (ATE11B7A; prominent in nearly every tissue except spleen and muscle), ATE1–4 (ATE11A7B; prominent in kidney, testis, and muscle), and ATE1–2 (ATE11B7B; prominent in brain, liver, and testis). These four isoforms showed the highest levels of arginylation activity based on the ability to target Asp-β-gal for degradation, with the following activity hierarchy: ATE1–3 (ATE11A7A) ≈ ATE1–1 (ATE11B7A) > ATE1–4 (ATE11A7B) ≈ ATE1–2 (ATE11B7B).51 In another study, it was reported that isoforms ATE1–3 (ATE11A7A) and ATE1–4 (ATE11A7B) were unable to arginylate substrates with N-terminal Asp and Glu residues but instead specifically arginylated proteins with N-terminal Cys residues;24 however, subsequent work has shown that ATE1–3 (ATE11A7A) is not Cys-specific,58 and thus Cys specificity remains open to debate. Intriguingly, the combined 7A7B isoforms are minor (expressed in lower quantities) and have much lower activity than the four isoforms containing only one copy of exon 7, but the molecular reasoning for this lowered activity is unknown.51 It is possible that differential expression, localization, and activity are achieved by the implementation of various ATE1 isoforms in other higher-order eukaryotes.
Structure.
While there currently exists no structure of an ATE1, insight can be garnered through comparison to other enzymes (chiefly prokaryotic) that are functionally similar. Despite a lack of sequence homology, ATE1 is predicted to have some structural homology to its prokaryotic counterparts, the L/F-transferases, as well as the FemABX enzymes, which are bacterial aminoacyl-tRNA peptide transferases involved in methicillin resistance.59 Both the L/F-transferase and FemABX enzymes belong to the extremely diverse Gcn5-related N-acetyl transferase (GNAT) superfamily of enzymes, which commonly acetylate a variety of different substrates using acetyl-coenzyme A (CoA) as a cofactor.60–62 Acetyl-CoA binding and acetyl transferase activity occur within a tertiary structural element known as the GNAT fold, which typically comprises a six-stranded β-sheet of mixed polarity surrounded by four α-helices of mixed orientations.62,63 Reflecting its function, the GNAT fold contains two binding sites for moieties of its acetyl-CoA cofactor (the pantothenate arm and the pyrophosphate), the latter site of which has a signature Q/RxxGxG motif.62 Proteins containing this signature fold are part of the larger GNAT superfamily, although many GNAT-containing enzymes use a different cofactor.
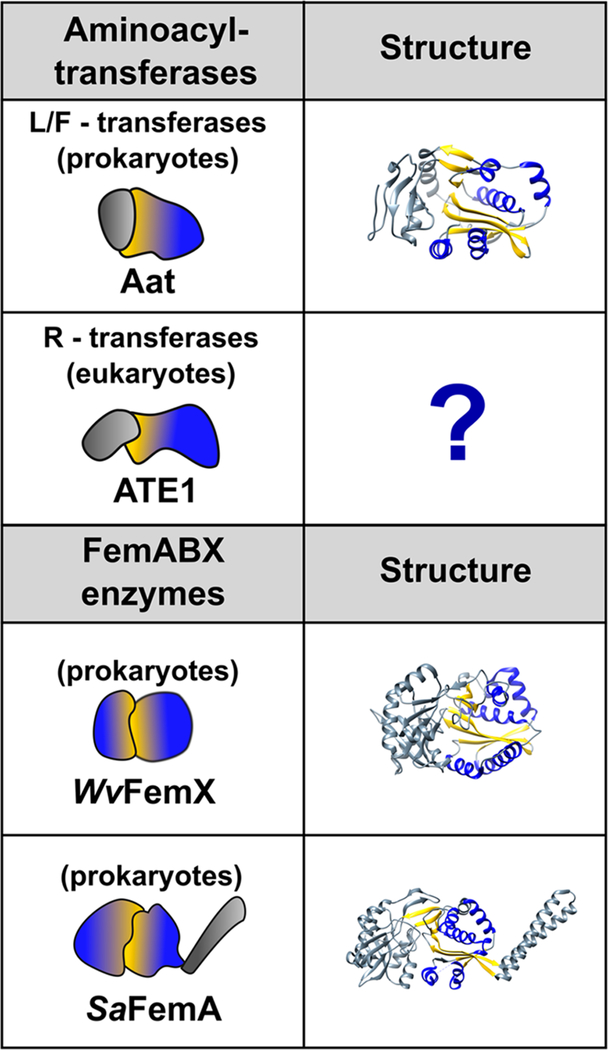
Within the GNAT superfamily, the GNAT fold of the FemABX enzymes and of the aminoacyl-tRNA transferases have both evolved to utilize aminoacylated tRNAs as cofactors rather than acetyl-CoA.61 Somewhat similarly to ATE1, the bacterial L/F-transferases transfer Leu or Phe from aminoacylated tRNAs (Leu-tRNALeu or Phe-tRNAPhe) to proteins bearing N-terminal Arg or Lys to mark for degradation. In contrast, FemABX enzymes are involved in the synthesis of interchain peptides of the peptidoglycan layer coating bacterial cell walls.61,64 The sugar components of the peptidoglycan, N-acetylglucosamine and N-acetylmuramic acid, are bound to linear pentapeptide chains that are subsequently cross-linked to each other via an interchain peptide, typically consisting of a chain of five Gly residues.64,65 The different Fem enzymes, FemA, FemB, and FemX, are aminoacyl-tRNA transferases that transfer Gly or Ala from aminoacylated tRNAs to synthesize the peptidoglycan interchain peptide.66 Representative structures of both the FemABX enzymes and the L/F transferases have been determined (Figure 3), providing insight into the mechanism, which may be similar to the ATE1 arginylation mechanism.
Figure 3.
GNAT folds of the FemABX enzymes and the aminoacyl tRNA transferases. The left-hand column depicts a cartoon representation of each protein, with domains that contain the GNAT fold colored with a blue-yellow gradient. The right-hand column contains ribbon depictions of the three-dimensional structures (if known). The α-helices and β-strands comprising the GNAT fold are colored blue and yellow, respectively. For clarity, only one of the GNAT folds is colored in the FemABX structures. PDB IDs are as follows: L/F transferase (2DPS); Weissella viridescens FemX (1P4N); and Staphylococcus aureus FemA (1LRZ). Notable is the lack of any three-dimensional structure of an ATE1 (R-transferase).
Of the FemABX enzymes, the X-ray crystal structures of Staphylococcus aureus (Sa) FemA and Weissella viridescens (Wv) FemX have been solved (Figure 3). FemX catalyzes the addition of the first residue of the peptidoglycan interpeptide chain, while FemA incorporates the second and third Gly residues to the peptidoglycan interpeptide chain.66 Structurally, both FemX and FemA comprise two nonidentical domains that each contain a GNAT fold, with the major difference being an additional coiled-coil domain in FemA (Figure 3).60,66 The coiled-coil domain has been suggested to orient and to position the tRNA 3′ acceptor end through contacts with the variable arm and TψC loop; however, this domain must not be essential for catalysis, as it is not present in FemX.65 The canonical substrate of FemX, UDP-MurNAc-pentapeptide, binds at the interface of domains 1 and 2, maintaining contacts mainly through domain 1.66 While no structure has been solved of FemX complexed with tRNA, docking studies suggest a potential binding site along a channel that runs through domain 2.66 Apo FemA, which has not been crystallized in the presence of any substrates, also contains a long L-shaped channel for putative tRNA binding within domain 2.65
Several X-ray crystal structures of the L/F-transferase have also been solved, in complex with various analogs of the tRNA 3′-end as well as a small product peptide.60,67 Like the FemABX enzymes, the L/F-transferase is also a bilobed protein; however, the GNAT fold of L/F-transferases is restricted only to the C-terminal domain (Figure 3). The first structure of an L/F-transferase with puromycin—an adenosine aminonucleoside representing an analog of the tRNA acceptor end—reveals puromycin binding in a hydrophobic pocket formed by the interface between the N- and C-terminal domains.60 Subsequent crystal structures of the transferase with Phe esterified to the 3′-hydroxyl of adenosine (rA-Phe) as well as one with a product peptide reveal binding in the same C-shaped hydrophobic pocket, although a ternary complex structure has yet to be determined.68 L/F-transferases have the highest selectivity for Leu-tRNALeu, and a preference for a single isoacceptor has been suggested based on the identification of key recognition nucleotides, although the structural basis for this recognition remains to be determined.69,70
Intriguingly, despite completely different sequence evolution, ATE1s are predicted to bear a GNAT fold as well. A 2006 NCBI database search with sensitive sequence comparison methods found that ATE1 was evolutionarily related to the L/F transferases and the Fem family of enzymes.61 The GNAT fold was suggested to have the highest sequence homology to the C-terminal half of ATE1, and secondary structural predictions suggested the presence of a GNAT-like β-sheet sandwiched by four α-helices.61 Sequence conservation with the N-terminal region of ATE1 was complicated due to various isoforms arising from the exon 1A/1B alternative splicing, but this region was speculated to aid in substrate determination.61 Several protein variants were generated based on sequence similarity to the FemABX enzymes and the L/F-transferases, and differences in in vivo arginylation among these variants were observed based on a β-gal assay.61 However, without a more robust structural model, the interpretation of the amino acid substitution effects on ATE1-catalyzed activity is difficult to parse. Thus, the determination of a three-dimensional structure of any ATE1 would be a major advancement in this field.
ATE1 MECHANISM
N-Terminal Arginylation.
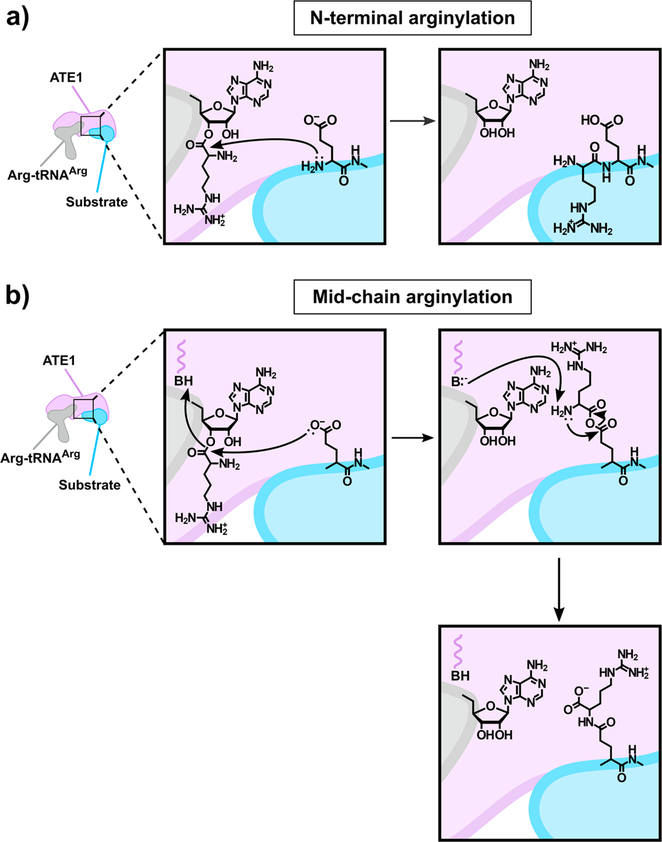
While the precise mechanism of ATE1-catalyzed arginylation is currently unknown, experiments on L/F transferases have given insight into a potentially analogous reaction pathway. The first major revelation into the L/F-transferase mechanism came from the crystal structures of the enzyme complexed with rA-Phe, which mimics the acceptor end of the tRNA, and complexed with a product peptide, the α-casein fragment H2N-RYLGYL-COOH.68 Superposition of the two structures revealed local changes in residue movement that were used to deduce a relay mechanism. Specifically, H-bonding interactions between Asp186 and Gln188 facilitate recognition of the positively charged N-terminal Arg guanidinium of the L/F substrate and promote the nucleophilic attack of the 3′-acyl group by the amino terminus. Tautomerization of the Gln side chain facilitates proton movement, resulting in the collapse of a tetrahedral intermediate, and release of the free 3′-OH.68 This inferred mechanism of peptide-bond formation uses protein-based chemistry, distinct from the canonically accepted ribosomal peptide bond formation that is chiefly RNA-driven.68 A similar ATE1-catalyzed mechanism as suggested by structural interrogation of the L/F transferases is shown in Figure 4A. It is known that the free amino terminus and the free side-chain carboxylate are both necessary for ATE1-catalyzed N-terminal arginylation, as blockage of either moiety inhibits arginylation, which occurs via the formation of a conventional peptide bond;45 however, further mechanistic details are unknown.
Figure 4.
Cartoon depiction of the proposed mechanism of ATE1-catalyzed arginylation. (a) The canonical ATE1-catalyzed N-terminal arginylation reaction involves the transfer of Arg from the aminoacylated Arg-tRNAArg to the N terminus of the acceptor substrate to form a peptide bond in a nonribosomal manner. The inferred mechanism for this process involves nucleophilic attack of the 3′-acyl group by the N-terminal amino group, with subsequent release of the unmodified tRNAArg and the arginylated substrate. (b) An alternative ATE1-catalyzed reaction involves the transfer of Arg from the aminoacylated Arg-tRNAArg to the internal Asp/Glu side chain of the acceptor substrate to form an isopeptide bond in a nonribosomal manner. The postulated mechanism for this process involves the attack of the Arg-tRNAArg 3′-acyl group by the Asp/Glu carboxylate side chain, proton transfer at an active-site catalytic acid, release of the 3′-OH of the tRNA, and the formation of an unstable anhydride-like intermediate. The active-site conjugate base could then deprotonate the Arg α amine, attacking the anhydride carbonyl internally, releasing the Arg α carboxylate, and forming the isopeptide bond. In both panels, ATE1 is represented as the lavender shape, tRNAArg is represented as the gray shape, and the acceptor substrate protein is represented as the blue shape.
Interestingly, subsequent activity assays with variants of the L/F-transferase mutated at the inferred key catalytic residues revealed that Asp186 and Gln188 appear to participate more significantly in the binding and the positioning of the substrate rather than nucleophilic relay, suggesting an alternative reaction pathway. Using quantitative MALDI, the authors were able to show that the L/F-transferase-catalyzed product was still observed, albeit at a much lower amount, when either Asp186 or Gln188 was altered, suggesting that the role of these amino acids is to position the substrate.71 Instead of a protein-based mechanism, the authors posited an RNA-based catalytic mechanism in which the 2′-OH of the tRNA performs a substrate-assisted catalytic role.71 The suggested mechanism, while also unverified, is more reminiscent of ribosome-catalyzed peptide bond formation.72 Application of a similar approach as has been taken with the L/F-transferases would significantly help to deduce the canonical mechanism of ATE1-catalyzed N-terminal arginylation and resolve whether a protein-based or RNA-based mechanism is more likely to be operative.
N-Terminal Recognition.
While it is widely accepted that ATE1 arginylates specifically at N-terminal Asp, Glu, and oxidized Cys residues, the exact consensus behind recognition of substrate is not well understood. Studies identifying proteins arginylated in mouse cells revealed that, as expected, arginylation was highly dependent on identity of the first residue of the polypeptide, but these experiments also found low specificity around the second residue, suggesting the possibility of a secondary or tertiary recognition element.57,73 There are also a few instances of arginylation occurring at amino acid residues other than Asp, Glu, and oxidized Cys, though it is unclear how common this occurrence is in vivo.58,73 Overall, while there are investigations into specificity of ATE1 and its recognition elements, more work is needed to identify the exact rules governing recognition elements that control arginylation.
Midchain Arginylation.
The 2005 revelation that ATE1 could catalyze arginylation at side chain carboxylates (Asp and Glu residues) within the interior of the polypeptide, a process which was subsequently termed midchain arginylation, remains both intriguing and puzzling.74 This first report of midchain arginylation used high-resolution mass spectrometry to demonstrate that neurotensin—a 13-residue peptide with multiple functional implications, including a link to several brain diseases such as schizophrenia, Huntington’s, and Parkinson’s—could be arginylated by forming an isopeptide bond with the oligopeptide Glu4 side chain. In follow-up studies, it was demonstrated that midchain arginylation did indeed take place in vivo, and dozens of other key cellular proteins such as β-actin, hemoglobin, tubulin, myosin, and several GPCR-associated proteins etc. were found to be midchain arginylated.45,73 More recently, a report has demonstrated that midchain arginylation could also occur on α-synuclein, another major protein linked to neurodegenerative disease.75 These results underscore the broad importance and potentially massive intracellular substrate pool of this alternative ATE1 catalytic pathway.
Despite this significance, the mechanism of midchain arginylation remains to be determined. Undoubtedly, a means for midchain arginylation distinct from N-terminal arginylation must exist, and a suggested mechanism has been proposed (Figure 4B).45 In this postulated scenario, the Asp/Glu side chain carboxylate would attack the 3′-acyl group of the Arg-tRNAArg. Facilitated by proton transfer at an active-site catalytic acid, the 3′-OH of the tRNA would be released, and an unstable anhydride-like intermediate would be formed. The active-site conjugate base could then deprotonate the Arg α amine, which would attack the anhydride carbonyl internally, releasing the Arg α carboxylate and forming the isopeptide bond. This mechanism is highly speculative and entirely protein-based rather than RNA-based, but there is some precedent based on other enzymatic acyltransfer reactions.45 Intriguingly, it has been found that some ATE1 isoforms have higher efficiency toward N-terminal arginylation, while others are more efficient at midchain arginylation.45,57,73 These observations raise the exciting possibility that different ATE1 isoforms may be expressed to control the type of arginylation (N-terminal or midchain) in a cellular-specific context. However, more work is necessary to understand the mechanism driving this reaction and to corroborate the observed differences in activity between the various isoforms.
BIOLOGICAL CONSEQUENCES AND BIOCHEMICAL TARGETS
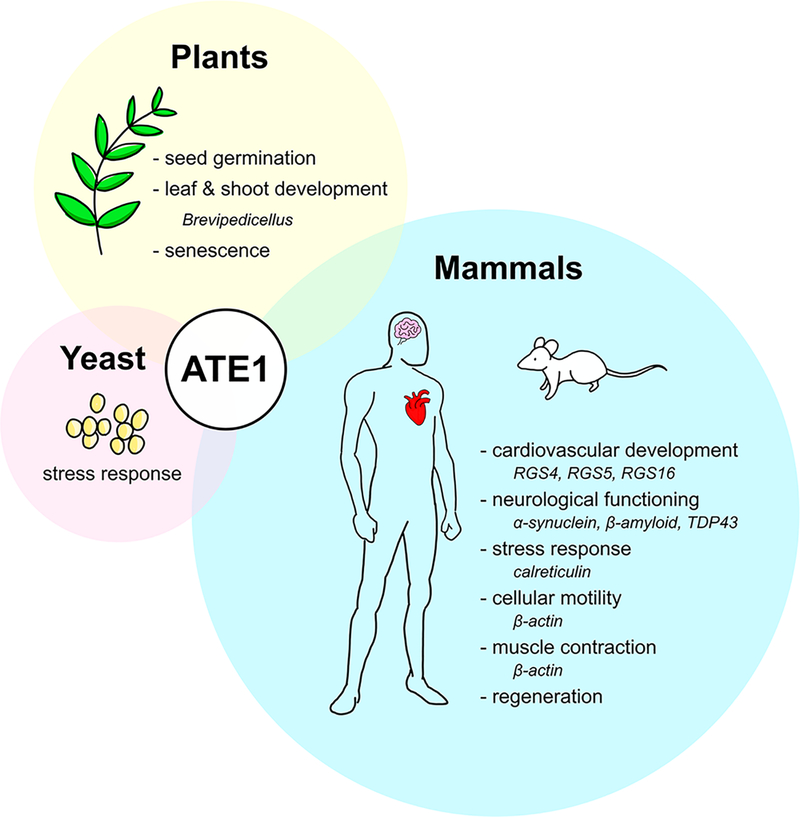
Despite unknowns regarding the structure and the mechanism of this nonribosomal peptide bond formation, multiple lines of evidence clearly demonstrate that ATE1-catalyzed arginylation has numerous biological consequences across the eukaryotic domain. While many of these effects are mediated by protein degradation via the UPS, there are also examples of arginylation affecting a protein’s function or localization, rather than acting as a degradation signal. Consequently, researchers have begun to compile the “arginylome,” a compendium of all arginylation targets within an organism.73 However, for a number of those proteins, it is not known how arginylation affects its fate. Moreover, for some biological processes affected by ATE1, the exact protein target(s) of arginylation remain(s) unknown. While much is still to be uncovered about the precise molecular details of these arginylated proteins, it is becoming increasingly evident that arginylation is a global regulator of most complex organisms, and the biological consequences and biochemical targets of arginylation are synopsized briefly below (Figure 5).
Figure 5.
ATE1 regulates a variety of biological processes across all eukaryotic organisms. In yeast (pink circle), ATE1 has a clear link to environmental stress. In plants (yellow circle), the lack of ATE1 affects germination, development, and senescence. In mammals (blue circle), the fidelity of post-translational arginylation is linked to general cellular homeostasis, with studies having demonstrated ATE1’s involvement in cardiovascular development, neurological functioning, the stress response, motility, muscle contraction, and cellular regeneration. Proteins that have been shown, or are surmised, to be arginylated by ATE1 are indicated in italics.
Yeast Development.
In 1983, researchers isolated a mutant S. cerevisiae strain defective in arginylation activity.76 Surprisingly, the ate1 null mutant did not display a phenotype differing greatly from wildtype, and this ate1 null strain was still viable.23 However, when ate1 null yeasts were exposed to environmental pressures such as oxidative stress, heavy metals, high salt and high temperature, the yeast stress response was disrupted.77 These results show that, in yeast, the loss of arginylation is not lethal, unlike in higher eukaryotes (vide infra), but the fidelity of arginylation is necessary for an adequate stress response in this organism (Figure 5).
Plant Growth Cycle.
In contrast to yeast, ATE1-catalyzed arginylation mediates effects throughout the entire growth cycle of plants (Figure 5). The first description of ATE1 function in plants was a report in 2002 in which researchers found a delayed leaf senescence mutant (dls1) of A. thaliana. In this work, the authors demonstrated that a lack of AtATE1 transcripts, caused by an insertion in intron 4 of the ATE1 gene, was linked to the observed phenotype in dls1 mutants. The manifestation of this phenotype gave rise to three indicators of senescence (age-related deterioration): a decrease in chlorophyll content, lowered soluble protein content, and a deficiency of photosystem II. Strikingly, all three phenotypes were rescued by transient expression of ATE1.53 Double mutants of A. thaliana lacking both arginine transferase genes displayed a variety of gross outward changes to the plant, including leaf deformities, loss of apical dominance, altered phyllotaxis (leaf arrangement), and internode elongation. These major deformities led researchers to conclude that both arginine transferase genes play an important role in normal leaf and shoot development.54 Additionally, ATE1s in plants have been shown to promote seed germination.78 Taken together, these observations underscore that ATE1 is a global regulator of plants and their development, which is a feature of ATE1 that is also echoed in mammals.
Mammalian Cardiovascular Development and Embryogenesis.
The importance of ATE1-catalyzed arginylation in mammalian cellular homeostasis and development has been tested using a number of murine models. One of the first major insights into the importance of ATE1 in mammalian cardiovascular maturation was a study that showed heterozygous knockout mice of ATE1 displayed no marked difference in phenotype from wildtype, while homozygous ATE1 knockouts were embryonically lethal.79 Careful examination of the homozygous ATE1 knockout embryos uncovered multiple signs of aberrant cardiovascular development, such as hemorrhages, ventricular and atrial septal defects (holes in the heart tissue), hypoplasia (underdevelopment) of left and right ventricles, and disruption of angiogenic remodeling.79 These physiological consequences were connected to perturbations in arginylation of RGS4, RGS5, and RGS16, which are all negative regulators of G-proteins that are involved in cardiovascular growth and angiogenesis.79,80 These results emphasize the importance of ATE1-catalyzed arginylation and cardiovascular development of the embryo.
Since homozygous ATE1 knockout murine models died during gestation, additional models were developed to probe the disruptive effects of ATE1 loss post birth. In a conditional knockout mouse model, postnatal deletion of ATE1 produced many health issues, occasionally culminating in early death. These issues included significant decreases in body weight due to a loss of white adipose tissue, enlarged brains leading to behavioral abnormalities, increased susceptibility to seizures, rounding of the spine, and infertility.81 When the biological phenotypes of both the homozygous and conditional ATE1 knockout murine models are considered together, these studies confirm the importance of arginylation and the Arg N-degron pathway in supporting global cellular homeostasis in mammals.
Mammalian Neurodegeneration.
Arginylation plays a critical role in maintaining the normal physiological functioning of significant contributors to neurodegeneration such as α-synuclein (α-syn), β-amyloid, and TDP43 proteolytic fragments.82 Thought to act as a chaperone in the controlled release of neurotransmitters during synaptic transmission, α-syn is a protein that is localized to presynaptic neurons.82,83 When dysregulated, its aggregation and formation of Lewy bodies in the brain is believed to be a contributing factor to development of Parkinson’s disease.75,83 Arginylation of α-syn occurs at two midchain Glu residues, and observations suggest that this PTM assists in the regular folding of the protein. Consistent with this hypothesis, when α-syn is not arginylated, its early misfolding then leads to its abnormal accumulation, contributing to the development of Parkinson’s disease. A similar example of arginylation stabilizing a protein is evident in β-amyloid, a cleavage product of the amyloid precursor protein, which is widely believed to cause Alzheimer’s disease by forming aggregates in the brain known as plaques. β-amyloid has an ordered β-strand in its hydrophobic C-terminal domain, while its hydrophilic N-terminal domain can adopt multiple conformations depending on its chemical environment. It was shown that β-amyloid was able to be arginylated in vitro, and it was speculated that the arginylated β-amyloid preferentially adopted the soluble α-helical conformation due to the addition of the charged Arg amino acid.46 Aggregation of the TAR DNA-binding protein (TDP43) has implications in amyotrophic lateral sclerosis (ALS) and a number of other neuropathies. Distinct from the stabilizing mechanisms seen with α-syn and β-amyloid, arginylation of TDP43 protein fragments seems to promote its removal by the UPS, preventing plaque formation.84
Links to Cellular Stress and Cancer in Mammals.
Cancer is a complicated group of diseases that can arise through the dysregulation of many essential cellular processes, so it is perhaps unsurprising that arginylation is one of those cellular processes that can contribute to carcinogenesis. In 2016, ATE1’s link to cancer was established when its mRNA levels were demonstrated to be downregulated in certain types of kidney and colon cancers.85 Moreover, ATE1 knockdown in mouse fibroblasts drove prostate cancer to pro-metastatic phenotypes, such as contact independent growth, loss of cell–cell contacts, and an increase in chromosomal aberrations.85 Additionally, ATE1 levels have been shown to be inversely correlated with rates of metastasis.85 A molecular explanation of such a phenomenon lies in ATE1’s regulation of proteins involved with several of the hallmarks of cancer, such as evading apoptosis, insensitivity to antigrowth signals, and tissue invasion.77,86 These proteins regulated by ATE1 arginylation include calreticulin, which is arginylated and associated with stress granules in a Ca2+-dependent manner, and actin, which is important in the cell motility needed for metastasis.44,50,87 While unlikely to be a cancer panacea, arginylation could be another targetable pathway in synergy with existing chemotherapy drugs for a more efficient treatment option. Consistent with this notion, there are two studies that have suggested that targeting the Arg N-degron pathway can sensitize cancers to certain chemotherapeutics such as bortezomib and doxorubicin, both apoptosis-inducing drugs.88,89 Researchers discovered that arginylated calreticulin enhanced bortezomib’s effect on tumor cells and that siRNA knockdowns of the Arg N-degron pathway-specific ubiquitin ligases amplified the response of cancer cells to doxorubicin.88,89 When taken together, these studies indicate that the Arg N-degron pathway plays an essential role in apoptosis, a cellular process that is often disrupted in cancerous cells.
POTENTIAL REGULATORY MODES
As the Arg N-degron pathway is linked to a number of essential biological processes ranging from cardiovascular maturation to chromosomal segregation to the stress response, regulation of ATE1 function is likely critical. In the context of the intracellular milieu, ATE1 has the capacity to encounter many different biological macromolecules, including its target substrates, off-target polypeptides, aminoacylated-tRNAs of different kinds, and other potential binding partners. Discrimination among the cellular targets of arginylation is unlikely to be stochastic; rather, it is anticipated to be exquisitely controlled. In addition to regulation at the genetic level via alternative splicing (vida supra), ATE1 is also likely to be regulated by interactions with various macromolecules (including partner proteins), cofactors, and even small molecules. In this section, we outline what is currently known regarding regulatory mechanisms of ATE1s.
Nitric Oxide.
The signaling molecule nitric oxide (NO) regulates a variety of cellular functions through both covalent and noncovalent interactions with proteins or protein prosthetic groups.37 The covalent attachment of NO to the thiol/thiolate side chain of a Cys residue, a post-translational modification known as S-nitrosylation, yields protein S-nitrosothiols, which may differ from their non-nitrosylated counterparts in conformation, chemistry, or interaction capabilities.90 As discussed earlier, the role of NO in the Arg N-degron pathway is to transform Cys, a tertiary destabilizing residue in mammalian cells, to a secondary destabilizing residue Cys* (either Cys-sulfinic acid or Cys-sulfonic acid), to be recognized directly by ATE1. To our knowledge, however, this process does not occur directly through S-nitrosylation but rather oxidation via NO, which is likely to be recapitulated in the presence of other oxidants. In this way, the Arg N-degron pathway acts as a sensor of NO and, potentially, oxidative stress, which would necessitate the degradation of oxidized (and potentially misfolded) polypeptides.37 There may even exist a higher-level cellular connection to this effector molecule, as it is well-known that NO is derived from catabolism of Arg via nitric oxide synthase (NOS).91 A delicate balance of the intracellular levels of free Arg may be linked to the formation of Arg-tRNAArg, its consumption by protein translation levels, and its consumption by the Arg N-degron pathway in protein degradation. Further work is necessary to decipher the extent of these connections.
Heme.
A 2008 study proposed that the Arg N-degron pathway is a sensor of iron protoporphyrin IX (heme b).92 Heme is one of the most versatile cofactors found in biology, with roles ranging from catalysis to electron transport to small molecule sensing.93,94 Spectroscopic analysis of heme titrated in vitro into purified mouse ATE11B7A suggested a protein–heme interaction, and arginylation assays demonstrated inhibition of mouse and yeast ATE1 in a heme-dependent manner.92 This inhibition was linked to the oxidation of two adjacent Cys residues to form a strained disulfide, which could be partially rescued by the addition of β-mercaptoethanol (a disulfide reducing agent). Heme was also found to interact with yeast and mouse UBR1, one of the downstream ubiquitin ligases of the Arg N-degron pathway. The study concluded that the Arg N-degron pathway was therefore a sensor of labile intracellular heme.
One intriguing aspect of this finding was the observation that heme interacted with the side chain of a Cys residue (Cys411 of mouse ATE11B7A). Cys-bound hemoproteins often come in two classes: those in which the heme is a catalytic site and those in which the heme is a sensory site.93 A catalytic function is wholly unnecessary here, whereas the sensory heme-Cys (thiolate) proteins often interact with and sense small gaseous molecules, such as carbon monoxide (CO), NO, and hydrogen sulfide (H2S). Given that the Arg N-degron pathway is a known global sensor of NO,37 it is tempting to speculate that heme in ATE1 is meant to sense directly cellular levels of NO. It is possible that the in vitro effects observed by oxidized heme to create a Cys–Cys disulfide may be spurious, as it is generally accepted that free heme does not exist in the cell under homeostatic conditions. More work on heme-bound ATE1 would aid in understanding the role of this cofactor in ATE1 function.
Protein Binding Partners.
In addition to being regulated by small molecules and cofactors, ATE1 and the Arg N-degron pathway may be regulated by protein–protein interactions. The first observation of a major ATE1 interacting partner was a 2014 study where the identification of a previously uncharacterized mouse protein that bound to ATE1, but was not an arginylation target, was reported.95 Termed Liat1 (ligand of ATE1), this small protein (228 residues) was demonstrated to interact with ATE1 in yeast-based two-hybrid (Y2H) and coimmunoprecipitation (co-IP) assays. Liat1 was shown to modestly increase (ca. 2-fold) the N-terminal arginylation efficacy of the four major mouse ATE1 isoforms using recombinant mouse dihydrofolate reductase with Asp or Cys at the N-terminus as the acceptor substrate,95 but the molecular reasoning behind this increase is unknown. Subsequently, a 2016 study used mass spectrometry to expand the knowledge of interacting partners of Physocomitrella patens (moss) ATE1.96 Using two immuno-affinity strategies, the authors of this study identified numerous new arginylation targets, as well as a small heat shock protein (sHP17.2a) that was found to coimmunoprecipitate with ATE1 but was not a target of arginylation. In vivo Förster resonance energy transfer (FRET) studies confirmed the protein–protein interaction, which is suggested to occur during the normal development of moss.96 While the function of this interaction is unknown, it was speculated that sHP17.2a may aid in the folding of ATE1 or in arginylation activity.96 The authors did not observe a Liat1-ATE1 interaction, as Liat1 appears to be absent from all plants and fungi, such as P. patens.
ATE1 binding partners are not limited to single proteins at one time but may include protein complexes. A very recent report demonstrated that five human enzymes of the Arg N-degron pathway (NTAN1, NTAQ1, UBR1/UBR2, and ATE1) form a targeting complex.97 Through a series of orthogonal Y2H and co-IP assays, pairwise interactions of the Nt-amidases, E3 ligases, and ATE1 were shown to be mutual, and several interactions were recapitulated using GST pulldowns of recombinant proteins.97 A similar complex was shown to form using the analogous S. cerevisiae enzymes.97 Surprisingly, the authors also ruled out that human cytosolic arginine synthetase bound to ATE1.97 The authors rationalized these interactions through a concept termed superchanneling, in which Arg N-degron substrates bearing Asn/Gln as tertiary destabilizing residues would be processed more efficiently by direct handoff from Nt-amidase (converting tertiary to secondary destabilizing residues via deamidation) to ATE1 (converting secondary to primary destabilizing residues via arginylation) to Ub ligase (to ubiquitinate the substrate polypeptide), bypassing dissociation and relocation throughout the cell. This postulate is both very logical and parsimonious, and analogous complexes has been observed for other metabolic processes.98,99
tRNA Levels.
Finally, regulation of ATE1 may be accomplished by interaction with either intact or fragmented tRNA. As demonstrated, tRNAArg and its five isoacceptors function as the Arg donor for ATE1-catalyzed arginylation, and depletion of either tRNAArg, or the requisite Arg tRNA synthetase, has a negative impact on arginylation efficacy. However, this biomolecule is not the only Arg donor that exists in the cell. A very recent study has shown that tRNA-derived fragments (tRFs) may also function as Arg donors for protein arginylation.100 tRFs are typically small, noncoding RNAs with several biological roles, and the authors explored the ability of ATE1 to use these fragmented RNAs for arginylation. Using mass spectrometry, it was shown that the Arg-charged stem-like fragments could function as Arg donors to arginylate angiotensin II.100 Intriguingly, the deletion of ATE1 decreased the cellular abundance of tRFArgs, suggesting a potential interaction between the machinery that creates tRFs and ATE1.100 The structural basis for the interaction between ATE1 and any tRNA molecule, intact or fragmented, remains unknown.
OUTLOOK
In this work, we have outlined the current research on the essential functions and roles of ATE1, as well as potential modes of action based on its relation to other homologues, but many questions remain to be answered. Major missing pieces to the ATE1 puzzle include both its tertiary and oligomeric structures. Determination of an ATE1 three-dimensional structure would provide clues to its interactions with the aminoacyl-tRNA as well as the diverse pool of substrate proteins that are arginylated. A structure would also provide clues as to the mechanism of arginylation, and whether or not the mechanism resembles a protein-based peptide bond formation or an RNA-based peptide bond formation. Furthermore, with ATE1 essential to so many different biological processes, it is imperative that we understand how this enzyme is regulated, what cofactors are necessary to its function, and whether binding partners can direct its specificity. These are important avenues of future research that will shed light onto an underappreciated but essential PTM.
ACKNOWLEDGMENTS
This work was supported by NIH-NIGMS grant R35 GM133497 (A.T.S.).
ABBREVIATIONS
- ATE1
arginyl-tRNA transferase
- α-syn
α-synuclein
- co-IP
coimmunoprecipitation
- dls1
delayed leaf senescence mutant
- GNAT
Gcn5-related N-acetyl transferase
- HECT
homologous to the E6AP carboxy-terminus
- Liat1
ligand of ATE1
- NO
nitric oxide
- Nt
N-terminal
- NTAN
N-terminal Asn amidohydrolase
- NTAQ
N-terminal Gln amidohydrolase
- PTM
post-translational modification
- rA-Phe
phenylalanyl adenosine
- RING
really interesting new gene
- TDP43
TAR DNA-binding protein
- tRNA
transfer ribonucleic acid
- Ub
ubiquitin
- UPS
ubiquitin proteasome system
- Y2H
yeast-based two hybrid
Footnotes
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acschembio.0c00677
REFERENCES
- (1).Mann M, and Jensen ON (2003) Proteomic analysis of posttranslational modifications. Nat. Biotechnol 21, 255–261. [DOI] [PubMed] [Google Scholar]
- (2).Varland S, Osberg C, and Arnesen T (2015) N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities, and biological effects. Proteomics 15, 2385–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Khoury GA, Baliban RC, and Floudas CA (2011) Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep 1, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Spoel SH (2018) Orchestrating the proteome with posttranslational modifications. J. Exp. Bot 69, 4499–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Deribe YL, Pawson T, and Dikic I (2010) Post-translational modifications in signal integration. Nat. Struct. Mol. Biol 17, 666–672. [DOI] [PubMed] [Google Scholar]
- (6).Wang Y-C, Peterson SE, and Loring JF (2014) Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 24, 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Xu H, Wang Y, Lin S, Deng W, Peng D, Cui Q, and Xue Y (2018) PTMD: A database of human disease-associated posttranslational modifications. Genomics, Proteomics Bioinf 16, 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Santos AL, and Lindner AB (2017) Protein posttranslational modifications: roles in aging and age-related disease. Oxid. Med. Cell. Longevity 2017, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Christensen DG, Baumgartner JT, Xie X, Jew KM, Basisty N, Schilling B, Kuhn ML, and Wolfe AJ (2019) Mechanisms, detection, and relevance of protein acetylation in prokaryotes. mBio 10, e02708–e02718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ree R, Varland S, and Arnesen T (2018) Spotlight on protein N-terminal acetylation. Exp. Mol. Med 50, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Paik WK, Paik DC, and Kim S (2007) Historical review: the field of protein methylation. Trends Biochem. Sci 32, 146–152. [DOI] [PubMed] [Google Scholar]
- (12).Humphrey SJ, James DE, and Mann M (2015) Protein phosphorylation: a major switch mechanism for metabolic regulation. Trends Endocrinol. Metab 26, 676–687. [DOI] [PubMed] [Google Scholar]
- (13).Michalak EM, Burr ML, Bannister AJ, and Dawson MA (2019) The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol 20, 573–589. [DOI] [PubMed] [Google Scholar]
- (14).Eberharter A, and Becker PB (2002) Histone acetylation: a switch between repressive and permissive chromatin. EMBO Rep. 3, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Momose K, and Kaji A (1966) Soluble amino acid-incorporating system III. Further studies on the product and its relation to the ribosomal system for incorporation. J. Biol. Chem 241, 3294–3307. [PubMed] [Google Scholar]
- (16).Kaji H, Novelli GD, and Kaji A (1963) A soluble amino acid-incorporating system from rat liver. Biochim. Biophys. Acta, Spec. Sect. Nucleic Acids Relat. Subj 76, 474–477. [PubMed] [Google Scholar]
- (17).Kaji H (1968) Further studies on the soluble amino acid incorporating system from rat liver. Biochemistry 7, 3844–3850. [DOI] [PubMed] [Google Scholar]
- (18).Soffer RL (1968) The arginine transfer reaction. Biochim. Biophys. Acta, Nucleic Acids Protein Synth 155, 228–240. [DOI] [PubMed] [Google Scholar]
- (19).Soffer RL, and Mendelsohn N (1966) Incorporation of arginine by a soluble system from sheep thyroid. Biochem. Biophys. Res. Commun 23, 252–258. [DOI] [PubMed] [Google Scholar]
- (20).Chakraborty G, and Ingoglia NA (1993) N-terminal arginylation and ubquitin-mediated proteolysis in nerve regeneration. Brain Res. Bull 30, 439–445. [DOI] [PubMed] [Google Scholar]
- (21).Manahan CO, and App AA (1973) An arginyl-transfer ribonucleic acid protein transferase from cereal embryos. Plant Physiol. 52, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lock RA, Harding HWJ, and Rogers GE (1976) Arginine transferase activity in homogenates from guinea-pig hair follicles. J. Invest. Dermatol 67, 582–586. [DOI] [PubMed] [Google Scholar]
- (23).Balzi E, Choder M, Chen WN, Varshavsky A, and Goffeau A (1990) Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J. Biol. Chem 265, 7464–7471. [PubMed] [Google Scholar]
- (24).Rai R, and Kashina A (2005) Identification of mammalian arginyltransferases that modify a specific subset of protein substrates. Proc. Natl. Acad. Sci. U. S. A 102, 10123–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bachmair A, Finley D, and Varshavsky A (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186. [DOI] [PubMed] [Google Scholar]
- (26).Bachmair A, and Varshavsky A (1989) The degradation signal in a short-lived protein. Cell 56, 1019–1032. [DOI] [PubMed] [Google Scholar]
- (27).Varshavsky A (1996) The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. U. S. A. 93, 12142–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Varshavsky A (2019) N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. U. S. A. 116, 358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kim J-M, Seok O-H, Ju S, Heo J-E, Yeom J, Kim D-S, Yoo J-Y, Varshavsky A, Lee C, and Hwang C-S (2018) Formylmethionine as an N-degron of a eukaryotic N-end rule pathway. Science 362, eaat0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Dong C, Chen S-J, Melnykov A, Weirich S, Sun K, Jeltsch A, Varshavsky A, and Min J (2020) Recognition of nonproline N-terminal residues by the Pro/N-degron pathway. Proc. Natl. Acad. Sci. U. S. A 117, 14158–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Timms RT, Zhang Z, Rhee DY, Harper JW, Koren I, and Elledge SJ (2019) A glycine-specific N-degron pathway mediates the quality control of protein N-myristoylation. Science 365, eaaw4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Hwang C-S, Shemorry A, and Varshavsky A (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Varshavsky A (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Harwood SM, Yaqoob MM, and Allen DA (2005) Caspase and calpain function in cell death: bridging the gap between apoptosis and necrosis. Ann. Clin. Biochem 42, 415–431. [DOI] [PubMed] [Google Scholar]
- (35).Cantor JR, Stone EM, and Georgiou G (2011) Expression and biochemical characterization of the human enzyme N-terminal asparagine amidohydrolase (hNTAN1). Biochemistry 50, 3025–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Park MS, Bitto E, Kim KR, Bingman CA, Miller MD, Kim H-J, Han BW, and Phillips GN Jr. (2014) Crystal structure of human protein N-terminal glutamine amidohydrolase, an initial component of the N-end rule pathway. PLoS One 9, e111142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hu R-G, Sheng J, Qi X, Xu Z, Takahashi TT, and Varshavsky A (2005) The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986. [DOI] [PubMed] [Google Scholar]
- (38).Sriram SM, and Kwon YT (2010) The molecular principles of N-end rule recognition. Nat. Struct. Mol. Biol 17, 1164–1165. [DOI] [PubMed] [Google Scholar]
- (39).Lilienbaum A (2013) Relationship between the proteasomal system and autophagy. Int. J. Biochem. Mol. Biol 4, 1–26. [PMC free article] [PubMed] [Google Scholar]
- (40).Dikic I, and Robertson M (2012) Ubiquitin ligases and beyond. BMC Biol. 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hwang C-S, Shemorry A, Auerbach D, and Varshavsky A (2010) The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat. Cell Biol 12, 1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, and Kwon YT (2005) A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol 25, 7120–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Metzger MB, Pruneda JN, Klevit RE, and Weissman AM (2014) RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta, Mol. Cell Res 1843, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Kashina A (2014) Protein arginylation, a global biological regulator that targets actin cytoskeleton and the muscle. Anat. Rec 297, 1630–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Wang J, Han X, Wong CCL, Cheng H, Aslanian A, Xu T, Leavis P, Roder H, Hedstrom L, Yates JR, and Kashina A (2014) Arginyltransferase ATE1 Catalyzes Midchain Arginylation of Proteins at Side Chain Carboxylates In Vivo. Chem. Biol 21, 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Bongiovanni G, Fidelio GD, Barra HS, and Hallak ME (1995) The post-translational incorporation of arginine into a beta-amyloid peptide increases the probability of alpha-helix formation. NeuroReport 7, 326–328. [PubMed] [Google Scholar]
- (47).Karakozova M, Kozak M, Wong CCL, Bailey AO, Yates JR III, Mogilner A, Zebroski H, and Kashina A (2006) Arginylation of β-actin regulates actin cytoskeleton and cell motility. Science 313, 192–196. [DOI] [PubMed] [Google Scholar]
- (48).Saha S, Mundia MM, Zhang F, Demers RM, Korobova F, Svitkina T, Perieteanu AA, Dawson JF, and Kashina A (2010) Arginylation regulates intracellular actin polymer level by modulating actin properties and binding of capping and severing proteins. Mol. Biol. Cell 21, 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Batsios P, Ishikawa-Ankerhold HC, Roth H, Schleicher M, Wong CCL, and Müller-Taubenberger A (2019) ATE1-mediated posttranslational arginylation affects substrate adhesion and cell migration in Dictostelium discoideum. Mol. Biol. Cell 30, 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Carpio MA, Decca MB, Lopez Sambrooks C, Durand ES, Montich GG, and Hallak ME (2013) Calreticulin-dimerization induced by post-translational arginylation is critical for stress granules scaffolding. Int. J. Biochem. Cell Biol 45, 1223–1235. [DOI] [PubMed] [Google Scholar]
- (51).Hu R-G, Brower CS, Wang H, Davydov IV, Sheng J, Zhou J, Kwon YT, and Varshavsky A (2006) Arginyltransferase, Its Specificity, Putative Substrates, Bidirectional Promoter, and Splicing-derived Isoforms. J. Biol. Chem 281, 32559–32573. [DOI] [PubMed] [Google Scholar]
- (52).Kwon YT, Reiss Y, Fried VA, Hershko A, Yoon JK, Gonda DK, Sangan P, Copeland NG, Jenkins NA, and Varshavsky A (1998) The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc. Natl. Acad. Sci. U. S. A 95, 7898–7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Yoshida S, Ito M, Callis J, Nishida I, and Watanabe A (2002) A delayed leaf senescence mutant is defective in arginyl-tRNA: protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 32, 129–137. [DOI] [PubMed] [Google Scholar]
- (54).Graciet E, Walter F, Ó’Maoiléidigh DS, Pollmann S, Meyerowitz EM, Varshavsky A, and Wellmer F (2009) The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc. Natl. Acad. Sci. U. S. A 106, 13618–13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Kwon YT, Kashina AS, and Varshavsky A (1999) Alternative Splicing Results in Differential Expression, Activity, and Localization of the Two Forms of Arginyl-tRNA-Protein Transferase, a Component of the N-End Rule Pathway. Mol. Cell. Biol 19, 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Brower CS, Veiga L, Jones RH, and Varshavsky A (2010) Mouse Dfa is a repressor of TATA-box promoters and interacts with the Abt1 activator of basal transcription. J. Biol. Chem 285, 17218–17234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Wang J, Han X, Saha S, Xu T, Rai R, Zhang F, Wolf YI, Wolfson A, Yates JR, and Kashina A (2011) Arginyltransferase is an ATP-Independent Self-Regulating Enzyme that Forms Distinct Functional Complexes In Vivo. Chem. Biol 18, 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Wang J, Pejaver VR, Dann GP, Wolf MY, Kellis M, Huang Y, Garcia BA, Radivojac P, and Kashina A (2018) Target site specificity and in vivo complexity of the mammalian arginylome. Sci. Rep 8, 16177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Graciet E, Hu R-G, Piatkov K, Rhee JH, Schwarz EM, and Varshavsky A (2006) Aminoacyl-transferases and the N-end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen. Proc. Natl. Acad. Sci. U. S. A 103, 3078–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Suto K, Shimizu Y, Watanabe K, Ueda T, Fukai S, Nureki O, and Tomita K (2006) Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog. EMBO J. 25, 5942–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Rai R, Mushegian A, Makarova K, and Kashina A (2006) Molecular dissection of arginyltransferases guided by similarity to bacterial peptidoglycan synthases. EMBO Rep. 7, 800–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Jakubowski H (2017) Homocysteine Editing, Thioester Chemistry, Coenzyme A, and the Origin of Coded Peptide Synthesis. Life (Basel, Switz.) 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Vetting MW, de Carvalho S, L. P., Yu M, Hegde SS, Magnet S, Roderick SL, and Blanchard JS (2005) Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys 433, 212–226. [DOI] [PubMed] [Google Scholar]
- (64).Hegde SS, and Shrader TE (2001) FemABX family members are novel nonribosomal peptidyltransferases and important pathogen-specific drug targets. J. Biol. Chem 276, 6998–7003. [DOI] [PubMed] [Google Scholar]
- (65).Benson TE, Prince DB, Mutchler VT, Curry KA, Ho AM, Sarver RW, Hagadorn JC, Choi GH, and Garlick RL (2002) X-Ray Crystal Structure of Staphylococcus aureus FemA. Structure 10, 1107–1115. [DOI] [PubMed] [Google Scholar]
- (66).Biarrotte-Sorin S, Maillard AP, Delettré J, Sougakoff W, Arthur M, and Mayer C (2004) Crystal Structures of Weissella viridescens FemX and Its Complex with UDP-MurNAc-Pentapeptide: Insights into FemABX Family Substrates Recognition. Structure 12, 257–267. [DOI] [PubMed] [Google Scholar]
- (67).Dong X, Kato-Murayama M, Muramatsu T, Mori H, Shirouzu M, Bessho Y, and Yokoyama S (2007) The crystal structure of leucyl/phenylalanyl-tRNA-protein transferase from Escherichia coli. Protein Sci. 16, 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Watanabe K, Toh Y, Suto K, Shimizu Y, Oka N, Wada T, and Tomita K (2007) Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature 449, 867–872. [DOI] [PubMed] [Google Scholar]
- (69).Fung AWS, Payoe R, and Fahlman RP (2016) Perspectives and insights into the competition for aminoacyl-tRNAs between the translational machinery and for tRNA dependent nonribosomal peptide bond formation. Life (Basel, Switz.) 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Fung AWS, Leung CCY, and Fahlman RP (2014) The determination of tRNALeu recognition nucleotides for Escherichia coli L/F transferase. RNA 20, 1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Fung AW, Ebhardt HA, Abeysundara H, Moore J, Xu Z, and Fahlman RP (2011) An Alternative Mechanism for the Catalysis of Peptide Bond Formation by L/F Transferase: Substrate Binding and Orientation. J. Mol. Biol 409, 617–629. [DOI] [PubMed] [Google Scholar]
- (72).Trobro S, and Åqvist J (2005) Mechanism of peptide bond synthesis on the ribosome. Proc. Natl. Acad. Sci. U. S. A 102, 12395–12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Wong CCL, Xu T, Rai R, Bailey AO, Yates JR, Wolf YI, Zebroski H, and Kashina A (2007) Global Analysis of Posttranslational Protein Arginylation. PLoS Biol. 5, e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Eriste E, Norberg Å, Nepomuceno D, Kuei C, Kamme F, Tran D-T, Strupat K, Jörnvall H, Liu C, Lovenberg TW, and Sillard R (2005) A Novel Form of Neurotensin Post-translationally Modified by Arginylation. J. Biol. Chem 280, 35089–35097. [DOI] [PubMed] [Google Scholar]
- (75).Wang J, Han X, Leu NA, Sterling S, Kurosaka S, Fina M, Lee VM, Dong DW, Yates JR, and Kashina A (2017) Protein arginylation targets alpha synuclein, facilitates normal brain health, and prevents neurodegeneration. Sci. Rep 7, 11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Savage M, Soffer RL, and Leibowitz MJ (1983) A mutant of Saccharomyces cerevisiae defective in arginyl-tRNA-protein transferase. Curr. Genet 7, 285–288. [DOI] [PubMed] [Google Scholar]
- (77).Kumar A, Birnbaum MD, Patel DM, Morgan WM, Singh J, Barrientos A, and Zhang F (2016) Posttranslational arginylation enzyme Ate1 affects DNA mutagenesis by regulating stress response. Cell Death Dis. 7, e2378–e2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Holman TJ, Jones PD, Russell L, Medhurst A, Ubeda Tomas S, Talloji P, Marquez J, Schmuths H, Tung S-A, Taylor I, Footitt S, Bachmair A, Theodoulou FL, and Holdsworth MJ (2009) The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A 106, 4549–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Kwon YT, Kashina AS, Davydov IV, Hu R-G, An JY, Seo JW, Du F, and Varshavsky A (2002) An Essential Role of N-terminal Arginylation in Cardivascular Development. Science 297, 96–99. [DOI] [PubMed] [Google Scholar]
- (80).Lee MJ, Tasaki T, Moroi K, An JY, Kimura S, Davydov IV, and Kwon YT (2005) RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. U. S. A 102, 15030–15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Brower CS, and Varshavsky A (2009) Ablation of arginylation in the mouse N-end rule pathway: loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS One 4, e7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Galiano MR, Goitea VE, and Hallak ME (2016) Posttranslational protein arginylation in the normal nervous system and in neurodegeneration. J. Neurochem 138, 506–517. [DOI] [PubMed] [Google Scholar]
- (83).Stefanis L (2012) α-Synuclein in Parkinson’s Disease. Cold Spring Harbor Perspect. Med 2, a009399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Brower CS, Piatkov KI, and Varshavsky A (2013) Neurodegeneration-Associated Protein Fragments as Short-Lived Substrates of the N-End Rule Pathway. Mol. Cell 50, 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Rai R, Zhang F, Colavita K, Leu NA, Kurosaka S, Kumar A, Birnbaum MD, Győrffy B, Dong DW, Shtutman M, and Kashina A (2016) Arginyltransferase suppresses cell tumorigenic potential and inversely correlates with metastases in human cancers. Oncogene 35, 4058–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Colotta F, Allavena P, Sica A, Garlanda C, and Mantovani A (2009) Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30, 1073–1081. [DOI] [PubMed] [Google Scholar]
- (87).Birnbaum MD, Zhao N, Moorthy BT, Patel DM, Kryvenko ON, Heidman L, Kumar A, Morgan WM, Ban Y, Reis IM, Chen X, Gonzalgo ML, Jorda M, Burnstein KL, and Zhang F (2019) Reduced Arginyltransferase 1 is a driver and a potential prognostic indicator of prostate cancer metastasis. Oncogene 38, 838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Comba A, Bonnet LV, Goitea VE, Hallak ME, and Galiano MR (2019) Arginylated Calreticulin Increases Apoptotic Response Induced by Bortezomib in Glioma Cells. Mol. Neurobiol 56, 1653–1664. [DOI] [PubMed] [Google Scholar]
- (89).Leboeuf D, Abakumova T, Prikazchikova T, Rhym L, Anderson DG, Zatsepin TS, and Piatkov KI (2020) Downregulation of the Arg/N-degron Pathway Sensitizes Cancer Cells to Chemotherapy in Vivo. Mol. Ther 28, 1092–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Hess DT, Matsumoto A, Kim S-O, Marshall HE, and Stamler JS (2005) Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol 6, 150–166. [DOI] [PubMed] [Google Scholar]
- (91).Griffith OW, and Stuehr DJ (1995) Nitric oxide synthases: properties and catalytical mechanism. Annu. Rev. Physiol 57, 707–736. [DOI] [PubMed] [Google Scholar]
- (92).Hu R-G, Wang H, Xia Z, and Varshavsky A (2008) The N-end rule pathway is a sensor of heme. Proc. Natl. Acad. Sci. U. S. A 105, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Smith AT, Pazicni S, Marvin KA, Stevens DJ, Paulsen KM, and Burstyn JN (2015) Functional Divergence of Heme-Thiolate Proteins: A Classification Based on Spectroscopic Attributes. Chem. Rev 115, 2532–2558. [DOI] [PubMed] [Google Scholar]
- (94).Sono M, Roach MP, Coulter ED, and Dawson JH (1996) Heme-containing oxygenases. Chem. Rev 96, 2841–2887. [DOI] [PubMed] [Google Scholar]
- (95).Brower CS, Rosen CE, Jones RH, Wadas BC, Piatkov KI, and Varshavsky A (2014) Liat1, an arginyltransferase-binding protein whose evolution among primates involved changes in the numbers of its 10-residue repeats. Proc. Natl. Acad. Sci. U. S. A 111, E4936–E4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Hoernstein SNW, Mueller SJ, Fiedler K, Schuelke M, Vanselow JT, Schuessele C, Lang D, Nitschke R, Igloi GL, Schlosser A, and Reski R (2016) Identification of targets and interaction partners of arginyl-tRNA protein transferase in the moss Physcomitrella patens. Mol. Cell. Proteomics 15, 1808–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Oh J-H, Hyun J-Y, Chen S-J, and Varshavsky A (2020) Five enzymes of the Arg/N-degron pathway form a targeting complex: The concept of superchanneling. Proc. Natl. Acad. Sci. U. S. A 117, 10778–10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).An S, Kumar R, Sheets ED, and Benkovic SJ (2008) Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320, 103–106. [DOI] [PubMed] [Google Scholar]
- (99).Kohnhorst CL, Kyoung M, Jeon M, Schmitt DL, Kennedy EL, Ramirez J, Bracey SM, Luu BT, Russell SJ, and An S (2017) Identification of a multienzyme complex for glucose metabolism in living cells. J. Biol. Chem 292, 9191–9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Avcilar-Kucukgoze I, Gamper H, Polte C, Ignatova Z, Kraetzner R, Shtutman M, Hou Y-M, Dong DW, and Kashina A (2020) tRNAArg-Derived Fragments Can Serve as Arginine Donors for Protein Arginylation. Cell Chem. Biol 27, 839. [DOI] [PMC free article] [PubMed] [Google Scholar]