Abstract
Flaviviruses are a group of important emerging and re-emerging human pathogens that cause worldwide epidemics with thousands of deaths annually. Flaviviruses are small, enveloped, positive-sense, single-stranded RNA viruses that are obligate intracellular pathogens, relying heavily on host cell machinery for productive replication. Proteomic approaches have become an increasingly powerful tool to investigate the mechanisms by which viruses interact with host proteins and manipulate cellular processes to promote infection. Here, we review recent advances in employing quantitative proteomics techniques to improve our understanding of the complex interplay between flaviviruses and host cells. We describe new findings on our understanding of how flaviviruses impact protein-protein interactions, protein-RNA interactions, protein abundance, and post-translational modifications to modulate viral infection.
Keywords: Virus-host interactions, Flavivirus, Proteomics, Mass spectrometry, West Nile virus, dengue virus, Zika virus, Japanese encephalitis virus
Introduction
Flaviviruses are a large group of medically relevant viruses that cause significant disease in humans and animals. These include dengue virus (DENV), Japanese encephalitis virus (JEV), West Nile virus (WNV), and Zika virus (ZIKV) [1,2]. Flavivirus virions contain the structural proteins, an envelope as well as a positive-sense, single-stranded RNA genome of approximately 11kb in length. The incoming flavivirus genome encodes a single open reading frame that is translated on the endoplasmic reticulum (ER) into a single polyprotein. This polyprotein is subsequently cleaved into three structural proteins (capsid, prM and envelope) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) by viral and host proteases. The structural proteins form the viral particles, while the non-structural proteins are required for intracellular viral propagation and immune evasion [1,2].
Given their limited protein repertoire, flaviviruses rely on the host cell machinery for many steps in their life cycle. Identification of host proteins that are required for viral replication can inform the development of effective host-directed therapeutics and new innate antiviral mechanisms. Recent advances in mass spectrometry (MS)-based proteomics have increased the sensitivity and specificity and allow for the systematic identification and quantification of proteins in a high-throughput manner (Box 1) [3]. This technology is being widely used in many fields, and has been used to identify host proteins that are involved in viral replication including flaviviral infection (Figure 1). Here, we review how proteomic approaches have improved our understanding of flavivirus-host interactions, highlighting the mechanisms by which flaviviruses manipulate host cellular processes to promote infection.
Box1. Different types of proteomics approaches discussed in this review.
Affinity purification-mass spectrometry (AP-MS):
AP-MS is a powerful technique to identify binding partners of a target protein. Affinity-tagged protein of interest is purified as bait and its associated prey proteins are analyzed by mass spectrometry.
Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS):
ChIRP-MS is novel technique to study RNA-protein interactions. RNA-protein complexes are crosslinked and purified by biotinylated anti-sense oligonucleotides against RNA of interest. The RNA-associated proteins are subsequently identified by mass spectrometry.
RNA affinity chromatography combined with MS:
RNA affinity chromatography combined with MS is a widely used method to identify RNA-binding proteins that interact with specific RNA sequences. Tagged RNA (e.g., biotin tag, tobramycin and streptomycin aptamer tag) is used to isolate RNA-protein complexes and the bound proteins are eluted for mass spectrometry analysis.
Stable isotope labeling by amino acids in cell culture (SILAC)-based MS:
SILAC is a proteomic technique for quantification of proteins from different samples using non-radioactive isotopic labeling. Two cell populations are labeled with normal amino acids (light label) or isotopic modified amino acids (heavy label), respectively. Heavy and light cell lysates are combined and subjected to mass spectrometry analysis. The ratio of peak intensities in the mass spectrum corresponds to the protein abundance.
MS-based phosphoproteomics:
MS-based phosphoproteomics is a powerful tool for identification of proteins with phosphorylation as a post-translational modification. The protein samples are digested into peptides and phosphopeptides with trypsin. Phosphopeptides are enriched via the affinity of phosphate groups towards metals (e.g., Fe3+, Ti4+ ions) on carrier resins. The enriched phosphopeptides are then quantified by mass spectrometry.
MS-based proteomics for identification of ubiquitination sites:
Protein ubiquitination sites can be identified by MS through enrichment of the diglycine remnant that is generated following trypsin digestion of ubiquitinated proteins. The resultant ubiquitin-remnant-containing peptides can be enriched by a diglycine-specific antibody and the ubiquitination sites are identified by mass spectrometry. It is important to note that this method also enriches for the peptide remnants of proteins modified by Nedd8 and ISG15, which cannot be distinguished by this method.
Figure 1.
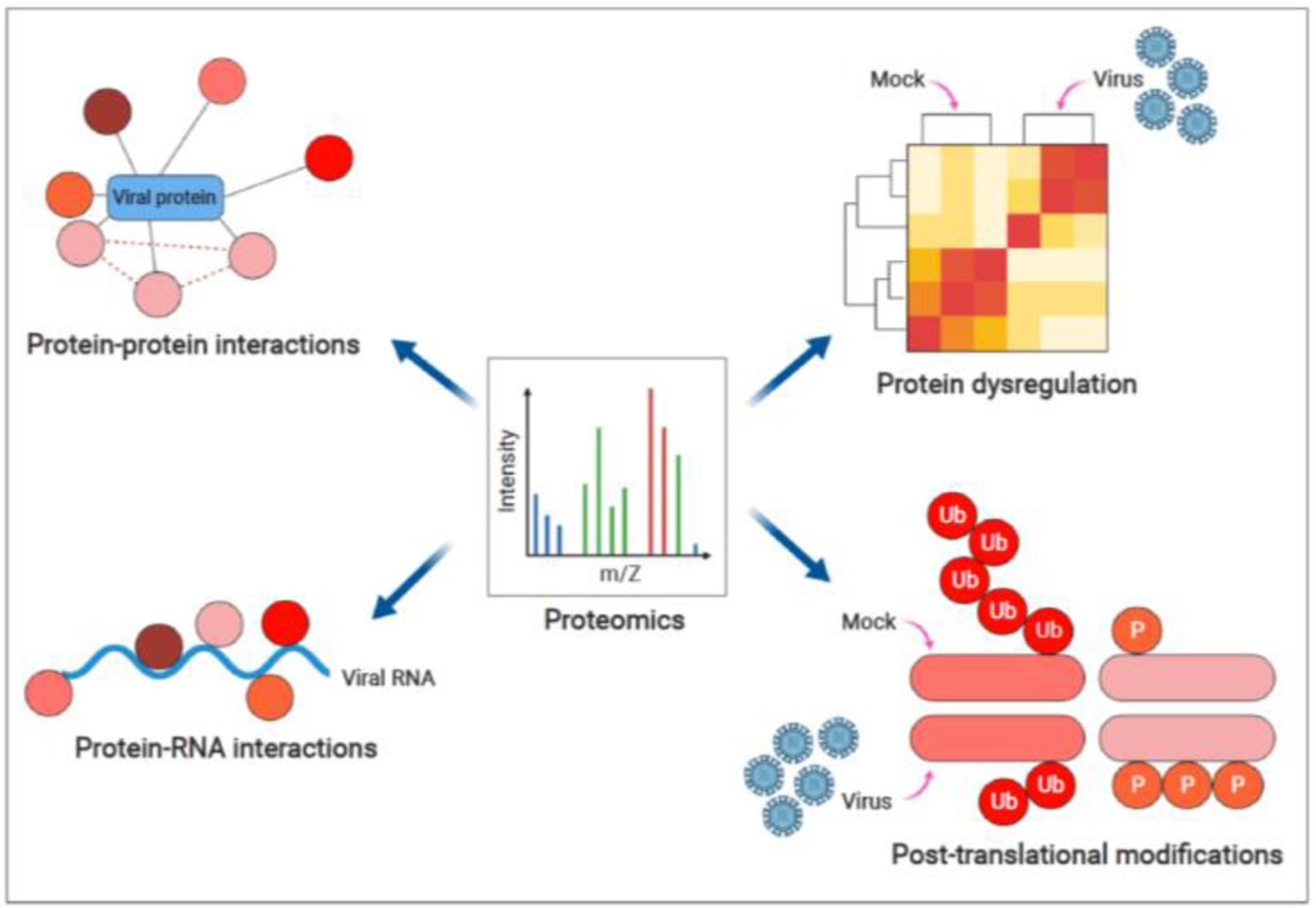
Applications of proteomics in investigating virus-host interactions (created with BioRender.com)
Mapping virus-host protein-protein interactions (PPIs)
Viruses hijack host machinery to ensure efficient viral replication. This is often achieved via physical interactions between viral and host proteins. By employing affinity purification-mass spectrometry (AP-MS) coupled with RNA interference (RNAi) screening, Li et al. generated WNV-host PPIs map and identified 26 virus-interacting host proteins that impact WNV infection [4••]. In particular, it was shown that WNV capsid interacts with PYM1, a host protein involved in the exon-junction complex (EJC) and nonsense-mediated decay (NMD) [5,6]. The interaction of WNV capsid diminishes the association of PYM1 with its canonical binding partners, the EJC proteins MAGOH and RBM8A. Further, the authors showed that viral RNA is targeted for degradation by NMD, and disrupting the interactions of PYM1 with EJC components can subvert this antiviral response [4••] (Figure 2). Interestingly, the role of NMD in flaviviral infection was further confirmed by the findings of Fontaine et al., which demonstrated that ZIKV infection is also restricted by NMD. Further, they showed that ZIKV capsid interacts with and downregulates the central NMD component, UPF1, to antagonize NMD restriction [7•] (Figure 2). Together, these papers suggest that NMD is an antiviral pathway that is subverted by flaviviruses via multiple mechanisms.
Figure 2.
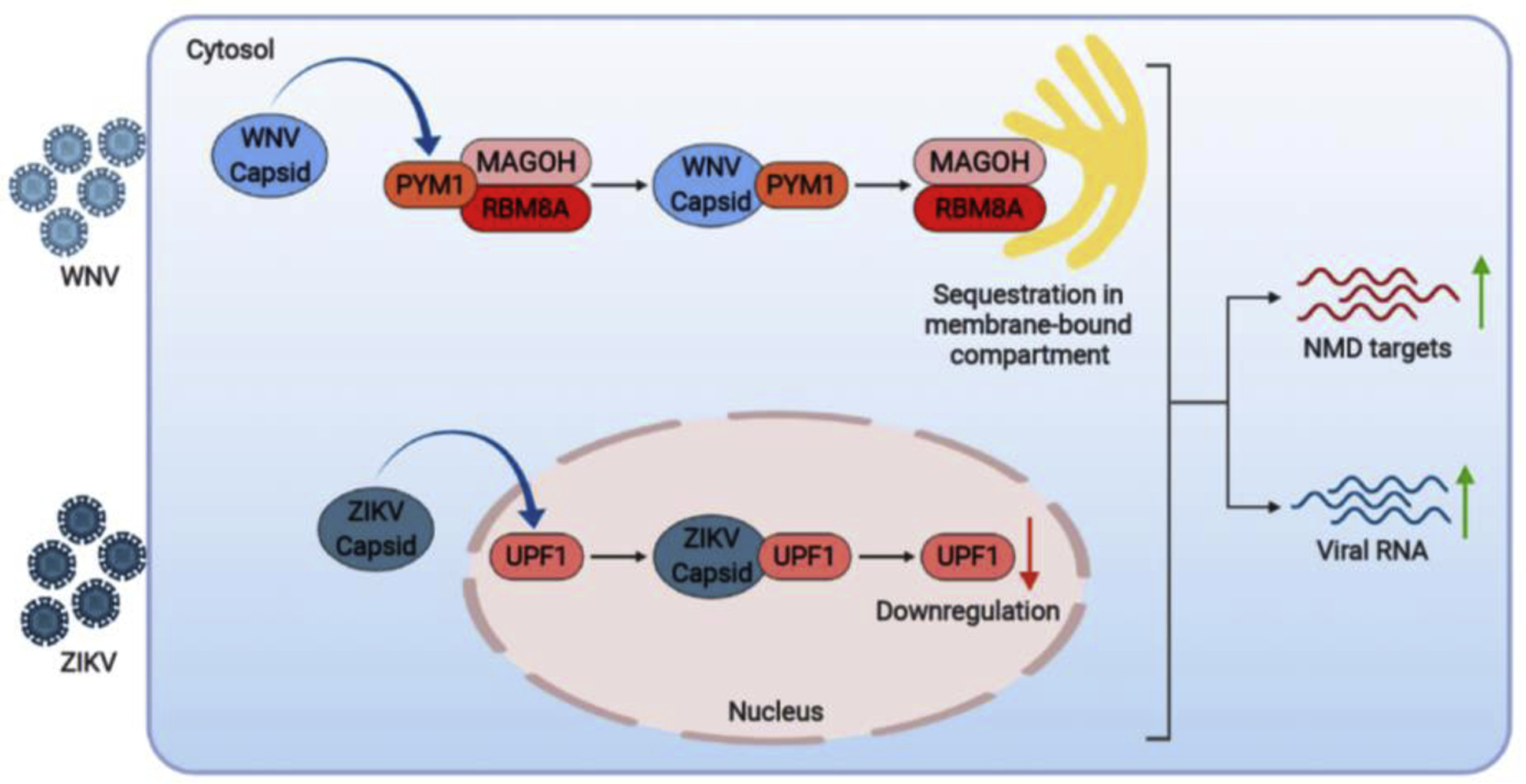
WNV and ZIKV antagonize host NMD-mediated restriction via protein-protein interactions. See text for further details. (created with BioRender.com)
To compare flavivirus-host PPIs, Shah et al. used AP-MS to create DENV-human and ZIKV-human PPIs network in HEK293T cells and found 28 shared human interactors [8••]. The authors suggest a role of flaviviral NS5 in counteracting host type I IFN response via interaction with the PAF1C complex, a chromatin-associated complex involved in transcriptional elongation [9]. Additionally, the authors generated a DENV-mosquito PPIs network using Aag2 mosquito cells to identify shared virus-host interactions and pathways between human and mosquito hosts. The authors found that ZIKV NS4A, but not DENV NS4A, impaired Drosophila brain development in an ANKLE2-dependent manner, suggesting a possible role for this interaction in ZIKV-induced microcephaly [8••].
Since ZIKV infection is associated with neurological defects during development, Scaturro et al. performed AP-MS to identify 386 ZIKV-interacting host proteins in neuronal cell lines and showed that ZIKV capsid and NS4B interact with host proteins related to neuronal development [10••]. Ectopic expression of NS4B alone or infection of differentiated neural progenitor cells (NPCs) with ZIKV led to the downregulation of host proteins related to neuronal differentiation. By using MS-based phosphoproteomics, the authors identified cellular factors whose phosphorylation status changes upon infection including cellular pathways previously associated with neurological diseases, such as Akt-mTOR signaling pathway [10••]. These findings are supported by another study showing that ZIKV NS4A and NS4B suppress Akt-mTOR pathway to inhibit neurogenesis of human fetal neural stem cells [11]. Collectively, these studies provide mechanistic insights on how ZIKV interacts with host Akt-mTOR pathway to induce neurological disorders.
Identifying the interactomes of individual viral proteins
While some groups have taken a broad approach to identify a comprehensive flavivirus-host interactome, others have defined the interactors of an individual viral protein critical for infection. The non-structural proteins of flaviviruses play diverse roles in viral replication and assembly and in antagonizing the host immune response, including type I IFN signaling and RNA interference (RNAi). The flaviviral non-structural protein, NS2A, was shown to suppress RNAi in both mammals and mosquitoes, highlighting the conserved role of flaviviral non-structural proteins in counteracting the innate immunity in different hosts [12]. Using a DENV replicon construct encoding an affinity-tagged NS1 protein but lacking the structural proteins, Hafirassou et al. performed AP-MS to identify DENV NS1-interacting host proteins and found an interaction and requirement for the oligosaccharyltransferase (OST) complex upon infection [13]. Depletion of components of the OST or pharmacological inhibition of this complex was shown to dampen NS1 glycosylation and inhibit DENV infection. The glycosylation deficient mutants of NS1 exhibited reduced stability, highlighting the importance of NS1 glycosylation during DENV infection [13]. The role of the OST complex in flavivirus infection was also confirmed in two CRISPR screens [14,15]. Two additional independent studies combined immunoprecipitation and MS to identify host proteins associated with DENV NS1 in human hepatic cell lines [16,17]. One of these studies found a large number of ribosomal proteins that interact with DENV NS1. These included the ribosomal protein RPL18, which was recruited to the perinuclear region to promote DENV infection [17].
While flavivirus NS4B is a part of the viral replication complex, its specific role in viral replication is largely unknown. Chatel-Chaix et al. showed that DENV NS4B interacts with mitochondrial proteins and induces mitochondrial elongation by inhibiting phosphorylation of the mitochondrial fission factor DRP1 at Ser616 and its subsequent translocation into mitochondria [18]. Furthermore, mitochondria elongation was shown to associate with decreased IFN response and increased viral titer, suggesting that DENV suppresses the host type I IFN response through the interaction between NS4B and mitochondria [18]. Peroxisomes are also associated with antiviral signaling [19]. Coyaud et al. found that ZIKV proteins perturb multiple host organelles, including ER and peroxisome [20]. This is further supported by the finding that WNV and DENV capsids bind to the peroxisome biogenesis regulator Pex19 to interfere with peroxisome biogenesis [21]. Whether this impacts peroxisome-dependent antiviral signaling is unknown.
Flavivirus NS5 is a multi-functional protein involved in viral RNA replication and host immune evasion. De Maio et al. took advantage of AP-MS technology to construct a DENV NS5-host PPIs map and identified a role of NS5 in the modulation of cellular splicing [22]. DENV NS5 has been shown to interact with spliceosome complex U5 snRNP to interfere with the splicing efficiency of host pre-mRNAs, including RIG-I. Interestingly, depletion of U5 snRNP, but not U2 snRNP components promotes DENV replication, suggesting that DENV manipulates specific splicing machinery to provide a favorable environment for infection [22]. Consistent with this finding, Poyomtip et al. used MS to identify a subset of splicing-related hnRNPs as major binding partners of DENV NS5 [23]. In addition to DENV NS5, hnRNPs were also found to associate with DENV RNA and enhance viral infection [24,25]. These studies suggest that DENV interacts with and manipulates host splicing machinery for efficient replication. The full spectrum of host genes whose alternative splicing is regulated by these flaviviral-host interactions and resultant outcomes related to viral pathogenesis remain unknown.
In addition to the non-structural proteins, several studies have utilized MS to identify the interactions between flavivirus structural proteins and host proteins. The cellular receptors required for flavivirus entry remain unclear. Studies found that domain III of DENV envelope interacts with human plasma proteins [26] and vimentin [27]. The transmembrane proteins PLVAP and GKN3 [28] as well as GRP78 [29] were shown to associate with JEV envelope and promote viral entry. MS was also used to identify host proteins that interact with flaviviral particles. For example, Annexin II and HSP70 were found to associate with DENV and ZIKV particles, respectively, to enhance viral infection [30,31].
Protein-protein interactions govern a wide range of cellular biological processes. As discussed above, MS-based proteomics is a robust tool to construct the virus-host interactomes and investigate the mechanisms by which viruses hijack or suppress host cellular pathways via protein-protein interactions.
Identifying host proteins that associate with viral RNA
By performing comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS), Ooi et al. identified 464 host proteins that interact with DENV or ZIKV genomic RNA [32••]. Complementary CRISPR and haploid genetic screens with multiple clinical isolates of DENV and ZIKV revealed overlap between these approaches. Indeed, a subset of ER-associated proteins that play an important role during viral infection including RRBP1 and vigilin, were shown to bind to the viral genomic RNA. Furthermore, RRBP1 and vigilin increase viral RNA stability, promote viral replication and protein translation [32••]. The ER is the major site for flavivirus replication. The critical roles of ER-associated proteins in flaviviral infection have been demonstrated by several independent studies [33–36].
The flaviviral 3’ UTR is critical for viral translation. Liao et al. applied RNA chromatography and MS to identify host proteins that bind to 3’ UTRs of DENV serotypes 1–4 [37]. The RNA binding protein Quaking (QKI) was shown to interact with DENV-4 3’ UTR and inhibit DENV-4 production, likely by suppressing viral translation. Interestingly, there is specificity as QKI does not interact with DENV-2 3’ UTR and has no impact on DENV-2 replication [37]. Ward et al. used the same strategy to identify ERI3, a Golgi-associated protein, as a binding partner of DENV-2 RNA. ERI3 interacts with the dumbbell structures within the DENV-2 3’ UTR and relocalizes to viral replication sites upon infection. Silencing of ERI3 reduced viral infection in both human cells and mosquitoes, indicating a conserved role of ERI3 in two hosts [38].
Subgenomic flaviviral RNA (sfRNA) is produced by the incomplete degradation of viral genomic RNA by the cellular 5’−3’ exoribonuclease XRN1, and has been shown to play an important role in counteracting the host immune response [39]. By using RNA affinity chromatography combined with MS, Soto-Acosta et al. identified that the fragile X mental retardation protein (FMRP) interacts with ZIKV sfRNA in human cells and mouse testes, and functions as an antiviral factor for ZIKV [40•]. Silencing of FMRP was found to increase viral translation and promote ZIKV infection. Expression of ZIKV sfRNA has been shown to upregulate endogenous FMRP targets, highlighting a role of ZIKV sfRNA in antagonism of FMRP function upon infection [40•]. Michalski et al. identified 28 host RNA-binding proteins that associate with sfRNAs of both DENV and ZIKV and several sfRNA interactors involved in RNA decay, RNA splicing and RNA editing were shown to inhibit ZIKV infection. Moreover, expression of ZIKV sfRNA was found to interfere with mRNA decay and RNA splicing pathways [41]. In summary, the development of novel RNA-centric techniques, such as ChIRP-MS, facilitates the use of proteomic approaches to further our understanding of how host machineries are engaged by viral RNA to promote viral infection.
Detecting host protein dysregulation upon viral infection
While we have a deep understanding of the changes in the gene expression landscape during infection, we have a much poorer understanding of how the proteome changes. Several groups have employed MS to define the changes in host protein abundance during flaviviral infection. Dong et al. used MS to quantify differential regulation of host proteins during DENV infection in 293T cells and found that DDX21 levels are decreased in infected cells. Upon infection, DDX21 translocates from the nucleus to the cytoplasm and inhibits DENV replication, likely by enhancing the type I IFN response. To overcome DDX21 restriction, DENV utilizes the NS2B/3 protease to degrade DDX21 [42]. Rabelo et al. employed MS to identify host proteins that are altered upon expression of DENV NS1, and they found a role for DENV NS1 in regulating the expression of host proteins related to apoptosis and cellular stress [43].
JEV infects neurons to cause pathogenesis. Mukherjee et al. identified 13 host proteins that are differentially regulated in JEV-infected human neural stem cells. GRP78, a protein involved in ER stress and apoptosis, was shown to be upregulated upon JEV infection and interact with viral RNA. Depletion of GRP78 led to the suppression of JEV infection and caspase 3 activation, suggesting a role of ER stress in JEV-induced apoptosis [44]. By performing stable isotope labeling by amino acids in cell culture (SILAC)-based MS in 293T cells, Tabata et al. identified several ESCRT proteins that are recruited to JEV replication sites on ER and play a role in viral particle formation [45]. These studies continue to highlight an important role of ER in flaviviral infection.
Most human flaviviruses are mosquito-transmitted, and Aedes aegypti and Aedes albopictus are major vectors for many viruses. Xin et al. utilized MS to identify 200 host proteins that are dysregulated in ZIKV-infected Aedes albopictus C6/36 cells. CHCHD2 was found to be upregulated in ZIKV-infected mosquito C6/36 and human HeLa cells. CHCHD2 has been shown to promote ZIKV infection and suppress virus-induced IFN-β production in human HeLa cells, indicating that ZIKV may antagonize host immune response by upregulation of CHCHD2 [46]. Flavivirus-induced suppression of the host immune response was also shown in mosquitoes. For example, the capsid proteins of yellow fever virus (YFV) and ZIKV were found to inhibit RNAi by suppression of Dicer processing in Aedes aegypti, suggesting a co-evolutionary arms race between virus and host [47]. Other studies demonstrated that ZIKV infection dysregulates host proteins involved in neuronal differentiation, IFN response, and energy production [48–53]. Flaviviruses can be also transmitted by ticks. In Langat virus (LGTV)-infected tick ISE6 cells, Grabowski et al. used MS to identify 68 host proteins that are upregulated upon infection. These proteins are associated with translation, amino acid metabolism, and protein folding/sorting/degradation pathways [54]. Further studies are required to understand the mechanisms by which these factors are controlled by flaviviral infection.
MS-based proteomic approaches can also be a valuable tool to identify host proteins that exhibit differential expression in virus-infected patients. Manchala et al. identified APO A-1 as an upregulated host protein in DENV patients co-infected with multiple DENV serotypes [55]. This virus-induced host protein dysregulation may associate with disease severity, serving as a fingerprint of disease progression. For example, serum levels of OLFM4, PF4, A2M, CFD and TPM4 are being developed as potential biomarkers to differentiate between two manifestations of DENV infection--dengue fever and dengue hemorrhagic fever [56,57].
Discovering post-translational modifications of host proteins in virus-infected cells
Post-translational modifications (PTMs) can regulate protein function, localization and stability. One strategy utilized by viruses to manipulate host proteins is to alter PTMs. Protein ubiquitylation is one of the most common PTMs that can play important roles in regulating protein stability and function. Following immunoprecipitation and MS analysis, Zhang et al. identified host proteins that are differentially ubiquitylated in DENV-infected cells [58•]. AUP1, a lipid droplet-localized type-III membrane protein, was shown to be deubiquitylated upon DENV infection. DENV-induced deubiquitylation increases AUP1 levels and leads to its accumulation in autophagosomes. AUP1 also interacts with DENV NS4A and promotes viral production. Ubiquitin-modified mutant of AUP1 was shown to suppress the AUP1-NS4A interaction, leading to defective lipophagy and impaired viral production [58•]. Further studies are needed to fully elucidate the ubiquitylation changes induced by flaviviral infection.
Phosphorylation is another well-characterized PTM. Ye et al. performed MS-based phosphoproteomics and identified 604 host proteins that are differentially phosphorylated in JEV-infected human U251 glial cells [59]. JEV infection activates JNK1 signaling and induces the phosphorylation of JNK1 substrates in human glial cells. JNK signaling is important as pharmacological inhibition of this pathway in mice led to decreased inflammatory cytokines secretion, reduced viral load and increased survival, indicating a critical role of JNK1 signaling in JEV-induced inflammation [59]. Using the same strategy, the host global phosphorylation profile was investigated for DENV [60], WNV [61] and ZIKV [10], showing the shared cellular pathways, whose phosphorylation status alters across different flaviviral infection, including RNA splicing and processing, and metabolic processes. These studies demonstrate how flaviviral infection perturbs host cellular pathways and provides initial mechanistic insights into virus-induced pathogenesis.
Concluding remarks
The interplay between viruses and their hosts is complex. To investigate such interactions, it is important to obtain a comprehensive view of both viral and host proteins during infection, and not just at the RNA level. MS-based proteomics approaches can be used to quantify protein abundance, interactions, and PTMs upon viral infection and provide researchers with opportunities to better understand the mechanisms by which viral infection alters the host cellular machinery.
AP-MS is a robust proteomics technique to investigate virus-host interactions. While AP-MS is widely used, a major challenge is that AP-MS can sometimes yield non-specific interactions, leading to false positives. Strategies such as tandem affinity purification-mass spectrometry (TAP-MS) and more sophisticated normalization algorithms are among the strategies employed to reduce the identification of false positives.
Many MS data sets have been generated by using transformed cell lines to analyze static protein-protein interactions in a non-physiological context. There is a growing interest in the field to increase our understanding of virus-host interactions in a more biologically relevant system, such as in the context of viral replication. The emergence of novel MS techniques such as cross-linking mass spectrometry (XL-MS) is a promising tool for identification of transient and dynamic virus-host interactions from different organelles and tissue samples. Moreover, the application of MS in proteomic studies of extracellular interactions that may occur between secreted viral proteins or virions and extracellular host factors will provide researchers the opportunity to investigate interactions that occur outside the host cell. With the development of new proteomics platforms, sample fractionation techniques, as well as bioinformatics tools, quantitative proteomics can greatly benefit both basic science and clinical research. This information can reveal the mechanisms by which viruses and host cells interact, informing the development of novel host-directed therapeutics.
Highlights.
Mass spectrometry-based proteomics is a powerful tool to investigate flavivirus-host interactions.
Affinity purification-mass spectrometry coupled with genetic screens can identify host proteins that physically interact with flaviviruses and impact viral replication.
Proteomics techniques can elucidate mechanisms of dysregulation and post-translational modifications of host proteins in flavivirus-infected cells.
Acknowledgements
This work was supported by grants from the National Institutes of Health to S.C. (5R01AI122749, 1R01AI140539, 1R01AI150246) and H.R. (1RO1AI143850). S.C. is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award. We apologize to all colleagues whose contributions were not cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared.
References
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Mukhopadhyay S, Kuhn RJ, Rossmann MG: A structural perspective of the flavivirus life cycle. Nature Reviews Microbiology 2005, 3:13–22. [DOI] [PubMed] [Google Scholar]
- 2.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R: Rewiring cellular networks by members of the Flaviviridae family. Nature Reviews Microbiology 2018, 16:125–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensimon A, Heck AJR, Aebersold R: Mass Spectrometry–Based Proteomics and Network Biology. Annual Review of Biochemistry 2012, 81:379–405. [DOI] [PubMed] [Google Scholar]
- 4.Li M, Johnson JR, Truong B, Kim G, Weinbren N, Dittmar M, Shah PS, Von Dollen J, Newton BW, Jang GM, et al. : Identification of antiviral roles for the exon–junction complex and nonsense-mediated decay in flaviviral infection. Nature Microbiology 2019, 4:985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This study generated WNV-host PPIs and demonstrated how WNV antagonizes EJC and NMD-mediated restriction.
- 5.Bono F, Gehring NH: Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biology 2011, 8:24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bono F, Ebert J, Unterholzner L, Güttler T, Izaurralde E, Conti E: Molecular insights into the interaction of PYM with the Mago–Y14 core of the exon junction complex. EMBO reports 2004, 5:304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontaine KA, Leon KE, Khalid MM, Tomar S, Jimenez-Morales D, Dunlap M, Kaye JA, Shah PS, Finkbeiner S, Krogan NJ, et al. : The Cellular NMD Pathway Restricts Zika Virus Infection and Is Targeted by the Viral Capsid Protein. mBio 2018, 9:e02126–02118. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study showed ZIKV capsid associates with and downregulates UPF1 to counteract antiviral activities of NMD.
- 8.Shah PS, Link N, Jang GM, Sharp PP, Zhu T, Swaney DL, Johnson JR, Von Dollen J, Ramage HR, Satkamp L, et al. : Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell 2018, 175:1931–1945.e1918. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This study compared virus-host PPIs in different hosts with DENV and ZIKV and highlighted a role of ZIKV NS4A and ANKLE2 interaction in microcephaly.
- 9.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. : The Paf1 Complex Is Required for Histone H3 Methylation by COMPASS and Dot1p: Linking Transcriptional Elongation to Histone Methylation. Molecular Cell 2003, 11:721–729. [DOI] [PubMed] [Google Scholar]
- 10.Scaturro P, Stukalov A, Haas DA, Cortese M, Draganova K, Płaszczyca A, Bartenschlager R, Götz M, Pichlmair A: An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 2018, 561:253–257. [DOI] [PubMed] [Google Scholar]; ••This study constructed ZIKV-host PPIs in neuronal cells and identified phosphorylation of host proteins upon ZIKV infection. Further investigation indicated that ZIKV infection causes dysregulation of cellular pathways related to neurological diseases.
- 11.Liang Q, Luo Z, Zeng J, Chen W, Foo S-S, Lee S-A, Ge J, Wang S, Goldman Steven A, Zlokovic Berislav V, et al. : Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell 2016, 19:663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu Y, Xu Y-P, Wang M, Miao M, Zhou H, Xu J, Kong J, Zheng D, Li R-T, Zhang R-R, et al. : Flavivirus induces and antagonizes antiviral RNA interference in both mammals and mosquitoes. Science Advances 2020, 6:eaax7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hafirassou ML, Meertens L, Umaña-Diaz C, Labeau A, Dejarnac O, Bonnet-Madin L, Kümmerer BM, Delaugerre C, Roingeard P, Vidalain P-O, et al. : A Global Interactome Map of the Dengue Virus NS1 Identifies Virus Restriction and Dependency Host Factors. Cell Reports 2017, 21:3900–3913. [DOI] [PubMed] [Google Scholar]
- 14.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, et al. : Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 2016, 535:159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin DL, Cherepanova NA, Bozzacco L, MacDonald MR, Gilmore R, Tai AW: Dengue Virus Hijacks a Noncanonical Oxidoreductase Function of a Cellular Oligosaccharyltransferase Complex. mBio 2017, 8:e00939–00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dechtawewat T, Paemanee A, Roytrakul S, Songprakhon P, Limjindaporn T, Yenchitsomanus P-t, Saitornuang S, Puttikhunt C, Kasinrerk W, Malasit P, et al. : Mass spectrometric analysis of host cell proteins interacting with dengue virus nonstructural protein 1 in dengue virus-infected HepG2 cells. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2016, 1864:1270–1280. [DOI] [PubMed] [Google Scholar]
- 17.Cervantes-Salazar M, Angel-Ambrocio AH, Soto-Acosta R, Bautista-Carbajal P, Hurtado-Monzon AM, Alcaraz-Estrada SL, Ludert JE, Del Angel RM: Dengue virus NS1 protein interacts with the ribosomal protein RPL18: This interaction is required for viral translation and replication in Huh-7 cells. Virology 2015, 484:113–126. [DOI] [PubMed] [Google Scholar]
- 18.Chatel-Chaix L, Cortese M, Romero-Brey I, Bender S, Neufeldt CJ, Fischl W, Scaturro P, Schieber N, Schwab Y, Fischer B, et al. : Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell Host & Microbe 2016, 20:342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixit E, Boulant S, Zhang Y, Lee ASY, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. : Peroxisomes Are Signaling Platforms for Antiviral Innate Immunity. Cell 2010, 141:668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coyaud E, Ranadheera C, Cheng D, Gonçalves J, Dyakov BJA, Laurent EMN, St-Germain J, Pelletier L, Gingras A-C, Brumell JH, et al. : Global Interactomics Uncovers Extensive Organellar Targeting by Zika Virus. Molecular & Cellular Proteomics 2018, 17:2242–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You J, Hou S, Malik-Soni N, Xu Z, Kumar A, Rachubinski RA, Frappier L, Hobman TC: Flavivirus Infection Impairs Peroxisome Biogenesis and Early Antiviral Signaling. Journal of Virology 2015, 89:12349–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, Mammi P, Mancini E, Yanovsky MJ, Andino R, et al. : The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLOS Pathogens 2016, 12:e1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poyomtip T, Hodge K, Matangkasombut P, Sakuntabhai A, Pisitkun T, Jirawatnotai S, Chimnaronk S: Development of viable TAP-tagged dengue virus for investigation of host–virus interactions in viral replication. Journal of General Virology 2016, 97:646–658. [DOI] [PubMed] [Google Scholar]
- 24.Viktorovskaya OV, Greco TM, Cristea IM, Thompson SR: Identification of RNA Binding Proteins Associated with Dengue Virus RNA in Infected Cells Reveals Temporally Distinct Host Factor Requirements. PLOS Neglected Tropical Diseases 2016, 10:e0004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips SL, Soderblom EJ, Bradrick SS, Garcia-Blanco MA: Identification of Proteins Bound to Dengue Viral RNA In Vivo Reveals New Host Proteins Important for Virus Replication. mBio 2016, 7:e01865–01815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huerta V, Ramos Y, Yero A, Pupo D, Martín D, Toledo P, Fleitas N, Gallien S, Martín AM, Márquez GJ, et al. : Novel interactions of domain III from the envelope glycoprotein of dengue 2 virus with human plasma proteins. Journal of Proteomics 2016, 131:205–213. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Zou L, Yang Y, Yuan J, Hu Z, Liu H, Peng H, Shang W, Zhang X, Zhu J, et al. : Superficial vimentin mediates DENV-2 infection of vascular endothelial cells. Scientific Reports 2016, 6:38372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee S, Sengupta N, Chaudhuri A, Akbar I, Singh N, Chakraborty S, Suryawanshi AR, Bhattacharyya A, Basu A: PLVAP and GKN3 Are Two Critical Host Cell Receptors Which Facilitate Japanese Encephalitis Virus Entry Into Neurons. Scientific Reports 2018, 8:11784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nain M, Mukherjee S, Karmakar SP, Paton AW, Paton JC, Abdin MZ, Basu A, Kalia M, Vrati S: GRP78 Is an Important Host Factor for Japanese Encephalitis Virus Entry and Replication in Mammalian Cells. Journal of Virology 2017, 91:e02274–02216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aliyu AI, Ling K-H, Md Hashim FN, Lam J-Y, Chee H-Y: Annexin II as a Dengue Virus Serotype 2 Interacting Protein Mediating Virus Interaction on Vero Cells. Viruses 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khachatoorian R, Cohn W, Buzzanco A, Riahi R, Arumugaswami V, Dasgupta A, Whitelegge JP, French SW: HSP70 Copurifies with Zika Virus Particles. Virology 2018, 522:228–233. [DOI] [PubMed] [Google Scholar]
- 32.Ooi YS, Majzoub K, Flynn RA, Mata MA, Diep J, Li JK, van Buuren N, Rumachik N, Johnson AG, Puschnik AS, et al. : An RNA-centric dissection of host complexes controlling flavivirus infection. Nature Microbiology 2019, 4:2369–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This study identified host proteins that interact with DENV or ZIKV RNA. Genetic screens discovered ER-associated proteins RRBP1 and vigilin that bind viral RNA and enhance viral replication.
- 33.Barrows NJ, Anglero-Rodriguez Y, Kim B, Jamison SF, Le Sommer C, McGee CE, Pearson JL, Dimopoulos G, Ascano M, Bradrick SS, et al. : Dual roles for the ER membrane protein complex in flavivirus infection: viral entry and protein biogenesis. Scientific Reports 2019, 9:9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ngo AM, Shurtleff MJ, Popova KD, Kulsuptrakul J, Weissman JS, Puschnik AS: The ER membrane protein complex is required to ensure correct topology and stable expression of flavivirus polyproteins. eLife 2019, 8:e48469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson RB, Ohlson MB, Eitson JL, Kumar A, McDougal MB, Boys IN, Mar KB, De La Cruz-Rivera PC, Douglas C, Konopka G, et al. : A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nature Microbiology 2018, 3:1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Muffat J, Omer Javed A, Keys HR, Lungjangwa T, Bosch I, Khan M, Virgilio MC, Gehrke L, Sabatini DM, et al. : Genome-wide CRISPR screen for Zika virus resistance in human neural cells. Proceedings of the National Academy of Sciences 2019, 116:9527–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao K-C, Chuo V, Ng WC, Neo SP, Pompon J, Gunaratne J, Ooi EE, Garcia-Blanco MA: Identification and characterization of host proteins bound to dengue virus 3′ UTR reveal an antiviral role for quaking proteins. RNA 2018, 24:803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward AM, Calvert MEK, Read LR, Kang S, Levitt BE, Dimopoulos G, Bradrick SS, Gunaratne J, Garcia-Blanco MA: The Golgi associated ERI3 is a Flavivirus host factor. Scientific Reports 2016, 6:34379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slonchak A, Khromykh AA: Subgenomic flaviviral RNAs: What do we know after the first decade of research. Antiviral Research 2018, 159:13–25. [DOI] [PubMed] [Google Scholar]
- 40.Soto-Acosta R, Xie X, Shan C, Baker CK, Shi P-Y, Rossi SL, Garcia-Blanco MA, Bradrick S: Fragile X mental retardation protein is a Zika virus restriction factor that is antagonized by subgenomic flaviviral RNA. eLife 2018, 7:e39023. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study identified host protein FMRP associates with ZIKV sfRNA and suppresses viral translation. Expression of ZIKV sfRNA led to upregulation of FMRP target genes, suggesting a role of ZIKV sfRNA in counteracting FMRP restriction.
- 41.Michalski D, Ontiveros JG, Russo J, Charley PA, Anderson JR, Heck AM, Geiss BJ, Wilusz J: Zika virus noncoding sfRNAs sequester multiple host-derived RNA-binding proteins and modulate mRNA decay and splicing during infection. Journal of Biological Chemistry 2019, 294:16282–16296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong Y, Ye W, Yang J, Han P, Wang Y, Ye C, Weng D, Zhang F, Xu Z, Lei Y: DDX21 translocates from nucleus to cytoplasm and stimulates the innate immune response due to dengue virus infection. Biochemical and Biophysical Research Communications 2016, 473:648–653. [DOI] [PubMed] [Google Scholar]
- 43.Rabelo K, Trugilho MRO, Costa SM, Pereira BAS, Moreira OC, Ferreira ATS, Carvalho PC, Perales J, Alves AMB: The effect of the dengue non-structural 1 protein expression over the HepG2 cell proteins in a proteomic approach. Journal of Proteomics 2017, 152:339–354. [DOI] [PubMed] [Google Scholar]
- 44.Mukherjee S, Singh N, Sengupta N, Fatima M, Seth P, Mahadevan A, Shankar SK, Bhattacharyya A, Basu A: Japanese encephalitis virus induces human neural stem/progenitor cell death by elevating GRP78, PHB and hnRNPC through ER stress. Cell Death & Disease 2018, 8:e2556–e2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabata K, Arimoto M, Arakawa M, Nara A, Saito K, Omori H, Arai A, Ishikawa T, Konishi E, Suzuki R, et al. : Unique Requirement for ESCRT Factors in Flavivirus Particle Formation on the Endoplasmic Reticulum. Cell Reports 2016, 16:2339–2347. [DOI] [PubMed] [Google Scholar]
- 46.Xin Q-L, Deng C-L, Chen X, Wang J, Wang S-B, Wang W, Deng F, Zhang B, Xiao G, Zhang L-K: Quantitative Proteomic Analysis of Mosquito C6/36 Cells Reveals Host Proteins Involved in Zika Virus Infection. Journal of Virology 2017, 91:e00554–00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samuel GH, Wiley MR, Badawi A, Adelman ZN, Myles KM: Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double-stranded RNA. Proceedings of the National Academy of Sciences 2016, 113:13863–13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sher AA, Glover KKM, Coombs KM: Zika Virus Infection Disrupts Astrocytic Proteins Involved in Synapse Control and Axon Guidance. Frontiers in Microbiology 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang X, Dong X, Li S-H, Zhou Y-P, Rayner S, Xia H-M, Gao GF, Yuan H, Tang Y-P, Luo M-H: Proteomic Analysis of Zika Virus Infected Primary Human Fetal Neural Progenitors Suggests a Role for Doublecortin in the Pathological Consequences of Infection in the Cortex. Frontiers in Microbiology 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosa-Fernandes L, Cugola FR, Russo FB, Kawahara R, de Melo Freire CC, Leite PEC, Bassi Stern AC, Angeli CB, de Oliveira DBL, Melo SR, et al. : Zika Virus Impairs Neurogenesis and Synaptogenesis Pathways in Human Neural Stem Cells and Neurons. Frontiers in Cellular Neuroscience 2019, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcez PP, Nascimento JM, de Vasconcelos JM, Madeiro da Costa R, Delvecchio R, Trindade P, Loiola EC, Higa LM, Cassoli JS, Vitória G, et al. : Zika virus disrupts molecular fingerprinting of human neurospheres. Scientific Reports 2017, 7:40780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glover KKM, Gao A, Zahedi-Amiri A, Coombs KM: Vero Cell Proteomic Changes Induced by Zika Virus Infection. PROTEOMICS 2019, 19:1800309. [DOI] [PubMed] [Google Scholar]
- 53.Diteepeng T, Khongwichit S, Paemanee A, Roytrakul S, Smith DR: Proteomic analysis of monkey kidney LLC-MK2 cells infected with a Thai strain Zika virus. Archives of Virology 2019, 164:725–737. [DOI] [PubMed] [Google Scholar]
- 54.Grabowski JM, Perera R, Roumani AM, Hedrick VE, Inerowicz HD, Hill CA, Kuhn RJ: Changes in the Proteome of Langat-Infected Ixodes scapularis ISE6 Cells: Metabolic Pathways Associated with Flavivirus Infection. PLOS Neglected Tropical Diseases 2016, 10:e0004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manchala NR, Dungdung R, Pilankatta R: Proteomic analysis reveals the enhancement of human serum apolipoprotein A-1(APO A-1) in individuals infected with multiple dengue virus serotypes. Tropical Medicine & International Health 2017, 22:1334–1342. [DOI] [PubMed] [Google Scholar]
- 56.Fragnoud R, Flamand M, Reynier F, Buchy P, Duong V, Pachot A, Paranhos-Baccala G, Bedin F: Differential proteomic analysis of virus-enriched fractions obtained from plasma pools of patients with dengue fever or severe dengue. BMC Infectious Diseases 2015, 15:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brasier AR, Zhao Y, Wiktorowicz JE, Spratt HM, Nascimento EJM, Cordeiro MT, Soman KV, Ju H, Recinos A, Stafford S, et al. : Molecular classification of outcomes from dengue virus −3 infections. Journal of Clinical Virology 2015, 64:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Lan Y, Li MY, Lamers MM, Fusade-Boyer M, Klemm E, Thiele C, Ashour J, Sanyal S: Flaviviruses Exploit the Lipid Droplet Protein AUP1 to Trigger Lipophagy and Drive Virus Production. Cell Host & Microbe 2018, 23:819–831.e815. [DOI] [PubMed] [Google Scholar]; •This study identified host proteins that are differentially ubiquitylated upon DENV infection and highlighted a role of lipid droplet protein AUP1 in DENV production.
- 59.Ye J, Zhang H, He W, Zhu B, Zhou D, Chen Z, Ashraf U, Wei Y, Liu Z, Fu ZF, et al. : Quantitative phosphoproteomic analysis identifies the critical role of JNK1 in neuroinflammation induced by Japanese encephalitis virus. Science Signaling 2016, 9:ra98–ra98. [DOI] [PubMed] [Google Scholar]
- 60.Miao M, Yu F, Wang D, Tong Y, Yang L, Xu J, Qiu Y, Zhou X, Zhao X: Proteomics Profiling of Host Cell Response via Protein Expression and Phosphorylation upon Dengue Virus Infection. Virologica Sinica 2019, 34:549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, Sun J, Ye J, Ashraf U, Chen Z, Zhu B, He W, Xu Q, Wei Y, Chen H, et al. : Quantitative Label-Free Phosphoproteomics Reveals Differentially Regulated Protein Phosphorylation Involved in West Nile Virus-Induced Host Inflammatory Response. Journal of Proteome Research 2015, 14:5157–5168. [DOI] [PubMed] [Google Scholar]


