Abstract
Characterized by impairments in brain and central nervous system development, neurodevelopmental diseases causes are highly heterogeneous. Although many of these diseases are individually rare, collectively more than 3% of the children are reported to be affected with a type of neurodevelopmental diseases worldwide, and many remain undiagnosed even with current genomic tools. Identifying the genetic causes of these diseases allows better clinical management and expands our understanding of human neurodevelopment. Over the last decade, expansion of genomic sequencing and some methodologic improvements have improved molecular diagnostic yield as well as the discovery of novel genetic causes for wide spectrum of neurodevelopmental diseases. Here we review the current diagnostic workflow and propose ways of improving the diagnostic yield.
Keywords: Neurodevelopmental diseases, Mendelian diseases, molecular diagnosis, novel gene discoveries, genomics
Introduction
Neurodevelopmental diseases (NDD) are due to disruptions in the complex process of central nervous system development and maintenance. NDD commonly presents as some form of developmental delay with or without variable syndromic or clinical diagnostic features such as autistic features, epilepsy and hypotonia. Many of the NDD are chronic and progressive with reduced reproductive fitness, imposing significant medical and economic burden on affected families and society. Worldwide, it is estimated that more than three percent of children are affected with a NDD [1–3]. Although both genetic and environmental factors can play a role in NDD, most of the severe forms are expected to originate from a genetic cause with a high penetrance pathogenic variant in a single gene [4,5]. Clarifying genetic causes of these early childhood onset diseases not only ends the diagnostic odyssey for families but also permits more focused genetic counseling, may reduce unnecessary tests, and allows physicians to have a clearer understanding of disease course, organs affected, and, in some instances, insights into therapy. However, while more common genetic diseases such as Rett syndrome, Neurofibromatosis type 1 (NF1) or Duchenne muscular dystrophy (DMD) have hallmark phenotypes, the initial clinical presentation of many NDD remains difficult to link to precise genetic diagnosis due to highly overlapping phenotypes and variable expressivity of syndromic components. Furthermore, NDD are genetically highly heterogeneous. In OMIM (Online Mendelian Inheritance in Man) [6], there are close to two thousand genes already identified for a form of NDD. Hence, the era of discovery of new traits by clinical presentation is much rarer now and superseded by our ability to observe causal variants through search across all genes. Here, we review the common clinical workup adopted in genetics for NDD that comprise the largest population referred for clinical exome sequencing (CES), the current obstacles to molecular diagnosis, recent methodologic improvements, and potential means to improve diagnostic rate. We also discuss how the Medical geneticists have already been practicing precision medicine, which became a popular term recently, for decades within the domain of rare genetic diseases including NDD.
Main Text
Review of molecular diagnostic rate using currently available genomic testing
Chromosomal Microarray and Clinical Exome Sequencing
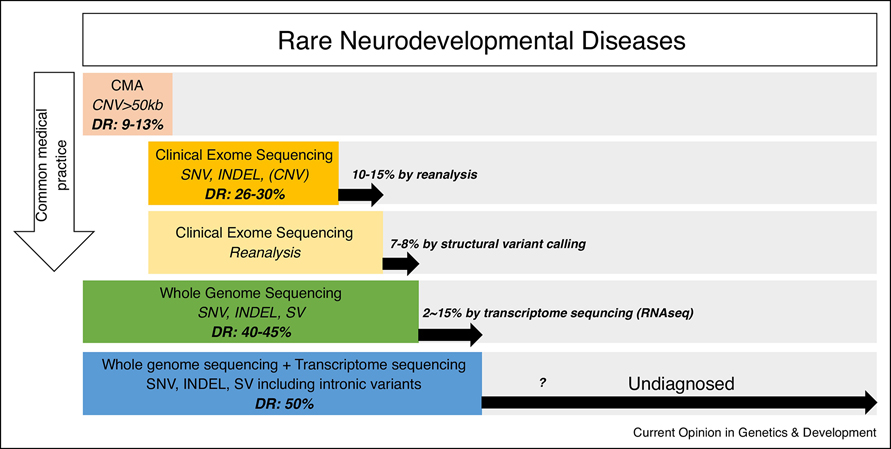
A common medical genetics approach to diagnosis of patients with NDD is to perform genomic testing in a sequential order starting with chromosomal microarray (CMA), and progressing to large gene panels and CES (Figure 1). Gene panels are often ordered instead of CES, mostly because CES is denied by insurance or in instances where repeat expansions are suspected. The diagnostic rates for CMA and CES for children with suspected genetic diseases are reported to be 9–13% [7–10] and 26–30% [11–14], respectively. Even though CES has the highest diagnostic yield for genetically heterogeneous diseases, it is clear that more than 60% of patients referred for CES remain undiagnosed after CMA and CES. Currently, reanalysis of CES can result in resolution of 10–15% of these unsolved cases, largely due to a new publication reporting a novel syndrome and genetic cause that allows clear reinterpretation of prior CES findings [15–19].
Figure 1.
Summary of available genomic tests for diagnosing rare neurodevelopmental diseases. The published diagnostic rate (DR) for each test is indicated. CMA: chromosomal microarray, CNV: copy number variant, SNV: single nucleotide variant, INDEL: small insertion and deletion, SV: structural variants including CNV.
Whole Genome Sequencing and Transcriptome Sequencing
The most plausible next step for NDD patients who remain undiagnosed after CES reanalysis is to have whole genome sequencing (WGS) performed. However, only a small portion of patients will undergo WGS largely because approval by insurance is low. Thus, much of this experience is derived from large research studies such as Undiagnosed Diseases Network (UDN) [20,21], Genomics England [22,23] and Deciphering Developmental Disorders (DDD) [24,25] studies where individual unsolved cases are sequenced more thoroughly. Although many WGS studies have reported a diagnostic rate above 40%, this includes variants that could have been identified by CMA or CES alone. Augmentation of the diagnostic rate by WGS is rather modest at 7–8% [26,27] with most of the added value derived from structural variants (SVs) not identifiable by CMA or CES such as inversions, translocations and copy number variants (CNVs) that are smaller than what CMA can detect and larger than what CES can detect. Many WGS variants are within intronic, untranslated or intergenic regions that remain difficult to interpret. Some clinical laboratories offer WGS as a diagnostic test, but the test is nearly equivalent to CES test with even SV calling not included as part of the standard clinical pipeline.
Recently, several reports described how transcriptome sequencing (RNAseq) can be used to interpret some of these non-coding variants for diagnosing exome or genome negative patients. Earlier studies [28–30] have focused on diseases for which relevant tissues were accessible such as muscle for muscle diseases and patient-derived skin fibroblast for mitochondrial diseases. The diagnostic rate augmented by these studies ranged from 2–24% (adjusted to only include cases for which RNAseq was essential for the diagnosis). Two more recent studies [27,31] have used blood, patient-derived skin fibroblast or muscle for wide spectrum of rare diseases including NDD and showed diagnostic rates of 2% [31] and 15% [27] (adjusted to only include cases for which RNAseq was essential for the diagnosis). Only one of these studies [27] fully incorporated the WGS data with RNAseq data to interpret the DNA variants not identifiable by CES or interpretable without the transcriptome data. This study reported multiple cryptic deep intronic pathogenic variants altering splicing as opposed to variants near known splice junctions, suggesting integrating the two omics datasets yields higher diagnostic rate than using RNAseq data alone. However, even with this integration, only about 15% of the cases could be resolved.
Approaches to increase the molecular diagnostic rate
The overall diagnostic rate achieved for NDD after going through series of diagnostic workup from CMA to WGS and RNAseq is estimated to be approximately 50% (Figure 1). Taking examples from a few well-characterized and comprehensively studied genetic diseases with close to 100% diagnostic rate such as cystic fibrosis (CF), NF1, and DMD, we predict that the diagnostic rate of this large cohort of undiagnosed patients with NDD are likely to have substantial improvements in diagnostic rate. However, because some NDD may be environmentally caused or oligogenic, it is unlikely that we can ever reach a diagnostic rate of 100%. The biggest difference between these well-characterized diseases with high diagnostic rate and NDD is that the well-characterized diseases have clinical phenotypes distinct enough for the medical geneticists to focus on a single gene and exhaustively search for a pathogenic variant while most of the NDD have such a substantial genetic heterogeneity with significant overlapping and variable phenotypes, the search space has to vastly expand to thousands of genes. It is also reasonable to extrapolate that thousands of additional genes are yet to be identified as causal for Mendelian genetic forms of NDD. Thus, methods that provide a more comprehensive search for mutation types or locations within the genome that are relatively unexplored provides an attractive route forward for novel gene discovery.
Paths to novel gene discovery
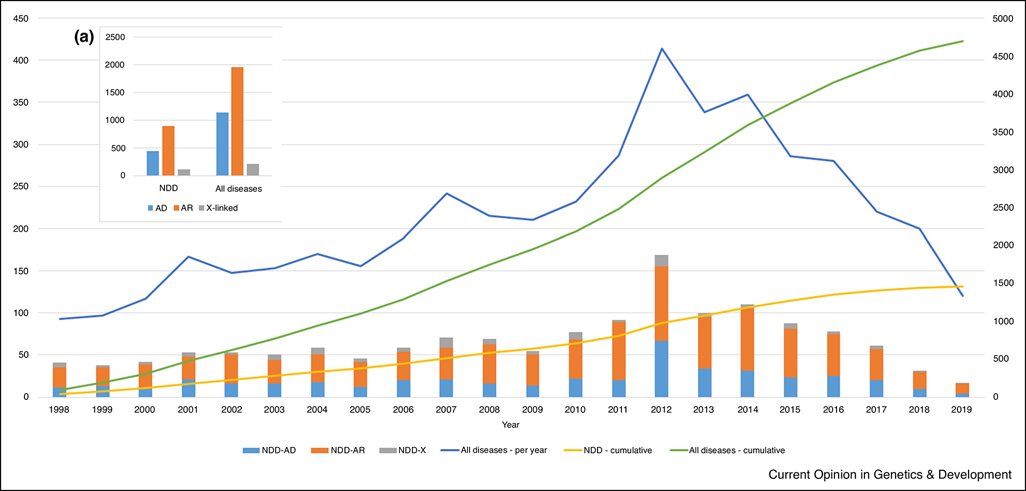
Positional cloning of novel disease genes using linkage and mutation screening resulted in a wave of steadily increasing novel genes discoveries since 1986 when the first positionally cloned disease genes were identified. However, this type of discovery was largely supplanted by exome sequencing, with or without positional information, starting in 2011 when exome sequencing could be applied to research cohorts and was implemented as a clinical diagnostic test. Novel discovery of genetic causes of NDD are still being made, but more recently, the discovery rate has decreased, showing a similar trajectory to the discovery rate of all Mendelian diseases (Figure 2) [32,33]. While large families and specifically collected individuals with similar phenotypes have led to substantial gene discovery [33,34], many additional discoveries have also occurred by post-hoc aggregation of individuals with rare genetic variants and similar phenotypes [35–37]. Tools to assist clinicians like GeneMatcher [38,39] are now commonly used to search for additional individuals with similar DNA variants, but can lead to artificially high ‘match rates’ and even with this accepted mechanism, the rate of detection/publication of genes associated with Mendelian diseases is decreasing. We infer that many of the remaining disease causing genes to be discovered are likely to be for ultra-rare diseases or lethal conditions that only present under rare circumstances such as somatic mosaicism or potentially with oligogenic causes. For these diseases, finding the second and third cases with the same molecular basis becomes more challenging, requiring significant functional analysis as supporting evidence, which is not always achievable. With prenatal CES more routinely performed, some of the genes causing lethal conditions may be detected. As shown in figure 2, there are approximately twice as many autosomal recessive genes as autosomal dominant genes for both NDD and all Mendelian diseases and the ratio has been consistent over the last 8 years for newly discovered genes except for 2012 when CES launched. This suggests that many of the to-be-discovered causal genes are likely to be recessive.
Figure 2.
Approximate number of novel genes discovered to be associated with an NDD (stacked bar) and Mendelian disease (blue line) per year (primary Y-axis) and cumulatively (secondary Y-axis and yellow and green curved line graph). For NDD, the genes were separately counted for autosomal dominant (NDD-AD), autosomal recessive (NDD-AR) and X-linked (NDD-X) disease genes. Estimation was made by counting the number of genes deposited in the Human Gene Mutation Database (HGMD) for the first time for each year. Only the disease-causing mutations (DM) were considered. (A) Number of autosomal dominant (AD), autosomal recessive (AR) and X-linked genes for NDD and all Mendelian disease genes cumulatively.
Increasing diagnostic rate from RNAseq
Augmentation of WGS with RNAseq is primarily limited by ready access to relevant tissues likely to express the mutant allele. Available tissues, such as blood or skin fibroblasts, for NDD will be inadequate if the mutant gene is mainly expressed in brain. The broad search space of known and unknown novel genes is in stark contrast to diseases like CF, NF1 or DMD, where the clinical phenotype is widely recognized. Of ~2,000 known potential genes for NDD, a portion are inadequately expressed in accessible tissues [27] but coverage improvement is likely possible by generating deeper RNAseq data. However, this will not improve observations of genes that are not expressed in the accessible tissues. Generating patient specific neuronal cells by reprograming patient-derived cells into induced pluripotent stem cells (iPSC) and differentiating into neurons [40,41] or directly converting patient-derived fibroblast into neuronal cells [42–44] are reasonable approaches to address this matter, but remain too inefficient to implement broadly on patient derived cells. Further, only a limited set of neuronal cell types are likely to be generated and thus one can reasonably project that some neuronal genes will still not be expressed. Alternate methods such as deregulating gene expression broadly may be helpful in order to generate spliced mRNA that may reveal splicing abnormalities caused by a DNA variant. These methods may be coupled with mechanisms to suppress nonsense-mediated decay (NMD) in patient-derived cells [45,46]. NMD can greatly challenge our ability to observe the mutant allele with a premature termination codon as the coverage of the mutant transcript is lower and thus less observed by RNA sequencing.
The most commonly used sequencing library preparation protocol is selection of polyadenylated (polyA) transcripts, which excludes non-coding RNAs and small RNAs. These provide an excellent assessment of the protein coding portion of the human genome, but may miss important functional RNA elements that may be observed with other methods. To date there has been little identification of causal mutations within these non-coding RNAs, and there remains no good estimate of how much variants in these RNAs contribute to the pathogenesis of rare NDD, but there is a growing literature of the importance of some long non-coding RNAs in the mechanism of pathogenesis of some of the rare diseases [47].
Identifying variants challenging to detect or interpret by current technologies
There will be pathogenic variants that are not detectable by short-read sequencing technologies that are most commonly used, although based on the examples of the diseases with near 100% diagnostic rate, we can predict that not a significant number of cases will have these types of variants. For example, the molecular diagnostic rate for patients clinically diagnosed with CF is 97–98% by conventional DNA sequencing test alone [48,49]. Significant portion of the 2–3% undiagnosed cases most likely have deep intronic variants affecting splicing but CFTR gene is not expressed in accessible tissues such as blood so it is not feasible to further explore. RNA or cDNA sequencing has been utilized more routinely for NF1. When the combined DNA and RNA (cDNA) sequencing analysis approach is used, ~97% of the cases receive a molecular diagnosis and RNA sequencing is estimated to be essential for ~10% of the diagnosed cases [50]. DMD has ~99% molecular diagnostic rate with most of the patients identified with pathogenic variants detectable by DNA sequencing test alone but ~80% of the variants are large deletions or duplication and only ~1% of the cases are solved by RNA sequencing with deep intronic variants altering splicing [51,52]. These examples suggest that the current technologies are capturing most of the pathogenic variants but they also show that the type of variants found to be pathogenic could vary by gene and a subset of genes can be enriched with pathogenic variants not readily detectable or interpretable by current technologies. Generating longer read genome and transcriptome data by newer technologies such as PacBio [53,54], Oxford Nanopore [55,56], BioNano [57] and 10X genomics [58] are some of the available alternate technologies. However, rare Mendelian disease patients diagnosed by pathogenic variants identified by these technologies, which could not have been detected by more routinely used technologies, are yet to be reported.
Improving variant calling and variant interpretation
Most genomic testing is performed on peripheral blood DNA and most of the variant callers are highly optimized to detect homozygous, hemizygous or heterozygous variants and not mosaic variants which can be an important contributor, especially for lethal diseases [59]. An example NDD caused by somatic variants is hemimegalencephaly for which only one hemisphere of the brain overgrows, often causing severe epilepsy and developmental delay [60]. Complementing the variant calling with somatic variant caller can be applied to resolve this limitation. However, collecting the appropriate tissue will be essential to detect some of these somatic mosaic variants and that could be challenging.
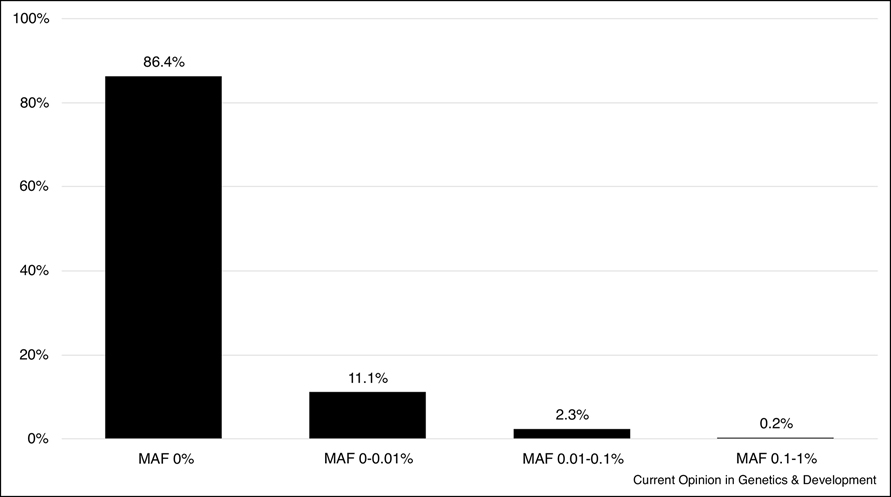
An important component of variant interpretation from exome, genome and transcriptome sequencing is filtering out common variants that cannot be causal for ultra-rare NDD. The establishment of ExAC (Exome Aggregation Consortium) [61] that merged into gnomAD (Genome Aggregation Database) browsers [62] made filtering out the common variants significantly more effective. As these dataset grows, especially with more data added from the under-sequenced minority populations and non-coding regions, it is expected that parental comparator sequencing may not even be necessary to identify de novo variants for autosomal dominant NDD as causal heterozygous variants for such a rare, severe diseases are not expected to be observed in the population although there could be exceptions due to factors such as incomplete penetrance, imprinting or mosaicism in the population. For recessive diseases, variants can be observed in the population but not at high frequency. ACMG-AMP (American College of Medical Genetics-Association for Molecular Pathology) joint consensus recommendation for the interpretation of sequence variants uses >5% minor allele frequency (MAF) threshold for classifying variants benign although in practice, a 1% threshold is more commonly used for most of the rare diseases [63–65]. The MAF threshold can be further lowered from 1% because as plotted in figure 3, with the exception of few genes, MAF of the pathogenic or likely pathogenic variants for recessive diseases are well under 0.1% with ~97% of the variants being under 0.01%, suggesting that MAF of 0.1% or greater are unlikely to be causal for most recessive NDD.
Figure 3.
Minor allele frequency distribution of pathogenic/likely pathogenic variants in autosomal recessive neurodevelopmental diseases. Neurodevelopmental disease gene list was created by querying OMIM for genes associated with autosomal recessive diseases involving developmental delay, intellectual disability, mental retardation, seizures or epilepsy. Variants that were in HGMD as disease causing and classified as pathogenic or likely pathogenic in ClinVar were selected. gnomAD was used to extract MAF information on these variants.
Lastly, databases such as ClinVar [66] or Human Gene Mutation Database [67] are helpful for classifying variants and identifying cohorts of patients with same or similar pathogenic variants. Along the line of making variant filtration more effective by growing the population database, having benign or likely benign variants catalogued in these variant classification databases are as essential as having pathogenic or likely pathogenic variants catalogued. A more rapid entry system into ClinVar is clearly needed to more promptly translate novel gene discovery into clinical impact [33].
Conclusion
In the past decade, the advent of next-generation sequencing (NGS) technology and its clinical application have made significant contribution to Medical Genetics by identifying the molecular basis of numerous known and a large number of new rare NDD. Not only has the novel gene discovery rate peaked during this period, but also the time and resources taken to molecular diagnosis for individual patients has substantially decreased with the NGS-based genomic tests becoming the first-tier test in the clinic. It is now possible to end the diagnostic odyssey earlier and provide more tailored clinical management. However, it is quite sobering that as we presented here, there are still about 50% of patients with highly suspected genetic diseases, referred for exome sequencing that remain without a molecular diagnosis, and the next decade will focus on improved methods to detect currently cryptic pathogenic variants. The workflow in place as described in figure 1 has been established quite stably in the current medical system and we expect that it will continue to stay the same for a while. However, we foresee and propose that in a near future, as sequencing price continues to decrease, WGS combined with RNAseq will replace CMA and CES and become the first-tier test performed in all NDD patients as soon as they start showing symptoms.
Precision health or precision medicine has been a buzzword in the past few years with the core of it being characterizing the genetic or molecular basis of a health condition to personalize healthcare. It is important to recognize that the rare disease community has been practicing precision medicine for a long time by tailoring clinical management based on the molecular basis of a disease and leading the effort in developing tools and standards for identifying and interpreting genetic information. It should also be noted that the vast majority of variants clearly causing a human phenotype are rare and cause a Mendelian phenotype despite massive efforts to reveal common variant contributions to common phenotypes. Thus, the rare disease community will continue to serve as the model for impactful translational genomic medicine.
Highlights.
Rate of diagnostic yield of Clinical Exome Sequencing is not increasing
Rate of new genetic disease discovery is slowing
Novel methods are needed to improve diagnostic and discovery rates
Acknowledgements
Hane Lee and Stanley F. Nelson are supported by awards from the National Institutes of Health [grant numbers U01HG007703, UL1TR001881].
Footnotes
Declaration of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baird PA, Anderson TW, Newcombe HB, Lowry RB: Genetic disorders in children and young adults: a population study. Am J Hum Genet 1988, 42:677–693. [PMC free article] [PubMed] [Google Scholar]
- 2.Shashi V, McConkie-Rosell A, Rosell B, Schoch K, Vellore K, McDonald M, Jiang YH, Xie P, Need A, Goldstein DB: The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med 2014, 16:176–182. [DOI] [PubMed] [Google Scholar]
- 3.Tarlungeanu DC, Novarino G: Genomics in neurodevelopmental disorders: an avenue to personalized medicine. Exp Mol Med 2018, 50:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boycott KM, Rath A, Chong JX, Hartley T, Alkuraya FS, Baynam G, Brookes AJ, Brudno M, Carracedo A, den Dunnen JT, et al. : International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. Am J Hum Genet 2017, 100:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niemi MEK, Martin HC, Rice DL, Gallone G, Gordon S, Kelemen M, McAloney K, McRae J, Radford EJ, Yu S, et al. : Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature 2018, 562:268–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Online Mendelian Inheritance in Man, OMIM®. Edited by. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; (Baltimore, MD: ). vol 2020.] [Google Scholar]
- 7.Coulter ME, Miller DT, Harris DJ, Hawley P, Picker J, Roberts AE, Sobeih MM, Irons M: Chromosomal microarray testing influences medical management. Genet Med 2011, 13:770–776. [DOI] [PubMed] [Google Scholar]
- 8.Henderson LB, Applegate CD, Wohler E, Sheridan MB, Hoover-Fong J, Batista DA: The impact of chromosomal microarray on clinical management: a retrospective analysis. Genet Med 2014, 16:657–664. [DOI] [PubMed] [Google Scholar]
- 9.Zilina O, Teek R, Tammur P, Kuuse K, Yakoreva M, Vaidla E, Molter-Vaar T, Reimand T, Kurg A, Ounap K: Chromosomal microarray analysis as a first-tier clinical diagnostic test: Estonian experience. Mol Genet Genomic Med 2014, 2:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho KS, Twede H, Vanzo R, Harward E, Hensel CH, Martin MM, Page S, Peiffer A, Mowery-Rushton P, Serrano M, et al. : Clinical Performance of an Ultrahigh Resolution Chromosomal Microarray Optimized for Neurodevelopmental Disorders. Biomed Res Int 2016, 2016:3284534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K, Weiss ME, Koster J, Marais A, Paknia O, Schroder R, Garcia-Aznar JM, Werber M, et al. : Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet 2017, 25:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, Vertino-Bell A, Smaoui N, Neidich J, Monaghan KG, et al. : Clinical application of whole-exome sequencing across clinical indications. Genet Med 2016, 18:696–704. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, et al. : Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014, 312:1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Das K, Toy T, Harry B, Yourshaw M, et al. : Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 2014, 312:1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu P, Meng L, Normand EA, Xia F, Song X, Ghazi A, Rosenfeld J, Magoulas PL, Braxton A, Ward P, et al. : Reanalysis of Clinical Exome Sequencing Data. N Engl J Med 2019, 380:2478–2480.*This paper describes the augmentation of molecular diagnosis by clinical exome sequencing reanalysis. Most of the newly diagnosed cases after reanalysis are driven by novel gene discoveries published in between the initial analysis and reanalysis.
- 16.Baker SW, Murrell JR, Nesbitt AI, Pechter KB, Balciuniene J, Zhao X, Yu Z, Denenberg EH, DeChene ET, Wilkens AB, et al. : Automated Clinical Exome Reanalysis Reveals Novel Diagnoses. J Mol Diagn 2019, 21:38–48. [DOI] [PubMed] [Google Scholar]
- 17.Shashi V, Schoch K, Spillmann R, Cope H, Tan QK, Walley N, Pena L, McConkie-Rosell A, Jiang YH, Stong N, et al. : A comprehensive iterative approach is highly effective in diagnosing individuals who are exome negative. Genet Med 2019, 21:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basel-Salmon L, Orenstein N, Markus-Bustani K, Ruhrman-Shahar N, Kilim Y, Magal N, Hubshman MW, Bazak L: Improved diagnostics by exome sequencing following raw data reevaluation by clinical geneticists involved in the medical care of the individuals tested. Genet Med 2019, 21:1443–1451. [DOI] [PubMed] [Google Scholar]
- 19.Wenger AM, Guturu H, Bernstein JA, Bejerano G: Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med 2017, 19:209–214. [DOI] [PubMed] [Google Scholar]
- 20.Ramoni RB, Mulvihill JJ, Adams DR, Allard P, Ashley EA, Bernstein JA, Gahl WA, Hamid R, Loscalzo J, McCray AT, et al. : The Undiagnosed Diseases Network: Accelerating Discovery about Health and Disease. Am J Hum Genet 2017, 100:185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Splinter K, Adams DR, Bacino CA, Bellen HJ, Bernstein JA, Cheatle-Jarvela AM, Eng CM, Esteves C, Gahl WA, Hamid R, et al. : Effect of Genetic Diagnosis on Patients with Previously Undiagnosed Disease. N Engl J Med 2018, 379:2131–2139.*This paper summarizes the outcome of a nationwide effort to perform comprehensive genetic/genomic search for undiagnosed diseases patients through the Undiagnosed Diseases Network.
- 22.Griffin BH, Chitty LS, Bitner-Glindzicz M: The 100 000 Genomes Project: What it means for paediatrics. Arch Dis Child Educ Pract Ed 2017, 102:105–107. [DOI] [PubMed] [Google Scholar]
- 23.Torjesen I: Genomes of 100,000 people will be sequenced to create an open access research resource. BMJ 2013, 347:f6690. [DOI] [PubMed] [Google Scholar]
- 24.Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K,Barrett DM, Bayzetinova T, et al. : Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 2015, 385:1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deciphering Developmental Disorders S: Large-scale discovery of novel genetic causes of developmental disorders. Nature 2015, 519:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stavropoulos DJ, Merico D, Jobling R, Bowdin S, Monfared N, Thiruvahindrapuram B, Nalpathamkalam T, Pellecchia G, Yuen RKC, Szego MJ, et al. : Whole Genome Sequencing Expands Diagnostic Utility and Improves Clinical Management in Pediatric Medicine. NPJ Genom Med 2016, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H, Huang AY, Wang LK, Yoon AJ, Renteria G, Eskin A, Signer RH, Dorrani N, Nieves-Rodriguez S, Wan J, et al. : Diagnostic utility of transcriptome sequencing for rare Mendelian diseases. Genet Med 2019.*This paper describes how integrating transcriptome sequencing data with whole genome sequencing data augments the diagnostic rate for rare Mendelian diseases by identifying crypting splice altering variants.
- 28.Gonorazky HD, Naumenko S, Ramani AK, Nelakuditi V, Mashouri P, Wang P, Kao D, Ohri K, Viththiyapaskaran S, Tarnopolsky MA, et al. : Expanding the Boundaries of RNA Sequencing as a Diagnostic Tool for Rare Mendelian Disease. Am J Hum Genet 2019, 104:466–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kremer LS, Bader DM, Mertes C, Kopajtich R, Pichler G, Iuso A, Haack TB, Graf E, Schwarzmayr T, Terrile C, et al. : Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat Commun 2017, 8:15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cummings BB, Marshall JL, Tukiainen T, Lek M, Donkervoort S, Foley AR, Bolduc V, Waddell LB, Sandaradura SA, O’Grady GL, et al. : Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med 2017, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fresard L, Smail C, Ferraro NM, Teran NA, Li X, Smith KS, Bonner D, Kernohan KD, Marwaha S, Zappala Z, et al. : Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nat Med 2019, 25:911–919.*This paper describes a method to detect outliers of blood transcriptome sequencing data to identify molecular diagnosis of rare Mendelian diseases.
- 32.Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, Harrell TM, McMillin MJ, Wiszniewski W, Gambin T, et al. : The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am J Hum Genet 2015, 97:199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bamshad MJ, Nickerson DA, Chong JX: Mendelian Gene Discovery: Fast and Furious with No End in Sight. Am J Hum Genet 2019, 105:448–455.*This paper is a commentary from one of the Centers for Mendelian Genomics summarizing paths to novel gene discoveries for Mendelian conditions and proposing improvements needed for further discoveries.
- 34.Ott J, Wang J, Leal SM: Genetic linkage analysis in the age of whole-genome sequencing. Nat Rev Genet 2015, 16:275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arboleda VA, Lee H, Dorrani N, Zadeh N, Willis M, Macmurdo CF, Manning MA, Kwan A, Hudgins L, Barthelemy F, et al. : De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am J Hum Genet 2015, 96:498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snijders Blok L, Madsen E, Juusola J, Gilissen C, Baralle D, Reijnders MR, Venselaar H, Helsmoortel C, Cho MT, Hoischen A, et al. : Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. Am J Hum Genet 2015, 97:343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephen J, Maddirevula S, Nampoothiri S, Burke JD, Herzog M, Shukla A, Steindl K, Eskin A, Patil SJ, Joset P, et al. : Bi-allelic TMEM94 Truncating Variants Are Associated with Neurodevelopmental Delay, Congenital Heart Defects, and Distinct Facial Dysmorphism. Am J Hum Genet 2018, 103:948–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobreira N, Schiettecatte F, Valle D, Hamosh A: GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 2015, 36:928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A: New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat 2015, 36:425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, et al. : Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 2015, 12:671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han SS, Williams LA, Eggan KC: Constructing and deconstructing stem cell models of neurological disease. Neuron 2011, 70:626–644. [DOI] [PubMed] [Google Scholar]
- 42.Caiazzo M, Giannelli S, Valente P, Lignani G, Carissimo A, Sessa A, Colasante G, Bartolomeo R, Massimino L, Ferroni S, et al. : Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Reports 2015, 4:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, Vierbuchen T, Hawkins JS, Geissler R, Barres BA, Wernig M: Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol 2013, 31:434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M: Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463:1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andreutti-Zaugg C, Scott RJ, Iggo R: Inhibition of nonsense-mediated messenger RNA decay in clinical samples facilitates detection of human MSH2 mutations with an in vivo fusion protein assay and conventional techniques. Cancer Res 1997, 57:3288–3293. [PubMed] [Google Scholar]
- 46.Bhuvanagiri M, Lewis J, Putzker K, Becker JP, Leicht S, Krijgsveld J, Batra R, Turnwald B, Jovanovic B, Hauer C, et al. : 5-azacytidine inhibits nonsense-mediated decay in a MYC-dependent fashion. EMBO Mol Med 2014, 6:1593–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sparber P, Filatova A, Khantemirova M, Skoblov M: The role of long non-coding RNAs in the pathogenesis of hereditary diseases. BMC Med Genomics 2019, 12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strom CM, Huang D, Chen C, Buller A, Peng M, Quan F, Redman J, Sun W: Extensive sequencing of the cystic fibrosis transmembrane regulator gene: assay validation and unexpected benefits of developing a comprehensive test. Genet Med 2003, 5:9–14. [DOI] [PubMed] [Google Scholar]
- 49.Moskowitz SM, Chmiel JF, Sternen DL, Cheng E, Gibson RL, Marshall SG, Cutting GR: Clinical practice and genetic counseling for cystic fibrosis and CFTR-related disorders. Genet Med 2008, 10:851–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans DG, Bowers N, Burkitt-Wright E, Miles E, Garg S, Scott-Kitching V, Penman-Splitt M, Dobbie A, Howard E, Ealing J, et al. : Comprehensive RNA Analysis of the NF1 Gene in Classically Affected NF1 Affected Individuals Meeting NIH Criteria has High Sensitivity and Mutation Negative Testing is Reassuring in Isolated Cases With Pigmentary Features Only. EBioMedicine 2016, 7:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laing NG, Davis MR, Bayley K, Fletcher S, Wilton SD: Molecular diagnosis of duchenne muscular dystrophy: past, present and future in relation to implementing therapies. Clin Biochem Rev 2011, 32:129–134. [PMC free article] [PubMed] [Google Scholar]
- 52.Nallamilli BR, Ankala A, Hegde M: Molecular diagnosis of Duchenne muscular dystrophy. Curr Protoc Hum Genet 2014, 83:9 25 21–29. [DOI] [PubMed] [Google Scholar]
- 53.Wenger AM, Peluso P, Rowell WJ, Chang PC, Hall RJ, Concepcion GT, Ebler J, Fungtammasan A, Kolesnikov A, Olson ND, et al. : Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol 2019, 37:1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. : Real-time DNA sequencing from single polymerase molecules. Science 2009, 323:133–138. [DOI] [PubMed] [Google Scholar]
- 55.Cretu Stancu M, van Roosmalen MJ, Renkens I, Nieboer MM, Middelkamp S, de Ligt J, Pregno G, Giachino D, Mandrile G, Espejo Valle-Inclan J, et al. : Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun 2017, 8:1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain M, Olsen HE, Paten B, Akeson M: The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol 2016, 17:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bocklandt S, Hastie A, Cao H: Bionano Genome Mapping: High-Throughput, Ultra-Long Molecule Genome Analysis System for Precision Genome Assembly and Haploid-Resolved Structural Variation Discovery. Adv Exp Med Biol 2019, 1129:97–118. [DOI] [PubMed] [Google Scholar]
- 58.Kitzman JO: Haplotypes drop by drop. Nat Biotechnol 2016, 34:296–298. [DOI] [PubMed] [Google Scholar]
- 59.Happle R: Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J Am Acad Dermatol 1987, 16:899–906. [DOI] [PubMed] [Google Scholar]
- 60.Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, Scott E, Bafna V, Hill KJ, Collazo A, et al. : De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet 2012, 44:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. : Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. : Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 2019.*This paper describes the genome aggregation database (gnomAD), the largest database that provides allele frequency information across the genome. gnomAD is a critical tool for variant filteration and interpretation.
- 63.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. : Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015, 17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, Berg JS, Biswas S, Bowling KM, Conlin LK, et al. : Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet 2016, 99:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jalali Sefid Dashti M, Gamieldien J: A practical guide to filtering and prioritizing genetic variants. Biotechniques 2017, 62:18–30. [DOI] [PubMed] [Google Scholar]
- 66.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. : ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018, 46:D1062–D1067.*This paper describes ClinVar, a publicly available cetralized databases of variants. Holding more than half a million submission record on more than 300,000 variants, Clinvar is an important tool for variant classification.
- 67.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN: Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat 2003, 21:577–581. [DOI] [PubMed] [Google Scholar]