Abstract
Study Objectives
Posttraumatic stress disorder (PTSD) is a common condition for military personnel and veterans. PTSD has been shown to impact gene expression, however, to date no study has examined comorbid conditions which may also impact gene expression, for example, excessive daytime sleepiness (EDS). As such, this study sought to examine gene expression using RNA sequencing across three group comparisons of military personnel and veterans: (1) PTSD with EDS (PTSDwEDS) versus PTSD without EDS (PTSDw/outEDS), (2) Controls (no PTSD or EDS) versus PTSDwEDS, and (3) Controls versus PTSDw/outEDS.
Methods
We performed experimental RNA-seq using Illumina’s HiSeq 2500 Sequencing System. We also used Ingenuity Pathway Analysis (IPA), a bioinformatics application, to identify gene pathways and networks which may be disrupted.
Results
There were only two genes that were significantly dysregulated between the Controls and PTSDw/outEDS, therefore IPA analysis was not conducted. However, comparisons revealed that there was significant gene dysregulation between Controls and the PTSDwEDS (251 genes), and the PTSDwEDS versus the PTSDw/outEDS (1,873 genes) groups. Four candidate networks were identified via the IPA software for analysis. Significantly dysregulated genes across the four candidate networks were associated with sleep and circadian function, metabolism, mitochondrial production and function, ubiquitination, and the glutamate system.
Conclusions
These results suggest that PTSD with concurrent EDS is associated with gene dysregulation. This dysregulation may present additional biological and health consequences for these military personnel and veterans. Further research, to track these gene changes over time and to determine the cause of the EDS reported, is vital.
Keywords: gene activity, PTSD, Epworth sleepiness scale, adiposity, metabolism, mitochondria
Statement of Significance.
This study sought to identify if excessive daytime sleepiness (EDS) was associated with gene dysregulation in military personnel and veterans with posttraumatic stress disorder (PTSD). RNA-sequencing analysis identified that when comparing personnel with EDS and PTSD with personnel with PTSD without EDS and Controls (no PTSD or EDS), there were a total of 1,973 differentially regulated genes. Using ingenuity pathway analysis, we identified significant gene networks linked to sleep and circadian regulation, mitochondrial functioning, and the glutamate system. Our results indicate that EDS with concurrent PTSD may have a biological impact on gene regulation and potentially on health. Although preliminary, these findings highlight a need for early interventions that focus on improving EDS symptoms in military personnel and veterans with PTSD.
Posttraumatic stress disorder (PTSD) is pervasive and debilitating, affecting an estimated 23% of US military personnel and veterans who served in Operations Enduring Freedom and Iraqi Freedom (OEF/OIF) [1, 2]. PTSD is associated with an increased incidence of psychiatric and medical comorbidities, disability, substance abuse, and suicide, alongside increased health care utilization [2]. Thus, identifying the underlying biological processes involved in PTSD pathophysiology is critical to allow for the identification of novel screening tools and to improve treatment and monitoring. Exploring alterations in gene expression and subsequent gene pathways have recently been posited as a promising avenue of investigation. Indeed, many transcriptome-wide studies indicate that dysregulation of genes associated with glucocorticoid receptor signaling and immune pathways are implicated in PTSD, for example, brain-derived neurotrophic factor (BDNF) [3] and the FK506 Binding Protein 51 (FKBP5) genes [4–6]. Both BDNF and FKBP5 have been found to influence glucocorticoid receptor sensitivity which has a major role in the regulation of the hypothalamic–pituitary–adrenal (HPA) axis. In turn, the HPA axis is intrinsically linked with our stress responses and the formation of emotional memories. One recent study also found that reductions in PTSD symptoms, following a cognitive behavioral therapy intervention, resulted in the downregulation of immune and metabolic networks with an NF-κB hub [7]. However, to date, no studies have accounted for comorbid conditions which may impact on both PTSD and gene expression, for example, sleepiness. Thus, the aim of this study was to examine whether PTSD with and without excessive daytime sleepiness (EDS) is associated with altered gene expression across the whole genome.
EDS is the primary symptom of chronic insufficient sleep [8] and of several sleep and circadian rhythm disorders [9, 10]. Sleep problems are a common complaint of OEF/OIF veterans [11, 12]. These sleep disturbances may be due to and exacerbated by a multitude of military-related factors such as frequent shift work, chronic sleep restriction/deprivation when out on missions, deployment across multiple time zones, maladaptive sleep practices, and potentially due to the physical and emotional stress of deployment [13]. Furthermore, sleep problems and PTSD commonly co-occur in this population. Up to 91% of military personnel and veterans with PTSD report co-occurring sleep disturbances and disorders, most commonly insomnia [14, 15]. Indeed, EDS and sleep disturbances, such as insomnia, commonly co-occur with PTSD [11, 16–18], especially in military populations. Also disordered sleep prior to and directly following a traumatic event (or events) has been implicated in the onset [19, 20], maintenance, and severity of PTSD symptomology [16, 21, 22]. Furthermore, EDS has been linked with increased risk of motor vehicle crashes, work-related accidents, greater negative ratings of quality of life, and declines in cognition and behavior [23, 24]. As such, daytime sleepiness is an important factor to consider and monitor in military populations.
Studies of rodents and drosophila have shown that chronic sleep deprivation has a drastic effect on gene expression, both within the brain and in peripheral, circadian-driven, organs, e.g. the liver [25, 26]. In humans, the genetic variants associated with sleep disorders such as narcolepsy (hypocretin/orexin system) [27], fatal familial insomnia [28], and period (PER) genes in the role of circadian regulation [29] have been established. However, our understanding of variations in gene expression that are associated with sleep disorders, such as obstructive sleep apnea (OSA), insomnia, EDS, or with fluctuations in sleep duration/quality more generally, is limited. Recent genome-wide association studies have found 42 genetic loci for daytime sleepiness which were enriched for genes expressed in brain tissues and in neuronal transmission pathways [10, 30]. However, whether or not EDS alters gene expression remains unknown. Current research does indicate that improving sleep may change gene expression. In a sleep intervention of military personnel and veterans with OSA and/or insomnia, it was found that personnel with improved sleep quality had significant downregulation in 113 genes, including a significant reduction in expression of genes associated with inflammatory cytokines; in comparison to no changes in gene expression postintervention in the group with no improvement in sleep quality [31]. Military personnel in this study who had improved sleep quality also showed significant reductions in depression and PTSD symptoms [31]. Taken together, current literature indicates that sleep and PTSD independently have significant implications for gene expression profiles and pathways. However, gene expression changes related to PTSD with concomitant EDS have not yet been examined.
The aim of this study was to examine gene expression in active-duty military personnel and veterans with PTSD, with and without EDS. Participants were categorized into three groups: (1) PTSD with EDS (PTSDwEDS), (2) PTSD without EDS (PTSDw/outEDS), or (3) Controls; no PTSD without EDS. Traumatic brain injury (TBI) and depression are also frequently comorbid with PTSD, especially in active-duty military personnel and veterans, as such we also assessed lifetime TBI history and depression symptoms in this study. Gene expression was measured using a transcriptome-wide, state-of-the-art RNA-seq approach that allows for an examination of gene activity across the genome.
Methods
This study has been detailed elsewhere [32], however briefly, nontreatment seeking, military personnel and veterans were enrolled in an ongoing recruitment and screening protocol for the Center for Neuroscience and Regenerative Medicine. Participants were recruited from the community via flyers and advertisements, with data collection taking place at two sites, Fort Belvoir Community Hospital and Walter Reed National Military Medical Center. The study was approved by Institutional Review Boards of the Health Science. Witnessed written informed consent was also obtained from each participant prior to data and sample collection. Participants were active-duty or veteran military personnel, the majority of them were from the OEF/OIF era. Exclusion criteria included severe psychiatric conditions (i.e. psychosis, schizophrenia, schizoaffective disorder, bipolar disorder, conversion disorder, or personality disorder). Participants completed a series of questionnaires, had their height and weight measured, provided a detailed account of their current medications, and provided a blood sample. Medications were classified in accordance to type, antidepressant/anxiety or sleep medications, and were subsequently dichotomized (yes/no) to indicate whether participants were taking this medication or not. As increased adiposity has been linked to an increase in sleep problems and disorders [12, 33], we calculated BMI for each participant.
PTSD symptoms were measured using the Posttraumatic Stress Disorder Checklist—Civilian Version (PCL-C). This is a self-report measure of PTSD symptoms [34] with scores ranging from 17 to 85. Participants were categorized into PTSD-Present (n = 46) or PTSD-Absent (n = 61) in accordance with the DSM IV-TR [35] criteria. Participants were classified into the PTSD-Present group based on the endorsement of moderate (≥3) or higher symptoms for (1) one or more Criterion B symptoms, (2) three or more Criterion C symptoms, and (3) two or more Criterion D symptoms.
EDS was assessed using the Epworth Sleepiness Scale (ESS). This is a widely used scale that assesses an individual’s likelihood of dozing or falling asleep during the day on a four-point Likert scale from “0” no chance of dozing to “3” high chance of dozing in eight situations of daily living [36]. Thus, the total score ranges from 0 to 24. A score of at least 13 was used to indicate moderate to severe EDS [36]. The ESS has been shown to reliably distinguish those with sleep disorders, such as narcolepsy, OSA, and idiopathic hypersomnia, from healthy controls [36].
Depression symptoms were self-reported using the Patient Health Questionnaire (PHQ-9). Scores range from 0 to 27, with higher scores indicating greater symptom severity [37]. The PHQ-9 is widely used to assess depression symptom severity in adults in clinical and research settings and has excellent sensitivity and good specificity and test–retest reliability when compared to the Structured Clinical Interview for DSM (Diagnostic and Statistical Manual of Mental Disorders) Disorders (SCID) [38].
Lifetime TBI history was determined by clinical researcher administration of the Ohio State University Traumatic Brain Injury Identification Method [39], which is a structured interview. History of TBI (present or absent) was assessed by a patient report of head injury which resulted in a period of alterations of consciousness and/or loss of consciousness.
Participants were categorized into three groups according to their PTSD symptom severity and presence of EDS: Controls (PTSD-Absent, without EDS; n = 57), PTSDw/outEDS (PTSD-Present, without EDS; n = 25), and PTSDwEDS (PTSD-Present, with EDS; n = 21). Note, there were four participants who identified as PTSD-Absent with concurrent EDS, therefore these participants were excluded from analyses.
Blood sampling
Peripheral blood samples for gene expression analysis were drawn, using PAX gene tubes. The tubes were processed per the manufacturers’ protocol, frozen at −80°C, and stored until analyzed.
Statistical methods
We performed RNA-seq with Illumina’s HiSeq 2500 Sequencing System, using paired-end sequencing. Paired-end sequencing is used to sequence both ends of a gene fragment, generating alignable sequence data. Each sample had at least 30 million reads: 15 million reads for read 1 and 15 million reads for read 2. Each read has 101 bp for its read length. For bioinformatics analysis, at first, we performed bioinformatics quality control using FastQC, version 0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). FastQC is a quality control tool that is used to assess the overall quality of a sequencing run. We then trimmed 15 bp from 5′-end, and 10 bp from 3′-end, to remove adapter contamination as well as low-quality base calls in 3′-end. We aligned to the human reference genome GRCh38, using STAR, version 2.5.3a. We counted the number of reads mapped to genes using the python package, htseq, version 0.6.1p1. Finally, we found differentially expressed genes with the cutoff of 0.10 on False Discovery Rate, this cutoff was chosen due to the exploratory nature of this investigation. This process was conducted using DESeq2, version 1.20.0 on R version 3.5.1 (2018-7-2), with the use of Bioconductor version 3.7 with BiocInstaller version 1.30.0. All significantly dysregulated genes were uploaded to QIAGEN’s Ingenuity Pathway Analysis (IPA) software (build version 389077M, content version 27821452; Qiagen, Redwood City, CA). IPA compares the imported gene list with the Ingenuity Knowledge Base, which is a list of relevant networks, upstream regulators, and algorithmically generated mechanistic networks. Identified networks are scored by significance using the IPA network score. The IPA network score is the p-value in log10. It is calculated from Fisher’s exact test of finding n1 of the focus molecules from the total number of n2 genes in the network. A score of greater than 30 was deemed as significant, which is equivalent to the p-value of 10−30. As all IPA networks were greater than 30 a two-step process of identifying candidate networks was undertaken. First, we examined each of the significant networks for overlap with the top upstream regulators identified through IPA; the networks with the most overlapping genes were considered more consistent. We also considered the current known pathology of PTSD and EDS and biological processes underlying these conditions, alongside available demographic information, for example, cancer, cardiovascular disease, and heredity disorder network, is commonly identified due to the common use of RNA expression in the cancer field providing more identifiable genes; however, the genes in this network are not necessarily associated with either PTSD, ESD, top upstream regulators, or the participants self-reported physical health. Analysis of variance (ANOVA) and chi-square (χ 2) models were utilized to determine group differences on demographic characteristics, analyzed using SPSS V24.0 (IBM Corp., Armonk, NY).
Results
Demographics
Participants were aged between 19 and 63 years (M = 37.6, SD = 11.16 years), with most being white (71.4%) and male (79.0%). The groups did not significantly differ on sex, race, military status, sleep medication use, or time since last injury (Table 1). However, the groups did significantly differ on age (p = .007), BMI (p = .010), and antidepressant/anxiety medication use (p < .001). Post hoc analysis (identified in Table 1) indicates that the PTSDwEDS group was significantly older than the Control participants; however, the PTSDw/outEDS group did not significantly differ in age compared to either the PTSDwEDS or Control groups. Similarly, the Control group had significantly lower BMI and were less likely to be using antidepressant/anxiety medication than either of the PTSD groups (with or without EDS); but the two PTSD groups did not differ significantly in BMI status or antidepressant/anxiety medication intake. The groups also significantly differed on the number of participants who reported they had sustained a TBI (p < .001). One-hundred percent of participants in the PTSDwEDS and PTSDw/outEDS groups reported having sustained a TBI, compared to only 40.4% of Control participants. Of those who had previously sustained a TBI, 84.5% were classified as mild TBI (loss of consciousness <30 min) and the groups did not significantly differ on TBI severity (χ 2 = 4.38, p = .357). Subsequent analysis was conducted using RNA-seq to determine if there were significant differences in gene dysregulation between those with and without a history of TBI in the Control group. There was a single gene that was dysregulated between these groups, and upon further analysis of the cohort, it was due to a single outlier and therefore not deemed interpretable (analysis not shown). As such, we determined that TBI did not have a significant effect on gene activity in this study and the Control group (with and without TBI) was combined.
Table 1.
Demographic Characteristics for the Sample
| Controls (n = 57) | PTSDw/outEDS (n = 26) | PTSDwEDS (n = 22) | F/χ 2 | p | |
|---|---|---|---|---|---|
| Age, years, M (SD) | 35.2 (11.2) | 37.7 (9.5)* | 43.9 (10.8)* | 5.20 | .007 |
| Sex, male, n (%) | 47 (82.5) | 20 (76.9) | 16 (72.7) | 1.00 | .606 |
| Race, n (%) | 9.68 | .469 | |||
| White | 44 (77.2) | 18 (69.2) | 13 (59.1) | ||
| Black or African American | 5 (8.8) | 5 (19.2) | 6 (27.3) | ||
| Asian | 4 (7.0) | 1 (3.8) | 1 (4.5) | ||
| Other/unknown | 4 (7.0) | 2 (7.7) | 1 (4.5) | ||
| Military status, n (%) | 14.97 | .060 | |||
| Active duty military | 46 | 14 | 14 | ||
| Reserve component | 2 | — | 1 | ||
| National guard | — | 1 | 1 | ||
| Retired from military | 8 | 6 | 5 | ||
| Veteran | 1 | 5 | 1 | ||
| Antidepressant/anxiety medications, yes, n (%) | 9 (15.7) | 19 (73.1)* | 18 (81.8)* | 40.14 | <.001 |
| Sleep medications, yes, n (%) | 5 (8.8) | 4 (15.4) | 5 (22.7) | 2.80 | .246 |
| Depression, yes, n (%) | 3 (5.3) | 23 (88.5)* | 21 (95.5)* | 78.91 | <.001 |
| BMI, M (SD) | 26.7 (3.3) | 29.2 (4.9)* | 29.0 (4.7)* | 4.78 | .010 |
| TBI | 23 (40.4) | 26 (100)* | 22 (100)* | 42.34 | <.001 |
| TSLI years, M (SD) | 16.0 (12.7) | 9.6 (9.6) | 9.9 (11.4) | 2.44 | .095 |
BMI, body mass index; TBI, traumatic brain injury; TSLI, time since the last injury.
*Denotes significant differences from the Control group in post hoc comparisons of p < .05. It is noted that there were no significant differences between the PTSD groups (with and without EDS) in any of the comparisons.
RNA-seq and IPA results
Three analyses were conducted to compare gene activity across the groups. The final gene lists from each of the three comparisons are provided in Supplementary Tables S1, S2, and S3. Quantile-quantile plots were also conducted to examine the difference between observed and expected p-values, transformed to the –log10 for the PTSDwEDS versus Controls and PTSDwEDS versus PTSDw/oEDS comparisons (Figure 1). As evidenced in Table 2, there were 153 genes shared across the three comparisons, thus there was a total of 1,973 differentially regulated genes. All of the differentially regulated genes for the PTSDwEDS versus Control (Supplementary Table S1) and the PTSDwEDS versus PTSDw/outEDS (Supplementary Table S2) comparisons were entered into IPA software. The analysis identified 10 significant networks between the PTSDwEDS versus Control (n = 5 networks) and PTSDwEDS versus PTSDw/outEDS (n = 5 networks) comparisons, here we will report on two of the top candidate networks from each of the comparisons (Table 3 provides the IPA results).
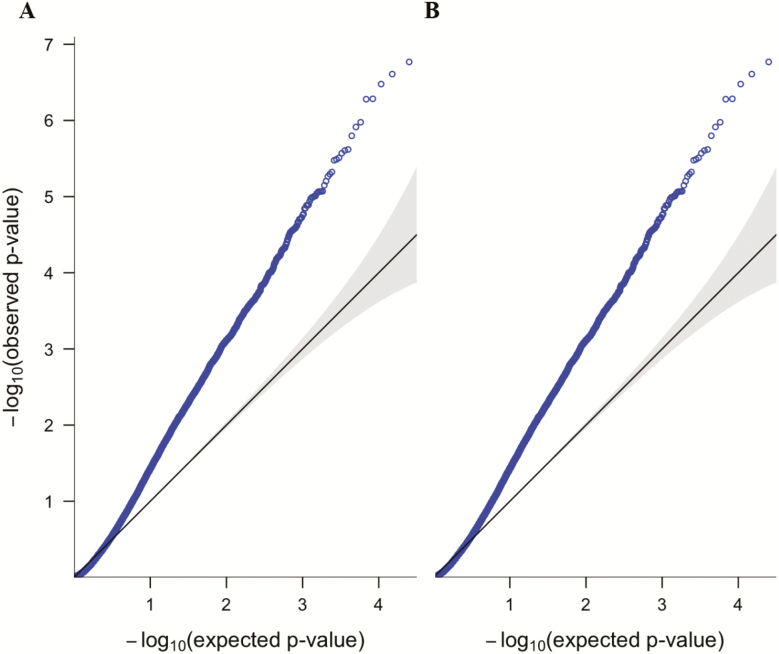
Figure 1.
Quantile-quantile (QQ) plots of observed −log10 transformed p-values, plotted to quantiles of the expected –log10 transformed p-values from uniform distributions for (A) PTSDwEDS versus Controls and (B) PTSDwEDS versus PTSDw/oEDS.
Table 2.
Number of Differentially Regulated Genes Identified by RNA-seq for Each Comparison
| Comparison | Up | Down | Total | No. of unique genes |
|---|---|---|---|---|
| PTSDw/outEDS vs Controls | 1 | 1 | 2 | 1 |
| PTSDwEDS vs Controls | 155 | 96 | 251 | 99 |
| PTSDwEDS vs PTSDw/outEDS | 1,016 | 857 | 1,873 | 1,873 |
| Total | 2,126 | 1,973 |
The number of unique genes is calculated by subtracting the genes that were shared in other analyses.
Table 3.
Networks Identified Through IPA Analysis
| Controls vs PTSDwEDS | PTSDwEDS vs PTSDw/outEDS | ||
|---|---|---|---|
| Network | IPA network score | Network | IPA network score |
| Organismal Injury and Abnormalities | 59 | Cancer, Cardiovascular disease, Hereditary Disorder | 40 |
| Cellular Function and Maintenance | 53 | Carbohydrate Metabolism | 40 |
| Gene Expression | 38 | RNA Posttranscriptional Modification | 38 |
| Neurological Disease | 33 | Molecular Transport | 38 |
| Immunological Disease | 31 | Cellular Function and Maintenance | 38 |
Bolded font identifies the four candidate networks selected for further analysis. The candidate networks were chosen firstly by assessing the significance of the networks (IPA network score). For network scores that were similar we also considered how consistent genes within the identified networks were to upstream regulators (identified through IPA) and known biological processes consistent with the pathology of PTSD and EDS.
PTSDw/outEDS versus Controls
IPA analysis was not possible for this comparison as the RNA-seq analysis identified only two genes which were dysregulated between the groups: RAP1 GTPase Activating Protein (RAP1GAP) which was upregulated (log-fold change [FClog] = 1.84, padj = .058) and TBC1 Domain Family Member 3E (TBC1D3E) which was downregulated (FClog = −3.19, padj = .059).
PTSDwEDS versus Controls
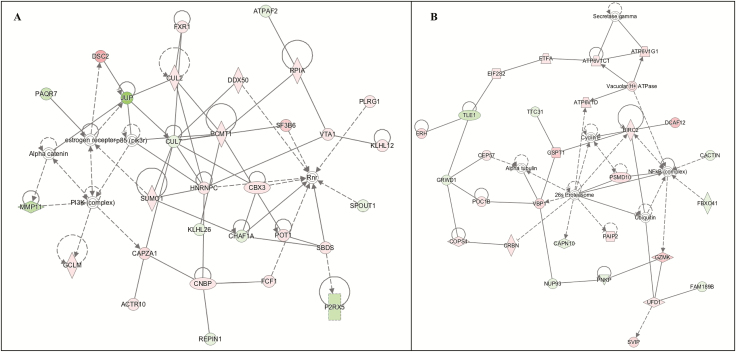
The two candidate networks and genes identified in this pathway are defined in Tables 4 and 5. The highest scoring pathways centered on organismal injury and abnormalities (Network 1; Figure 2, A). The highest log-fold changes in this network were observed for JUP (FClog = −.80, padj = .033) which was downregulated, and DISC2 (FClog = .70, padj = .072) which was upregulated in participants with PTSDwEDS when compared to Controls. The second network was centered on cellular function and maintenance (Network 2; Figure 2, B). In Network 2, the highest upregulated gene was DCAF12 (FClog = .53, padj = .097) and TLE1 (FClog = −.38, padj = .077) was the highest downregulated gene in the PTSDwEDS group compared to Controls. Genes in both the networks were associated with ubiquitination, immune response, ATP modulation, and modulation of the P13K–Akt pathway. Indeed, Network 2 (Figure 2, B) had both NF-κB (complex) and ubiquitin (complex) hubs.
Table 4.
IPA Network 1
| Gene | Gene name | Log-fold change | p adj |
|---|---|---|---|
| Upregulated | |||
| DSC2 | Desmocollin 2 | 0.70200 | .072 |
| SF3B6 | Splicing Factor 3b Subunit 6 | 0.44800 | .060 |
| CAPZA1 | Capping Actin Protein Of Muscle Z-Line Subunit Alpha 1 | 0.32800 | .098 |
| POT1 | Protection of Telomeres 1 | 0.26000 | .078 |
| SBDS | SBDS Ribosome Maturation Factor | 0.25900 | .072 |
| CBX3 | Chromobox 3 | 0.25400 | .072 |
| GCLM | Glutamate-Cysteine Ligase Modifier Subunit | 0.25300 | .097 |
| FXR1 | Fragile X Mental Retardation, Autosomal Homolog 1 | 0.22500 | .071 |
| DDX50 | DExD-Box Helicase 50 | 0.22200 | .072 |
| PLRG1 | Pleiotropic Regulator 1 | 0.21500 | .069 |
| CNBP | CCHC-Type Zinc Finger Nucleic Acid Binding Protein | 0.21000 | .072 |
| SUMO1 | Small Ubiquitin Like Modifier 1 | 0.20700 | .071 |
| HNRNPC | Heterogeneous Nuclear Ribonucleoprotein C (C1/C2) | 0.19200 | .078 |
| FCF1 | FCF1 RRNA-Processing Protein | 0.18000 | .078 |
| RPIA | Ribose 5-Phosphate Isomerase A | 0.16300 | .097 |
| ACTR10 | Actin Related Protein 10 | 0.16200 | .097 |
| VTA1 | Vesicle Trafficking 1 | 0.15800 | .098 |
| CUL2 | Cullin 2 | 0.14500 | .078 |
| KLHL12 | Kelch Like Family Member 12 | 0.14400 | .078 |
| Downregulated | |||
| JUP | Junction Plakoglobin | −0.8030 | .033 |
| P2RX5 | Purinergic Receptor P2X 5 | −0.4220 | .078 |
| PAQR7 | Progestin and AdipoQ Receptor Family Member 7 | −0.4080 | .071 |
| CHAF1A | Chromatin Assembly Factor 1 Subunit A | −0.2320 | .036 |
| ATPAF2 (ATP12) | ATP Synthase Mitochondrial F1 Complex Assembly Factor 2 | −0.2080 | .072 |
| SPOUT1 | SPOUT Domain Containing Methyltransferase 1 | −0.1860 | .071 |
| REPIN1 | Replication Initiator1 | −0.1860 | .071 |
| CUL7 | Cullin 7 | −0.1840 | .071 |
Table 5.
IPA Network 2
| Gene | Gene names | Log-fold change | p adj |
|---|---|---|---|
| Upregulated | |||
| DCAF12 | DDB1 And CUL4 Associated Factor 12 | 0.530853 | .097 |
| GZMK | Granzyme K | 0.454456 | .090 |
| GSPT1 | G1 To S Phase Transition | 0.450250 | .079 |
| SVIP | Small VCP Interacting Protein | 0.357731 | .084 |
| VBP1 | VHL Binding Protein 1 | 0.339924 | .084 |
| ATP6V1G1 | ATPase H+ Transporting V1 Subunit G1 | 0.304901 | .097 |
| PSMD10 | Proteasome 26S Subunit, Non-ATPase 10 | 0.292566 | .072 |
| PAIP2 | Poly(A) Binding Protein Interacting Protein 2 | 0.255587 | .071 |
| POC1B | POC1 Centriolar Protein B | 0.255553 | .062 |
| CRBN | Cereblon | 0.254111 | .080 |
| COPS4 | COP9 Signalosome Subunit 4 | 0.243402 | .081 |
| ATP6V1C1 | ATPase H+ Transporting V1 Subunit C1 | 0.234761 | .072 |
| CEP57 | Centrosomal Protein 57 | 0.232703 | .099 |
| BIRC2 | Baculoviral IAP Repeat Containing 2 | 0.209530 | .097 |
| ETFA | Electron Transfer Flavoprotein Subunit Alpha | 0.176101 | .080 |
| UFD1 | Ubiquitin Recognition Factor in ER Associated Degradation 1 | 0.164150 | .085 |
| EIF2S2 | Eukaryotic Translation Initiation Factor 2 Subunit Beta | 0.161477 | .073 |
| Downregulated | |||
| GRWD1 | Glutamate Rich WD Repeat Containing 1 | −0.382867 | .077 |
| TTC31 | Tetratricopeptide Repeat Domain 31 | −0.225393 | .071 |
| NUP93 | Nucleoporin 93 | −0.207949 | .072 |
| ATP6V1D | ATPase H+ Transporting V1 Subunit D | −0.201461 | .071 |
| PNKP | Polynucleotide Kinase 3′-Phosphatase | −0.177668 | .071 |
| FBXO41 | F-Box Protein 41 | −0.154091 | .097 |
| CACTIN | Cactin, Spliceosome C Complex Subunit | −0.142608 | .097 |
| CAPN10 | Calpain 10 | −0.289406 | .071 |
Figure 2.
The networks identified by IPA for PTSDwEDS versus Controls: (A) Network 1—Organismal Injury and Abnormalities and (B) Network 2—Cellular Function and Maintenance. Green indicates that the gene is downregulated and red indicates that the gene is upregulated, with increased color saturation representing more extreme measurement in the dataset. Solid lines represent interactions, nontargeting interactions, or correlations between chemicals, proteins, or RNA. Arrowed lines represent activation, causation, expression, localization, membership, modification, molecular cleavage, phosphorylation, protein–DNA interactions, protein–TNA interaction, regulation of binding, and transcription. Shapes represent molecule type (double circle = complex/group; square = cytokine; diamond = enzyme; inverted triangle = kinase; triangle = phosphatase; oval = transcription regulator; trapezoid = transporter; circle = other).
PTSDwEDS versus PTSDw/outEDS
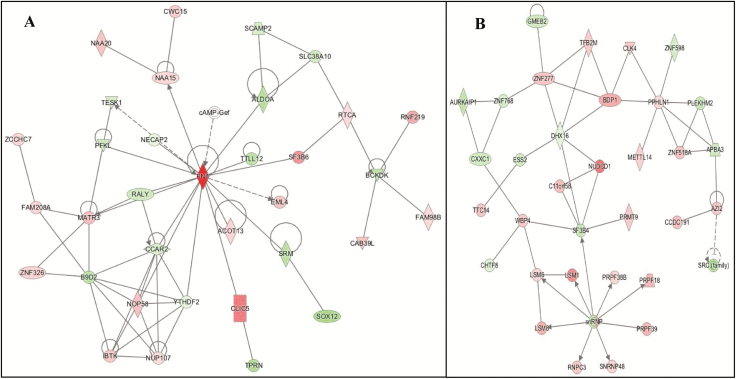
The networks identified by IPA were very similar in scores (Table 3), thus, to determine the top candidate pathways we utilized a two-step process. Firstly, we considered the top upstream regulators; in this analysis the top regulators included metabolic and mitochondrial genes. Then, after considering the known pathology and biological processes associated with PTSD and EDS, the Carbohydrate Metabolism (Network 3; Figure 3, A) and the RNA Posttranscriptional Modification (Network 4; Figure 3, B) networks were selected and they are identified and defined in Tables 6 and 7, respectively. Genes within these networks were associated with ubiquitination, neurodegeneration, mitochondrial function, glucocorticoid modulation, sleep disturbances (i.e. insomnia), and circadian regulation (both peripheral and central clock regulators). In Network 3, FN1 (FClog = .99. padj = .084), CLIC5 (FClog = .48, padj = .041), and SF3B6 (FClog = .41, padj = .048) were highly upregulated. Alongside, SOX12 (FClog = −.34, padj = .015) and TPRN (FClog = −.31, padj = .048) were significantly downregulated. For Network 4, the top significant upregulated genes were NUDCD1 (FClog = .51, padj = .013) and LSM1 (FClog = .43, padj = .025) and downregulated were TFB2M (FClog = −.25, padj = .047) and SF3B4 (FClog = −.23, padj = .028).
Figure 3.
The networks identified by IPA for PTSDwEDS versus PTSDw/outEDS: (A) Network 3—Carbohydrate Metabolism and (B) Network 4—RNA Posttranscriptional Modification. Green indicates that the gene is downregulated and red indicates that the gene is upregulated, with increased color saturation representing more extreme measurement in the dataset. Solid lines represent interactions, nontargeting interactions, or correlations between chemicals, proteins, or RNA. Arrowed lines represent activation, causation, expression, localization, membership, modification, molecular cleavage, phosphorylation, protein–DNA interactions, protein–TNA interaction, regulation of binding, transcription. Shapes represent molecule type (double circle = complex/group; square = cytokine; diamond = enzyme; inverted triangle = kinase; triangle = phosphatase; oval = transcription regulator; trapezoid = transporter; circle = other).
Table 6.
IPA Network 3
| Gene | Gene name | Log-fold change | p adj |
|---|---|---|---|
| Upregulated | |||
| FN1 | Fibronectin 1 | 0.988145 | .084 |
| RNF219 | Ring Finger Protein 219 | 0.615292 | 1.00 |
| CLIC5 | Chloride Intracellular Channel 5 | 0.480689 | .041 |
| SF3B6 | Splicing Factor 3b Subunit 6 | 0.411130 | .048 |
| MATR3 | Matrin 3 | 0.273653 | .025 |
| NOP58 | NOP58 Ribonucleoprotein | 0.254913 | .081 |
| NAA20 | N(Alpha)-Acetyltransferase 20, NatB Catalytic Subunit | 0.249584 | .055 |
| CAB39L | Calcium Binding Protein 39 Like | 0.246969 | .034 |
| EML4 | EMAP Like 4 | 0.244383 | .063 |
| IBTK | Inhibitor of Bruton Tyrosine Kinase | 0.230292 | .043 |
| CWC15 | CWC15 Spliceosome Associated Protein Homolog | 0.222963 | .102 |
| ZNF326 | Zinc Finger Protein 326 | 0.197428 | .042 |
| ACOT13 | Acyl-CoA Thioesterase 13 | 0.184483 | .038 |
| FAM208A | Transcription Activation Suppressor | 0.184261 | .040 |
| ZCCHC7 | Zinc Finger CCHC-Type Containing 7 | 0.170832 | .048 |
| NAA15 | N(Alpha)-Acetyltransferase 15, NatA Auxiliary Subunit | 0.170033 | .058 |
| RTCA | RNA 3′-Terminal Phosphate Cyclase | 0.163013 | .032 |
| FAM98B | Family With Sequence Similarity 98 Member B | 0.161480 | .094 |
| NUP107 | Nucleoporin 107 | 0.158365 | .029 |
| Downregulated | |||
| SOX12 | SRY-Box 12 | −0.345513 | .015 |
| TPRN | Taperin | −0.305638 | .047 |
| ALDOA | Aldolase, Fructose-Bisphosphate A | −0.280285 | .064 |
| BCKDK | Branched Chain Ketoacid Dehydrogenase Kinase | −0.268262 | .057 |
| TTLL12 | Tubulin Tyrosine Ligase Like 12 | −0.268135 | .003 |
| SRM | Spermidine Synthase | −0.267465 | .037 |
| B9D2 | B9 Domain Containing 2 | −0.261256 | .089 |
| PFKL | Phosphofructokinase, Liver Type | −0.202250 | .084 |
| RALY | RALY Heterogeneous Nuclear Ribonucleoprotein | −0.201685 | .077 |
| SLC38A10 | Solute Carrier Family 38 Member 10 | −0.198797 | .072 |
| TESK1 | Testis Associated Actin Remodeling Kinase 1 | −0.182732 | .050 |
| SCAMP2 | Secretory Carrier Membrane Protein 2 | −0.163219 | .055 |
| CCAR2 | Cell Cycle And Apoptosis Regulator 2 | −0.157864 | .090 |
| NECAP2 | NECAP Endocytosis Associated 2 | −0.111572 | .069 |
| YTHDF2 | YTH N6-Methyladenosine RNA Binding Protein 2 | −0.081420 | .076 |
Table 7.
IPA Network 4
| Gene | Gene name | Log-fold change | p adj |
|---|---|---|---|
| Upregulated | |||
| NUDCD1 | NudC Domain Containing 1 | 0.513907 | .012 |
| LSM1 | LSM1 Homolog, mRNA Degradation Associated | 0.439193 | .025 |
| BDP1 | B Double Prime 1, Subunit Of RNA Polymerase III Transcription Initiation Factor IIIB | 0.337677 | .070 |
| LSM8 | LSM8 Homolog, U6 Small Nuclear RNA Associated | 0.33675 | .027 |
| PRPF18 | Pre-mRNA Processing Factor 18 | 0.302657 | .056 |
| PRPF39 | Pre-mRNA Processing Factor 39 | 0.301832 | .020 |
| CCDC191 | Coiled-Coil Domain Containing 191 | 0.272445 | .064 |
| ZNF518A | Zinc Finger Protein 518A | 0.257776 | .081 |
| TTC14 | Tetratricopeptide Repeat Domain 14 | 0.247466 | .065 |
| ZNF277 | Zinc Finger Protein 277 | 0.241244 | .065 |
| AZI2 | 5-Azacytidine Induced 2 | 0.237125 | .086 |
| WBP4 | WW Domain Binding Protein 4 | 0.229413 | .072 |
| PRMT9 | Protein Arginine Methyltransferase 9 | 0.226234 | .059 |
| CLK4 | CDC Like Kinase 4 | 0.225283 | .094 |
| C11orf58 | Chromosome 11 Open Reading Frame 58 | 0.216792 | .014 |
| METTL14 | Methyltransferase Like 14 | 0.200934 | .042 |
| SNRNP48 | Small Nuclear Ribonucleoprotein U11/U12 Subunit 48 | 0.194575 | .063 |
| LSM6 | LSM6 Homolog, U6 Small Nuclear RNA And MRNA Degradation Associated | 0.187706 | .065 |
| PPHLN1 | Periphilin 1 | 0.167741 | .019 |
| ZNF598 | Zinc Finger Protein 598 | −0.1737 | .030 |
| TFB2M | Transcription Factor B2, Mitochondrial | −0.25012 | .046 |
| RNPC3 | RNA Binding Region (RNP1, RRM) Containing 3 | ||
| PRP38B | Pre-mRNA Processing Factor 38B | ||
| Downregulated | |||
| SRC (Family) | SRC Proto-Oncogene, Non-Receptor Tyrosine Kinase | −0.29017 | .071 |
| AURKAIP1 | Aurora Kinase A Interacting Protein 1 | −0.24699 | .083 |
| SF3B4 | Splicing Factor 3b Subunit 4 | −0.23496 | .027 |
| PLEKHM2 | Pleckstrin Homology And RUN Domain Containing M2 | −0.21903 | .020 |
| GMEB2 | Glucocorticoid Modulatory Element Binding Protein 2 | −0.20078 | .026 |
| APBA3 | Amyloid Beta Precursor Protein Binding Family A Member 3 | −0.19655 | .051 |
| CXXC1 | CXXC Finger Protein 1 | −0.19296 | .096 |
| ZNF768 | Zinc Finger Protein 768 | −0.17737 | .095 |
| CHTF8 | Chromosome Transmission Fidelity Factor 8 | −0.13347 | .042 |
| DHX16 | DEAH-Box Helicase 16 | −0.1141 | .084 |
| ESS2 | Ess-2 Splicing Factor Homolog |
Discussion
In this study, we report differential gene expression comparing military personnel and veterans with PTSD, with or without EDS, and Controls (no PTSD and no sleep problems). There were only two genes significantly dysregulated between PTSDw/outEDS and the Control group: RAP1GAP and TBC1D3E. This limited dysregulation between these groups does contradict prior results suggesting significant gene dysregulation due to the presence of PTSD [4]. One potential explanation for this is that there may be limited heterogeneity between the Control and PTSDw/outEDS groups in terms of PTSD symptoms. Total PCL-C score in the Control population ranged from 17 to 51, suggesting that some level of PTSD symptoms were experienced in all groups. Alternatively, this may be due to some other shared variance between these groups given the military context. Indeed, we found significant variation in the number of genes that were dysregulated between the Controls versus PTSDwEDS (251 genes; 98 genes were unique to this analysis) and the PTSDwEDS versus PTSDw/outEDS groups (1,873 genes). Given that we were unable to control for age and other demographic characteristics that differed significantly between the Controls compared to the PTSD groups in this analysis, we have decided to predominantly focus this discussion on the comparison between PTSDwEDS and PTSDw/outEDS. These groups did not significantly differ on any of the demographic characteristics measured, making the high number of genes that were differentially regulated between the groups even more compelling. Although these results present an explorative investigation, these results may present novel insights into the effects of EDS with concurrent PTSD on gene expression. Specifically, these findings indicate that when EDS co-occurs with PTSD there may be significant additional biological impact and gene dysregulation. Thus, additional monitoring and intervention may be warranted to improve outcomes for military personnel and veterans with PTSD and EDS.
In support of this, we identified several sleep, fatigue, and circadian-related genes across the networks that were dysregulated in military personnel and veterans with PTSD and EDS, compared to those with PTSD and no significant daytime sleepiness (Networks 3 and 4). In Network 3, FN1, a glycoprotein that encodes fibronectin, was strongly upregulated (FC log = .98). FN1 has been shown to be significantly altered in the serum of both humans with obstructive sleep apnea-hypopnea syndrome (OSAHS) and rats with chronic intermittent hypoxia (CIH) compared to samples with no OSAHS or CIH, indicating that both conditions cause significant alterations in protein composition [40]. In Network 4, RNA posttranscriptional modification network there were several genes associated with sleep problems, including insomnia and circadian regulation. Specifically, three genes that were significantly upregulated (TFB2M, WBP4, and SNRNP48) were found to be dysregulated in the thalamus of patients with Chinese fatal familial insomnia (postmortem) compared with Controls [28]. Importantly, TFB2M is required for the transcription of mitochondrial genes and thus mitochondrial function [41]. Given the importance of mitochondrial bioenergetics to both sleep and health, TFB2M has also been implicated as a candidate marker of chronic fatigue [42]. Therefore, EDS when experienced concurrently with PTSD is associated with the dysregulation of genes involved in sleep, fatigue, and circadian regulation. However, the cause of this reported EDS remains to be determined. Although the participants in this study were not treatment-seeking, we were unable to ascertain if these personnel had concurrent sleep problems such as insomnia or OSA. Sleep disorders such as OSA and insomnia have commonly been reported in military and veteran populations [12]. It is noted, however, that age, sex, military status, sleep and antidepressant/anxiety medication use, BMI, TBI, and other key participant demographics which may indicate a greater propensity to sleep problems did not differ between the PTSDwEDS and PTSDw/outEDS groups.
Genes associated with circadian regulation and functioning were downregulated in both Networks 3 and 4. In Network 4, PLEKHM2 was significantly downregulated (FC = −.21), which has been associated with both insomnia [43] and advanced sleep phase disorder [44], which is a circadian disorder characterized by an early wake-up phenotype. CCAR2 was also significantly downregulated (FC = −.16, N3) and this gene has been shown to be important for regulating the circadian clock (through BMAL1 and CLOCK expression) and metabolism [45]. Together, these findings may be indicative of dysregulation to the circadian clock in participants with both EDS and PTSD. As such, the use of habitual sleep monitoring devices such as actigraphy may be helpful to monitor and identify participants at risk of circadian rhythm problems in clinical settings, especially in patients presenting with PTSD and concurrent EDS.
Glutamate is an important neurotransmitter that has been linked to the regulation of sleep architecture (i.e. initiation of rapid eye movement [REM] sleep) and neurodegenerative disease. Together with gamma-aminobutyric acid (GABA), it plays a key role in memory formation and, particularly, the encoding of emotional and fear memories, which underlie anxiety disorders such as PTSD [46]. Thus, it is unsurprising that the glutamatergic and GABAergic systems have been implicated with the hypothesis that PTSD is initiated via memory processing in REM [47]. Across all four networks, there was significant dysregulation in genes that were associated with glutamate production and regulation, e.g. GMEB2 (N4). Thus, glutamate production and transport may be dysregulated in participants with EDS and PTSD. Calls for treatments that specifically target the glutamatergic system, beyond ketamine, are already underway for many mental health disorders including depression [48], alcohol use disorder [49], and schizophrenia [50]. As such, if this finding is able to be replicated across studies and populations of people with PTSD and concurrent EDS, this may provide initial insights for potential future pharmacological interventions for these patients.
Across all four candidate networks, we identified significant dysregulation in genes associated with ubiquitination. Ubiquitin has been shown to play a role in sleep regulation, including being implicated in an insomnia phenotype [51], and shown to disrupt circadian rhythms [52] in preclinical models. Therefore, these current findings may extend previous research suggesting ubiquitin is implicated in sleep disturbances and regulation, as it may also be associated with symptoms of EDS. The direction of this effect, i.e. if ubiquitin is disrupted due to factors associated with EDS or EDS in participants with PTSD is precipitated, in part, by dysregulation of ubiquitination, remains to be determined. Longitudinal studies that track gene activity in cohorts with PTSD and sleep disturbances over time are critically needed.
Disturbed sleep has consistently been identified as a risk factor of obesity and metabolic syndromes. An important determination of energy, weight, and metabolism within the body is adenosine triphosphate (ATP). ATP-related genes were dysregulated throughout Networks 1, 2, and 3. Thus, participants with both EDS and PTSD may be at an increased risk of metabolic disturbances, such as insulin resistance. Longitudinal tracking of this relationship and screening for metabolic diseases is needed to determine the directionality of these associations.
Limitations
This study is an exploration of potential genes that may be differentially regulated in patients with PTSD with comorbid EDS. Our study has a number of strengths, including the use of RNA-seq technology on a relatively young cohort of active-duty military personnel and veterans. However, there are limitations, including a relatively small number of participants, the majority were male, and white/Caucasian military personnel and veterans of the OIF/OEF era. Thus, our findings are not necessarily generalizable and require replication in other populations. Another limitation is our reliance on the self-reported measures of both sleepiness and PTSD. The ESS is an indicator of an increased daytime propensity for sleep and consequently means that we are unable to determine which factors, or potentially which sleep disorders, underlie the EDS experienced by these participants. As such, objective measurement of sleep, using polysomnography and/or actigraphy, is needed to validate and extend these findings. We are also unable to determine whether the EDS preceded or was a consequence of PTSD or some other unmeasured feature within this population, therefore longitudinal tracking is recommended. It is also noted that almost all personnel with PTSD in this study reported concomitant depression symptoms. This finding supports a significant body of epidemiological research reporting high comorbidity rates in military populations [53–55] which may be partially due to overlap in clinical presentation. In this study, there is some overlap between symptom items in the PHQ-9 and the PCL-C. Therefore, further research that parses out the effects of PTSD and depression symptoms, using clinical assessments, may be necessary, especially in nonmilitary populations. Although preliminary, the results of this study can be utilized to frame future research to validate these findings in other community-based populations. If validated in further investigations, the findings from this study indicate that PTSD with comorbid EDS is associated with differential gene regulation in pathways related to sleep and circadian disturbances, glutamate and ubiquitin regulation, and metabolism; consequently, health may be affected. Thus, military personnel, presenting with PTSD and concurrent EDS, may need additional monitoring and could potentially benefit from early intervention to prevent long-term health and sleep disturbances. In clinical settings, exploring the route cause for a patient’s EDS and consequent treatment either through cognitive behavioral therapy for insomnia, sleep hygiene education, pharmacological interventions, or other individualized treatment may help to both improve PTSD symptom severity and mitigate long-term health risks.
Conclusions
The results of this preliminary investigation indicate that EDS with concurrent PTSD may be associated with dysregulation of genes across many networks which compromise health and exacerbate PTSD symptoms. Further research with prospective longitudinal tracking of gene dysregulation in the wider community with PTSD and EDS is vital. However, these findings highlight that there may be a need for early interventions that focus on improving EDS symptoms, in military personnel and veterans with PTSD.
Supplementary Material
Acknowledgments
We would like to thank the participants, without whom, this research would not be possible. Also, we would like to thank Center for Nueroscience and Regenerative Medicine for their support of this research.
Financial Disclosure: None.
Nonfinancial Disclosures: CLP, JMG, MJR, VAG, KE, H-SK, CL, KD, PT, and SM have no conflicts to report. SY reports being a director of Yotta Biomed.
References
- 1. U.S. Department of Veterans Affairs. How common is PTSD in veterans?Washington, DC; 2018. https://www.ptsd.va.gov/understand/common/common_veterans.asp. Accessed December 3, 2019. [Google Scholar]
- 2. Fulton JJ, et al. . The prevalence of posttraumatic stress disorder in Operation Enduring Freedom/Operation Iraqi Freedom (OEF/OIF) veterans: a meta-analysis. J Anxiety Disord. 2015;31:98–107. [DOI] [PubMed] [Google Scholar]
- 3. Cattaneo A, et al. . The human BDNF gene: peripheral gene expression and protein levels as biomarkers for psychiatric disorders. Transl Psychiatry. 2016;6(11):e958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuan PF, et al. . Gene expression associated with PTSD in world trade center responders: an RNA sequencing study. Transl Psychiatry. 2017;7(12):1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daskalakis NP, et al. . Recent genetics and epigenetics approaches to PTSD. Curr Psychiatry Rep. 2018;20(5):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castro-Vale I, et al. . Genetics of glucocorticoid regulation and posttraumatic stress disorder—what do we know? Neurosci Biobehav Rev. 2016;63:143–157. [DOI] [PubMed] [Google Scholar]
- 7. Rusch HL, et al. . Gene expression differences in PTSD are uniquely related to the intrusion symptom cluster: a transcriptome-wide analysis in military service members. Brain Behav Immun. 2019;80:904–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen DA, et al. . Uncovering residual effects of chronic sleep loss on human performance. Sci Transl Med. 2010;2(14):14ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Slater G, et al. . Excessive daytime sleepiness in sleep disorders. J Thorac Dis. 2012;4(6):608–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang H, et al. . Genome-wide association analysis of self-reported daytime sleepiness identifies 42 loci that suggest biological subtypes. Nat Commun. 2019;10(1):3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McLay RN, et al. . Insomnia is the most commonly reported symptom and predicts other symptoms of post-traumatic stress disorder in U.S. service members returning from military deployments. Mil Med. 2010;175(10):759–762. [DOI] [PubMed] [Google Scholar]
- 12. Mysliwiec V, et al. . Sleep disorders in US military personnel: a high rate of comorbid insomnia and obstructive sleep apnea. Chest. 2013;144(2):549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mysliwiec V, et al. . Sleep disorders and associated medical comorbidities in active duty military personnel. Sleep. 2013;36(2):167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Plumb TR, et al. . Sleep disturbance is common among servicemembers and veterans of Operations Enduring Freedom and Iraqi Freedom. Psychol Serv. 2014;11(2):209–219. [DOI] [PubMed] [Google Scholar]
- 15. Neylan TC, et al. . Sleep disturbances in the Vietnam generation: findings from a nationally representative sample of male Vietnam veterans. Am J Psychiatry. 1998;155(7):929–933. [DOI] [PubMed] [Google Scholar]
- 16. Westermeyer J, et al. . Correlates of daytime sleepiness in patients with posttraumatic stress disorder and sleep disturbance. Prim Care Companion J Clin Psychiatry. 2010;12(2):e1–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Williams SG, et al. . Sleep disorders in combat-related PTSD. Sleep Breath. 2015;19(1):175–182. [DOI] [PubMed] [Google Scholar]
- 18. Khazaie H, et al. . Sleep disturbances in veterans with chronic war-induced PTSD. J Inj Violence Res. 2016;8(2):99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pace-Schott EF, et al. . Sleep and REM sleep disturbance in the pathophysiology of PTSD: the role of extinction memory. Biol Mood Anxiety Disord. 2015;5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Liempt S, et al. . Impact of impaired sleep on the development of PTSD symptoms in combat veterans: a prospective longitudinal cohort study. Depress Anxiety. 2013;30(5):469–474. [DOI] [PubMed] [Google Scholar]
- 21. Mellman TA, et al. . A relationship between REM sleep measures and the duration of posttraumatic stress disorder in a young adult urban minority population. Sleep. 2014;37(8):1321–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Straus LD, et al. . Sleep variability in military-related PTSD: a comparison to primary insomnia and healthy controls. J Trauma Stress. 2015;28(1):8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwartz JRL, et al. . Recognition and management of excessive sleepiness in the primary care setting. Prim Care Companion J Clin Psychiatry. 2009;11(5):197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Institute of Medicine (US) Committee on Sleep Medicine and Research. Functional and Economic Impact of Sleep Loss and Sleep-Related Disorders. In: Colten H, Altevogt B, eds. Sleep Disorders and Sleep Deprivation: An Unmet Public Health Problem. Washington DC: National Academies Press (US); 1996:137–172. https://www.ncbi.nlm.nih.gov/books/NBK19958/. Accessed December 3, 2019. [PubMed] [Google Scholar]
- 25. Cirelli C, et al. . Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem. 2005;94(5):1411–1419. [DOI] [PubMed] [Google Scholar]
- 26. Davis CJ, et al. . P2X7 receptors in body temperature, locomotor activity, and brain mRNA and lncRNA responses to sleep deprivation. Am J Physiol Regul Integr Comp Physiol. 2016;311(6):R1004–R1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bernardini C, et al. . Genome-wide gene expression profiling of human narcolepsy. Gene Expr. 2012;15(4):171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tian C, et al. . Comparative analysis of gene expression profiles between cortex and thalamus in Chinese fatal familial insomnia patients. Mol Neurobiol. 2013;48(1):36–48. [DOI] [PubMed] [Google Scholar]
- 29. Zhang R, et al. . A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111(45):16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lane JM, et al. . Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49(2):274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Livingston WS, et al. . Improved sleep in military personnel is associated with changes in the expression of inflammatory genes and improvement in depression symptoms. Front Psychiatry. 2015;6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pattinson CL, et al. . Elevated tau in military personnel relates to chronic symptoms following traumatic brain injury. J Head Trauma Rehabil. 2020;35(1):66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moraes W, et al. . Association between body mass index and sleep duration assessed by objective methods in a representative sample of the adult population. Sleep Med. 2013;14(4):312–318. [DOI] [PubMed] [Google Scholar]
- 34. Weathers FW, et al. . The PTSD checklist (PCL): reliability, validity, and diagnostic utility. Presented at: Annual Meeting of the International Society for Traumatic Stress Studies; October, 1993; San Antonio, TX.
- 35. American Psychiatric Association. DSM-IV: Diagnostic and Statistical Manual of Mental Disorders. In: Widiger T, Frances A, Pincus H, First M, Ross R, Davis W, eds. Washington, DC: American Psychiatric Press Inc.; 1994. [Google Scholar]
- 36. Johns MW. A new method for measuring daytime sleepiness: the Epworth Sleepiness Scale. Sleep J Sleep Res Sleep Med. 1991;14(6):540–545. [DOI] [PubMed] [Google Scholar]
- 37. Kroenke K, et al. . The PHQ-9: validity of a brief depression severity measure. J Gen Intern Med. 2001;16(9):606–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Steel JL, et al. . Measuring depression and PTSD after trauma: common scales and checklists. Injury. 2011;42(3):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Corrigan JD, et al. . Initial reliability and validity of the Ohio State University TBI Identification Method. J Head Trauma Rehabil. 2007;22(6):318–329. [DOI] [PubMed] [Google Scholar]
- 40. Zhang H, et al. . The contribution of chronic intermittent hypoxia to OSAHS: from the perspective of serum extracellular microvesicle proteins. Metabolism. 2018;85:97–108. [DOI] [PubMed] [Google Scholar]
- 41. Litonin D, et al. . Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J Biol Chem. 2010;285(24):18129–18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Presson AP, et al. . Integrated weighted gene co-expression network analysis with an application to chronic fatigue syndrome. BMC Syst Biol. 2008;2:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ding M, et al. . Integrative analysis of genome-wide association study and brain region related enhancer maps identifies biological pathways for insomnia. Prog Neuropsychopharmacol Biol Psychiatry. 2018;86:180–185. [DOI] [PubMed] [Google Scholar]
- 44. Li J, et al. . Predicting and analyzing early wake-up associated gene expressions by integrating GWAS and eQTL studies. Biochim Biophys Acta Mol Basis Dis. 2018;1864(6 Pt B):2241–2246. [DOI] [PubMed] [Google Scholar]
- 45. Magni M, et al. . Cell cycle and apoptosis regulator 2 at the interface between DNA damage response and cell physiology. Mutat Res. 2018;776:1–9. [DOI] [PubMed] [Google Scholar]
- 46. Riaza Bermudo-Soriano C, et al. . New perspectives in glutamate and anxiety. Pharmacol Biochem Behav. 2012;100(4):752–774. [DOI] [PubMed] [Google Scholar]
- 47. Murkar ALA, et al. . Consolidative mechanisms of emotional processing in REM sleep and PTSD. Sleep Med Rev. 2018;41:173–184. [DOI] [PubMed] [Google Scholar]
- 48. Gerhard DM, et al. . Emerging treatment mechanisms for depression: focus on glutamate and synaptic plasticity. Drug Discov Today. 2016;21(3):454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goodwani S, et al. . Metabotropic and ionotropic glutamate receptors as potential targets for the treatment of alcohol use disorder. Neurosci Biobehav Rev. 2017;77:14–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mouchlianitis E, et al. . Treatment-resistant schizophrenia patients show elevated anterior cingulate cortex glutamate compared to treatment-responsive. Schizophr Bull. 2016;42(3):744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stavropoulos N, et al. . Insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron. 2011;72(6):964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pfeffer M, et al. . Disturbed sleep/wake rhythms and neuronal cell loss in lateral hypothalamus and retina of mice with a spontaneous deletion in the ubiquitin carboxyl-terminal hydrolase L1 gene. Neurobiol Aging. 2012;33(2):393–403. [DOI] [PubMed] [Google Scholar]
- 53. Colston MJ, et al. . Posttraumatic stress disorder, depression, and other comorbidities: clinical and systems approaches to diagnostic uncertainties. Fed Pract. 2016;33(11):37–45. [PMC free article] [PubMed] [Google Scholar]
- 54. Marshall BD, et al. . Coincident posttraumatic stress disorder and depression predict alcohol abuse during and after deployment among Army National Guard soldiers. Drug Alcohol Depend. 2012;124(3):193–199. [DOI] [PubMed] [Google Scholar]
- 55. Belsher BE, et al. . Population impact of PTSD and depression care for military service members: reach and effectiveness of an enhanced collaborative care intervention. Psychiatry. 2018;81(4):349–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.