Abstract
Introduction
Smoking is a leading cause of death, and genetic variation contributes to smoking behaviors. Identifying genes and sets of genes that contribute to risk for addiction is necessary to prioritize targets for functional characterization and for personalized medicine.
Methods
We performed a gene set–based association and heritable enrichment study of two addiction-related gene sets, those on the Smokescreen Genotyping Array and the nicotinic acetylcholine receptors, using the largest available GWAS summary statistics. We assessed smoking initiation, cigarettes per day, smoking cessation, and age of smoking initiation.
Results
Individual genes within each gene set were significantly associated with smoking behaviors. Both sets of genes were significantly associated with cigarettes per day, smoking initiation, and smoking cessation. Age of initiation was only associated with the Smokescreen gene set. Although both sets of genes were enriched for trait heritability, each accounts for only a small proportion of the single nucleotide polymorphism-based heritability (2%–12%).
Conclusions
These two gene sets are associated with smoking behaviors, but collectively account for a limited amount of the genetic and phenotypic variation of these complex traits, consistent with high polygenicity.
Implications
We evaluated evidence for the association and heritable contribution of expert-curated and bioinformatically identified sets of genes related to smoking. Although they impact smoking behaviors, these specifically targeted genes do not account for much of the heritability in smoking and will be of limited use for predictive purposes. Advanced genome-wide approaches and integration of other ‘omics data will be needed to fully account for the genetic variation in smoking phenotypes.
Introduction
Smoking is one of the most prominent causes of death in the United States, leading to numerous diseases and shortened life expectancy.1 Although the majority of smokers report a desire to quit, very few are able.2 Furthermore, although smoking rates have decreased, other forms of nicotine consumption are rapidly growing, such as adolescent vaping,3 demonstrating a pressing need to characterize the underlying biology of nicotine use and smoking to reduce subsequent premature death.
Abundant evidence from twin, adoption, and family studies4–8 indicates that up to 50% of the phenotypic variance in nicotine dependence (from either structured clinical interviews or Fagerstrom Test for Nicotine Dependence scores), as well as specific smoking behaviors such as smoking initiation and quantity of smoking, is due to genetic factors. In addition to this family study evidence, genome-wide association studies (GWAS) have begun to identify variants associated with smoking behaviors,9,10 providing insights into the genetic etiology of smoking and nicotine dependence. A key finding from such studies has been the high polygenicity of these traits—Liu et al.9 found over 200 conditionally independent loci throughout the genome that influenced smoking initiation, escalation, and cessation using over 1.2 million individuals, with additional loci expected to be identified as sample sizes increase.
Efficient genotyping and disease-specific arrays have been developed with the aim of identifying particular variants to use in individualized therapies through predictive genetic models.11 To this end, the Smokescreen Genotyping Array was developed to tag over 1000 addiction-related genes, identified through expert knowledge, bioinformatic databases, and previous studies.11 These genes are thought to be strongly associated with addiction, and specifically nicotine use, behaviors.
Nicotinic acetylcholine receptors (nAChRs), which are bound by nicotine, play a key role in smoking behaviors and have been extensively investigated both in human population samples and in functional mouse models. In particular, the nAChR alpha 5 subunit (CHRNA5) is one of the most widely studied genes related to addiction, with replicated GWAS associations9,10,12 and functional characterization in the mouse.13,14 Identifying the key genes that interact with these receptors and whether they contribute to heritable variation in smoking phenotypes can lead to biological insight and potentially novel therapeutic targets. Melroy-Greif et al.15 performed an extensive literature search, in collaboration with experts in the field of nAChR research, to identify a set of 107 such genes involved in the upregulation, function, processing, and downstream effects of nAChR signaling (only 36 of these overlap with the Smokescreen Array gene set). Melroy-Greif et al.15 posited that this set of genes that are involved in nAChR upregulation, known to occur in response to nicotine exposure, play a role in smoking behaviors. Although Melroy-Greif et al.15 did not find significant gene set associations with smoking phenotypes, GWAS sample sizes have since increased dramatically,9 leading to greater statistical power to detect such associations.
Understanding which genes and sets of genes are associated with smoking phenotypes may help prioritize future functional studies in model organisms16 and so remains an important goal. These two sets of genes provide a starting point to prioritize potential targets, but the high degree of polygenicity of smoking behaviors9 raises the question of whether such genes are more strongly associated or enriched than the rest of the genome.
We sought here to assess whether and the degree to which these two sets of genes (“Smokescreen” and “Nicotinic” receptor gene sets) are associated with four smoking phenotypes. Using the most recent and largest GWAS summary statistics (GSCAN9) for smoking behaviors, we applied gene set association and heritability enrichment analyses to specifically test the contribution of these particular genes to all four smoking behaviors examined in GSCAN: smoking initiation, age of initiation, cigarettes per day, and smoking cessation.
Materials and Methods
We used multi-marker analysis of genomic annotation17 (MAGMA) to test gene-level associations of the Smokescreen and Nicotinic gene sets with all four smoking behaviors with GWAS summary statistics from the GSCAN9 project after removing the 23andMe data: (1) smoking initiation (N = 632 802), defined as whether an individual had ever in their lifetime been a regular smoker, measured by GSCAN in multiple ways, as having smoked regularly, as ever smoked every day for at least a month, or as having smoked over 100 cigarettes over one’s lifetime; (2) age of smoking initiation (N = 262 990), defined in GSCAN as the age at which an individual began smoking regularly; (3) cigarettes per day (N = 263 954), defined in GSCAN as a five-bin variable based on responses of the number of cigarettes per day; and (4) smoking cessation (N = 312 812), defined as individuals who were not current smokers but had been regular smokers at one point. These phenotypes include key aspects of nicotine dependence.18 The final three phenotypes required an individual to be or to have been a regular smoker at some point. Full descriptions of the phenotypes can be found in reference.9 We included a control phenotype, alcoholic drinks per week (N = 537 349), which was the fifth GSCAN phenotype and defined by GSCAN as the number of alcoholic drinks per week an individual reported across several questions among component studies. Although it can be viewed as an addiction-related phenotype,19 and has single nucleotide polymorphism (SNP)-based genetic correlations9 with smoking phenotypes ranging from 0.1 to 0.34, it is distinct from smoking behaviors and thus provides an assessment of whether the particular gene sets relate to smoking phenotypes or more generally addiction-related behaviors.
We applied the competitive MAGMA test to determine whether the gene sets of interest were more strongly associated with these phenotypes than the rest of the genome. We tested two smoking-related gene sets. First, we tested the set of 1031 addiction genes included on the Smokescreen Genotyping Array11 (hereafter referred to as the “Smokescreen” gene set), of which 1009 were also tagged by variants with GWAS summary statistics from GSCAN. These genes were chosen based on bioinformatic databases and expert curation, being characterized as “addiction-relevant” genes; a full description of the Smokescreen array can be found in Bauerly et al.11 The second smoking-relevant gene set comprised 107 expert-curated nAChR-related genes that are directly involved in signaling through nAChRs as identified by Melroy-Greif et al.15 (hereafter the “Nicotinic” gene set). Thirty-six genes were present in both the Smokescreen and Nicotinic gene sets.
We also tested three control gene sets, which were predicted to show no association with smoking behaviors based on patterns of genetic correlation as presented by Liu et al.9: height (444 genes; rg ~ −0.10 to 0.04), Alzheimer’s (476 genes; rg ~ −0.06 to 0.08), and inflammatory bowel disease (234 genes; rg ~ −0.05 to 0.04). These served as negative controls, not expected to have substantial genetic overlap with smoking behaviors. Control sets of genes were identified by querying the GWAS Catalog (https://www.ebi.ac.uk/gwas/) for the terms “height,” “Alzheimer’s,” and “inflammatory bowel disease,” identifying all the gene names tagged by GWAS Catalog as being associated with these traits, and then aggregating these genes into gene sets. As the Nicotinic genes included only protein-coding genes, we excluded pseudogenes, ncRNA genes, lncRNA genes, and miRNA genes from all control gene sets.
In all analyses, variants were annotated to genes using a 25Kb window around the start and end point of each gene in MAGMA in an attempt to include variants that might exert close-range regulatory effects (eg, within promoter regions) as in previous gene set analyses.20 We included the default covariates in MAGMA (gene size, density, inverse MAC, per-gene sample size, plus the log value of each) to control for possible confounding factors. We tested five gene sets for each phenotype; therefore, we applied a Bonferroni multiple testing correction for significance for five tests as the within-trait significance threshold (α = .01).
Following the competitive tests described earlier, we performed conditional tests of within-trait significant gene sets. These assessed whether the association of a target gene set for a particular phenotype was significant conditional on the effects of additional gene sets. First, we tested the association of the Nicotinic genes conditional on the effects of the Smokescreen genes, which allowed us to determine whether the effects of the Nicotinic genes were simply the effect of genes shared between the two gene sets. Next, we examined the association of the Smokescreen gene set conditional on the Nicotinic gene set. Because no control gene set analysis passed our within-trait Bonferroni significance threshold, we did not evaluate conditional associations of those. In total, we performed seven conditional tests and used a Bonferroni correction for seven tests (α = .00714).
Finally, we performed an enrichment analysis of the heritable contribution of each gene set, relative to the number of markers in the gene set, using partitioned linkage disequilibrium (LD)-score regression21 (LDSC). We added annotations for each of the five gene sets listed above to the baseline with LD annotation model,22 as suggested in the LDSC documentation. We applied a within-trait Bonferroni correction for these enrichment analyses based on five gene sets for each phenotype (α = .01).
Results
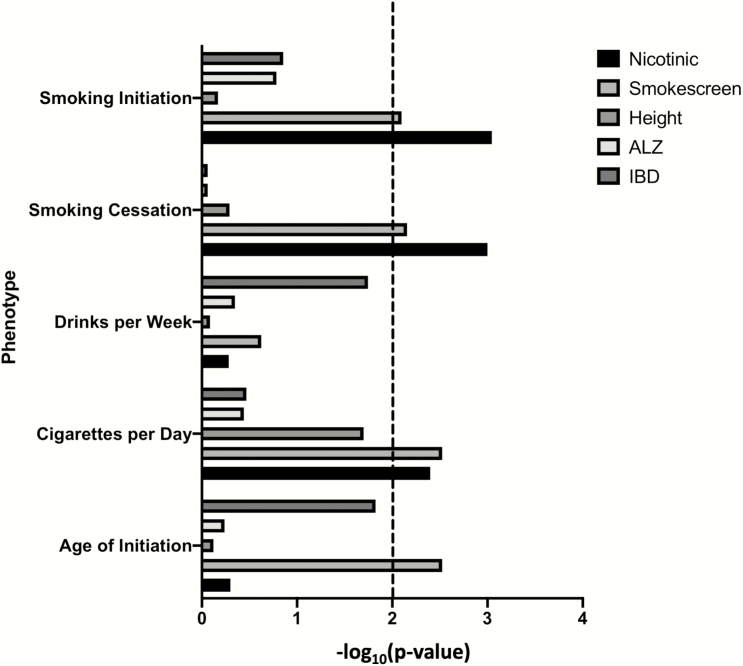
We found that the smoking gene sets (Smokescreen and Nicotinic) were significantly associated with the four smoking phenotypes, but not the control addiction-related phenotype, drinks per week (Figure 1, Supplementary Table 1). Although both smoking gene sets were significantly associated with cigarettes per day, smoking initiation, and smoking cessation, age of initiation was only associated with the Smokescreen data set. None of the negative control gene sets was significantly associated with any phenotype (all p > .01).
Figure 1.
−Log10(p) of MAGMA competitive tests the four smoking phenotypes and one control trait (drinks per week) across the two smoking gene sets and three control gene sets. Dashed line is the within-trait Bonferroni correction for multiple testing.
Many genes were present in more than one gene set (36 overlapped between the Nicotinic and Smokescreen gene sets), and 128 individual genes from one or more gene sets were individually significantly associated with one or more phenotypes (Supplementary Table S2). Nineteen of these were within the Nicotinic gene set, 61 were within the Smokescreen set, and 9 of these overlapped (Supplementary Table S2). Thus, though the trait genetic correlations are very weak, some of these genes are likely to influence multiple traits (Supplementary Table S2).
To test whether the smoking gene sets were independently associated with smoking phenotypes or whether the associations for each resulted from the genes overlapping genes in both gene sets, we performed conditional analyses (Table 1). For example, of the 107 Nicotinic genes, 36 overlapped with the Smokescreen gene set. In both cases, the association of Smokescreen and Nicotinic data sets, conditional on the other, remained significant for cigarettes per day (p < .01). The Nicotinic gene set, conditional on the Smokescreen gene set, was also significantly associated with smoking initiation and cessation, but the Smokescreen gene set, conditional on the Nicotinic gene set, was not (p > .01).
Table 1.
MAGMA Conditional Test Results for the Gene Sets That Were Significantly Associated With Particular Traits in the Competitive Test (Table 1)
| Trait | Target gene set | Conditional gene set | Beta | SE | p |
|---|---|---|---|---|---|
| Cigarettes per day | Nicotinic | Smokescreen | 0.222 | 0.090 | .007* |
| Smoking initiation | Nicotinic | Smokescreen | 0.332 | 0.102 | .001* |
| Smoking cessation | Nicotinic | Smokescreen | 0.250 | 0.087 | .002* |
| Age of initiation | Smokescreen | Nicotinic | 0.078 | 0.028 | .003* |
| Cigarettes per day | Smokescreen | Nicotinic | 0.079 | 0.031 | .005* |
| Smoking initiation | Smokescreen | Nicotinic | 0.076 | 0.036 | .017 |
| Smoking cessation | Smokescreen | Nicotinic | 0.061 | 0.028 | .015 |
MAGMA = multi-marker analysis of genomic annotation.
*p < .05/7 < .00714.
Next, we used LDSC to estimate the total SNP-heritability (h2SNP) for each smoking phenotype. Total h2SNP (SE) (on liability scale for the binary phenotypes, cessation, and smoking initiation) for each trait was smoking cessation: 0.074 (0.005); smoking initiation: 0.095 (0.003); age of initiation: 0.041 (0.002); cigarettes per day: 0.061 (0.002); and drinks per week: 0.049 (0.002). These estimates are similar to those estimated with the full (including 23andMe) data set reported by Liu et al.9
To assess the proportion of the estimated h2SNP attributable to each gene set, we applied partitioned LDSC. The heritable contribution of the Smokescreen gene set was significantly enriched for age of initiation, cigarettes per day, smoking cessation, and smoking initiation (enrichment, the proportion of h2SNP attributable to the gene set divided by the proportion of SNPs within the gene set, ranged from 1.4 to 2.3; Table 2). The Nicotinic gene set was significantly enriched only for smoking cessation. Although Nicotinic gene set enrichment estimates for all phenotypes were comparatively high (ranging from 1.59 to 3.27; Table 2), the relatively small number of genes, and therefore small number of variants as a proportion of all variants, led to high uncertainty in these estimates. The Nicotinic gene set enrichment SE were generally the largest and limited our statistical power to detect enrichment (Table 2). None of the control gene sets were significantly enriched for any trait. Overall, the enrichment analyses indicated that the genes present on the Smokescreen array are indeed enriched for smoking-relevant genetic variants and that at least some of the genetic signal in smoking phenotypes comes from variants within the nicotinic receptors and genes they interact with. However, these gene sets cumulatively explain a small percentage of h2SNP; less than 3% of h2SNP is attributable to Nicotinic genes, and less than 13% is attributable to all of the Smokescreen genes (Table 2).
Table 2.
LDSC Heritability Enrichment of Each Gene Set Annotation Within the Full Baseline + LD Model for the Four Smoking Phenotypes and One Control Trait Across the Two Smoking Gene Sets and Three Control Gene Sets
| Trait | Gene set | Proportion of SNPs in annotation | h 2 SNP proportion | h 2 SNP proportion SE | Enrichment | Enrichment SE | p |
|---|---|---|---|---|---|---|---|
| Age of initiation | Nicotinic | 0.008 | 0.016 | 0.004 | 1.840 | 0.467 | .075 |
| Cigarettes per day | Nicotinic | 0.008 | 0.028 | 0.009 | 3.274 | 1.092 | .045 |
| Drinks per week | Nicotinic | 0.008 | 0.013 | 0.002 | 1.593 | 0.240 | .014 |
| Smoking cessation | Nicotinic | 0.008 | 0.022 | 0.005 | 2.640 | 0.628 | .010* |
| Smoking initiation | Nicotinic | 0.008 | 0.020 | 0.005 | 2.375 | 0.601 | .024 |
| Age of initiation | Smokescreen | 0.054 | 0.098 | 0.013 | 1.811 | 0.242 | .001* |
| Cigarettes per day | Smokescreen | 0.054 | 0.121 | 0.019 | 2.238 | 0.356 | .001* |
| Drinks per week | Smokescreen | 0.054 | 0.079 | 0.013 | 1.471 | 0.236 | .049 |
| Smoking cessation | Smokescreen | 0.054 | 0.075 | 0.006 | 1.401 | 0.114 | <.001* |
| Smoking initiation | Smokescreen | 0.054 | 0.100 | 0.016 | 1.851 | 0.295 | .006* |
| Age of initiation | Height | 0.030 | 0.027 | 0.006 | 0.914 | 0.212 | .686 |
| Cigarettes per day | Height | 0.030 | 0.035 | 0.007 | 1.169 | 0.249 | .494 |
| Drinks per week | Height | 0.030 | 0.029 | 0.004 | 0.973 | 0.137 | .846 |
| Smoking cessation | Height | 0.030 | 0.030 | 0.004 | 1.020 | 0.135 | .884 |
| Smoking initiation | Height | 0.030 | 0.027 | 0.008 | 0.912 | 0.255 | .731 |
| Age of initiation | ALZ | 0.043 | 0.072 | 0.012 | 1.685 | 0.286 | .015 |
| Cigarettes per day | ALZ | 0.043 | 0.039 | 0.007 | 0.910 | 0.160 | .573 |
| Drinks per week | ALZ | 0.043 | 0.053 | 0.007 | 1.243 | 0.162 | .137 |
| Smoking cessation | ALZ | 0.043 | 0.050 | 0.005 | 1.171 | 0.128 | .183 |
| Smoking initiation | ALZ | 0.043 | 0.039 | 0.010 | 0.905 | 0.225 | .675 |
| Age of initiation | IBD | 0.012 | 0.016 | 0.007 | 1.350 | 0.606 | .564 |
| Cigarettes per day | IBD | 0.012 | 0.017 | 0.005 | 1.443 | 0.417 | .284 |
| Drinks per week | IBD | 0.012 | 0.018 | 0.004 | 1.565 | 0.341 | .104 |
| Smoking cessation | IBD | 0.012 | 0.020 | 0.007 | 1.713 | 0.575 | .217 |
| Smoking initiation | IBD | 0.012 | 0.021 | 0.010 | 1.841 | 0.888 | .345 |
LDSC = linkage disequilibrium–score regression.
*p < .01, the within-trait Bonferroni correction for multiple testing.
Discussion
We examined the genes on the Smokescreen Genotyping Array and those involved with nicotinic receptors to assess whether they are associated with smoking phenotypes, and the degree to which they contribute to variation in the behavior (h2SNP). Both sets of genes were significantly associated with at least some of the tested smoking phenotypes. This is in contrast to the previous study of nicotinic genes by Melroy-Greif et al.15. However, the GSCAN summary statistics utilized a sample an order of magnitude larger than was available earlier, suggesting that the previous lack of gene set associations was due to low power. Additional associations of the Nicotinic genes may be found with larger sample sizes, though the partitioned LDSC analyses suggest that the nicotinic receptors and the genes they interact with cumulatively account for only 1.6%–2.8% of h2SNP. This implies in combination with other studies (eg, Liu et al.9) that although the nicotinic receptors are influential in smoking behaviors and play a key physiological role in response to nicotine, much of the genetic variation in these phenotypes is due to other pathways, consistent with a highly polygenic model of these complex traits.
Smokescreen incorporated roughly 10 times the number of genes compared with the Nicotinic receptors, and accordingly accounted for a larger proportion of h2SNP, up to 12% of the genetic variance tagged by genome-wide markers. However, much of the genetic variance in smoking phenotypes remains unaccounted for by these variants and genes, and must be attributable to genes and pathways not tagged by this custom array. Together, these results suggest that the larger number of smoking-relevant genes in the Smokescreen gene set as a whole provide limited additional information over nicotinic receptor-related genes. However, because there were numerous individual genome-wide significant genes uniquely belonging to either the Smokescreen gene set or Nicotinic gene set (Supplementary Table S1), specific genes within each set are clearly still contributing to variation in smoking behaviors. Interestingly, the Smokescreen gene set was associated with age of initiation, but the Nicotinic gene set was not. Age of initiation is a phenotype that occurs early in the developmental trajectory toward dependence, and it may be related to a more general underlying behavioral disinhibition phenotype that predisposes certain individuals for risk.23–25 Perhaps a broad phenotype is the result of even more genes, given the known polygenicity of complex behaviors, which may be better captured by the much larger number of genes in the Smokescreen set compared with the Nicotinic set. In addition, the specific set of genes tagged in Smokescreen includes several that are individually associated with age of initiation (Supplementary Table S2), which are involved with behavioral, neuronal, and brain-related phenotypes: BDNF, GRK4, DCC, FOXP1, and MEF2C. Collectively, these genes play roles in neural development, neurodevelopmental disorders, intellectual disability, and/or postdevelopmental synaptic and cognitive processing, so it is plausible that these genes may contribute to smoking initiation via developmental changes predisposing individuals to nicotine use.26–36
For predictive purposes and individualized treatment for nicotine use, additional variants outside of classical “addiction genes,” including nicotinic receptors, will need to be considered. Genetically based individual variation exists in individuals’ ability to quit (h2 ~ 50%; ref.37) and in individuals’ responses to pharmacotherapies (summarized in Bauerly et al.11), highlighting the potential utility of individualized treatment plans. However, as neither Smokescreen gene set nor Nicotinic gene set accounted for even a majority of the genetic variance in smoking cessation, genes beyond these classic addiction genes will need to be incorporated into polygenic prediction for personalized treatments.
Our study was limited to the GSCAN sample,9 restricted to only those of European ancestry, and no independent replication samples are available at this time. Furthermore, although GSCAN contains the currently largest available sample for summary statistics, we were limited to those excluding 23andMe data and increasing the sample size would probably increase our power to detect gene set associations. However, this is unlikely to substantially change our conclusions, particularly as LDSC-based h2SNP estimates indicate that these genes collectively account for a small proportion of the genetic variance. Notably, the GSCAN performed GWAS of common variants only, and thus the gene set associations and h2SNP estimates presented here are restricted to the influence of common variants. Rare variants within genes of these gene sets may influence smoking behaviors and therefore these genes may yet have a larger role in smoking behavior than these analyses suggest. However, even if this is the case, it is unlikely that rare variant h2SNP would account for the large discrepancy between current total h2SNP estimates and twin-based h2.
Smoking is one of the leading cause of premature death in the United States,1 and new forms of nicotine use, such as e-cigarettes, have gained prominence.3 There remains, therefore, a critical need to understand the underlying biology of nicotine use, and identifying key genes and sets of genes is one possible avenue toward this goal. Nicotinic acetylcholine receptors play a key role in nicotine use and the genes with which they interact influence smoking behaviors, as do genes traditionally thought of as “addiction” genes. However, although these loci influence genetic variation in smoking behaviors, it is clear that these specifically targeted genes do not yet account for a large proportion of the heritability in smoking, and will therefore be of limited use for predictive purposes.38 Ever-larger association studies, beyond GSCAN,9 will be instrumental for this purpose, by examining genome-wide, unascertained markers in combination with improved statistical power as well as incorporating other ‘omics data sets and advanced methodologies to go beyond positional mapping, such as imputing genetically regulated gene expression39 or integrating information from animal models of addiction.
Supplementary Material
Acknowledgments
We thank Mengzhen Liu and Scott Vrieze for providing GSCAN summary statistics.
Funding
Funding was obtained from the University of Colorado Institute for Behavioral Genetics, (National Institute on Alcohol Abuse and Alcoholism F32 AA027435 [PI: ECJ], National Institute on Drug Abuse T32 DA017637, National Institute of Mental Health 2R01 MH100141 [PI: MCK], National Institute on Drug Abuse R01 DA044283 [PI: Scott I. Vrieze].
Declaration of Interests
The authors declare no conflict of interest.
References
- 1. US Department of Health and Human Services. Health Consequences of Smoking – 50 Years of Progress. A Report of the Surgeon General. Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control and Prevention; 2014:1081. [Google Scholar]
- 2. Centers for Disease Control and Prevention. Quitting smoking among adults – United States, 2001–2010. MMWR Morb Mortal Wkly Rep. 2011;60:1513–1519. [PubMed] [Google Scholar]
- 3. Cullen KA, Ambrose BK, Gentzke AS, Apelberg BJ, Jamal A, King BA. Notes from the field: use of electronic cigarettes and any tobacco product among middle and high school students – United States, 2011–2018. MMWR Morb Mortal Wkly Rep. 2018;67(45):1276–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haberstick BC, Ehringer MA, Lessem JM, Hopfer CJ, Hewitt JK. Dizziness and the genetic influences on subjective experiences to initial cigarette use. Addiction. 2011;106(2):391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haberstick BC, Zeiger JS, Corley RP, et al. Common and drug-specific genetic influences on subjective effects to alcohol, tobacco and marijuana use. Addiction. 2011;106(1):215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaprio J. Genetic epidemiology of smoking behavior and nicotine dependence. COPD. 2009;6(4):304–306. [DOI] [PubMed] [Google Scholar]
- 7. Rose JE, Broms U, Korhonen T, Dick DM, Kaprio J. Genetics of smoking behavior. In: Kim YK, ed. Handbook of Behavior Genetics. New York, NY: Springer; 2009. [Google Scholar]
- 8. Kendler KS, Schmitt E, Aggen SH, Prescott CA. Genetic and environmental influences on alcohol, caffeine, cannabis, and nicotine use from early adolescence to middle adulthood. Arch Gen Psychiatry. 2008;65(6):674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu M, Jiang Y, Wedow R, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baurley JW, Edlund CK, Pardamean CI, Conti DV, Bergen AW. Smokescreen: a targeted genotyping array for addiction research. BMC Genomics. 2016;17:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saccone NL, Culverhouse RC, Schwantes-An TH, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6(8):e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165(9):1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O’Neill HC, Wageman CR, Sherman SE, Grady SR, Marks MJ, Stitzel JA. The interaction of the Chrna5 D398N variant with developmental nicotine exposure. Genes Brain Behav. 2018;17(7):e12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Melroy-Greif WE, Simonson MA, Corley RP, Lutz SM, Hokanson JE, Ehringer MA. Examination of the involvement of cholinergic-associated genes in nicotine behaviors in European and African Americans. Nicotine Tob Res. 2017;19(4):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nadeau JH, Auwerx J. The virtuous cycle of human genetics and mouse models in drug discovery. Nat Rev Drug Discov. 2019;18(4):255–272. [DOI] [PubMed] [Google Scholar]
- 17. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heatherton TF, Kozlowski LT, Frecker RC, Fagerström KO. The Fagerström test for nicotine dependence: a revision of the Fagerström Tolerance Questionnaire. Br J Addict. 1991;86(9):1119–1127. [DOI] [PubMed] [Google Scholar]
- 19. Walters RK, Polimanti R, Johnson EC, et al. ; 23andMe Research Team Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018;21(12):1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson EC, Border R, Melroy-Greif WE, de Leeuw CA, Ehringer MA, Keller MC. No evidence that Schizophrenia candidate genes are more associated with Schizophrenia than noncandidate genes. Biol Psychiatry. 2017;82(10):702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finucane HK, Bulik-Sullivan B, Gusev A, et al. ; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47(11):1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gazal S, Finucane HK, Furlotte NA, et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat Genet. 2017;49(10):1421–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pihl RO. Personality disorders, behavioral disinhibition, and addiction: a commentary. Biol Psychiatry. 2007;62(6):551–552. [DOI] [PubMed] [Google Scholar]
- 24. Palmer RH, Knopik VS, Rhee SH, et al. Prospective effects of adolescent indicators of behavioral disinhibition on DSM-IV alcohol, tobacco, and illicit drug dependence in young adulthood. Addict Behav. 2013;38(9):2415–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hicks BM, Iacono WG, McGue M. Index of the transmissible common liability to addiction: Heritability and prospective associations with substance abuse and related outcomes. Drug Alcohol Depend. 2012;123(suppl 1):S18–S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goldman JS, Ashour MA, Magdesian MH, et al. Netrin-1 promotes excitatory synaptogenesis between cortical neurons by initiating synapse assembly. J Neurosci. 2013;33(44):17278–17289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kennedy TE, Serafini T, de la Torre JR, Tessier-Lavigne M. Netrins are diffusible chemotropic factors for commissural axons in the embryonic spinal cord. Cell. 1994;78(3):425–435. [DOI] [PubMed] [Google Scholar]
- 28. Knüsel B, Winslow JW, Rosenthal A, et al. Promotion of central cholinergic and dopaminergic neuron differentiation by brain-derived neurotrophic factor but not neurotrophin 3. Proc Natl Acad Sci USA. 1991;88(3):961–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eisch AJ, Bolaños CA, de Wit J, et al. Brain-derived neurotrophic factor in the ventral midbrain-nucleus accumbens pathway: a role in depression. Biol Psychiatry. 2003;54(10):994–1005. [DOI] [PubMed] [Google Scholar]
- 30. Sorensen SD, Conn PJ. G protein-coupled receptor kinases regulate metabotropic glutamate receptor 5 function and expression. Neuropharmacology. 2003;44(6):699–706. [DOI] [PubMed] [Google Scholar]
- 31. Bowers JM, Konopka G. The role of the FOXP family of transcription factors in ASD. Dis Markers. 2012;33(5):251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamura S, Morikawa Y, Iwanishi H, Hisaoka T, Senba E. Foxp1 gene expression in projection neurons of the mouse striatum. Neuroscience. 2004;124(2):261–267. [DOI] [PubMed] [Google Scholar]
- 33. Hamdan FF, Daoud H, Rochefort D, et al. De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am J Hum Genet. 2010;87(5):671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H, Radford JC, Ragusa MJ, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci USA. 2008;105(27):9397–9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rocha H, Sampaio M, Rocha R, Fernandes S, Leão M. MEF2C haploinsufficiency syndrome: report of a new MEF2C mutation and review. Eur J Med Genet. 2016;59(9):478–482. [DOI] [PubMed] [Google Scholar]
- 36. Leifer D, Golden J, Kowall NW. Myocyte-specific enhancer binding factor 2C expression in human brain development. Neuroscience. 1994;63(4):1067–1079. [DOI] [PubMed] [Google Scholar]
- 37. Xian H, Scherrer JF, Madden PA, et al. The heritability of failed smoking cessation and nicotine withdrawal in twins who smoked and attempted to quit. Nicotine Tob Res. 2003;5(2):245–254. [PubMed] [Google Scholar]
- 38. Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14(7):507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gusev A, Ko A, Shi H, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.