Significance
In untreated individuals, HIV-1 evolves rapidly to escape the host immune responses. Viral replication can be effectively suppressed by antiretroviral therapy (ART). However, ART is not curative due to persistence of latent virus in a stable reservoir in resting CD4+ T cells, and viral rebound occurs if ART is stopped. We show here that outgrowth of a significant fraction of the viruses persisting in the latent reservoir is effectively blocked by the host antibody response. It is important to find ways to induce antibodies or other immune responses to block outgrowth of the remaining viruses.
Keywords: HIV, reservoir, neutralizing antibody, QVOA, latency
Abstract
In untreated HIV-1 infection, rapid viral evolution allows escape from immune responses. Viral replication can be blocked by antiretroviral therapy. However, HIV-1 persists in a latent reservoir in resting CD4+ T cells, and rebound viremia occurs following treatment interruption. The reservoir, which is maintained in part by clonal expansion, can be measured using quantitative viral outgrowth assays (QVOAs) in which latency is reversed with T cell activation to allow viral outgrowth. Recent studies have shown that viruses detected in QVOAs prior to treatment interruption often differ from rebound viruses. We hypothesized that autologous neutralizing antibodies directed at the HIV-1 envelope (Env) protein might block outgrowth of some reservoir viruses. We modified the QVOA to reflect pressure from low concentrations of autologous antibodies and showed that outgrowth of a substantial but variable fraction of reservoir viruses is blocked by autologous contemporaneous immunoglobulin G (IgG). A reduction in outgrowth of >80% was seen in 6 of 15 individuals. This effect was due to direct neutralization. We established a phylogenetic relationship between rebound viruses and viruses growing out in vitro in the presence of autologous antibodies. Some large infected cell clones detected by QVOA carried neutralization-sensitive viruses, providing a cogent explanation for differences between rebound virus and viruses detected in standard QVOAs. Measurement of the frequency of reservoir viruses capable of outgrowth in the presence of autologous IgG might allow more accurate prediction of time to viral rebound. Ultimately, therapeutic immunization targeting the subset of variants resistant to autologous IgG might contribute to a functional cure.
HIV Type-1 (HIV-1) persists in a latent form in resting CD4+ T cells despite long-term treatment with combination antiretroviral therapy (ART) (1–3). In resting CD4+ T cells, the transcriptional environment is relatively nonpermissive for HIV-1 gene expression (reviewed in ref. 4), and thus the latently infected cells may not be recognized or eliminated by the immune response. The latent reservoir is established early during infection (5, 6) and is extremely stable, with an estimated half-life of 44 mo (7–9). The stability and persistence of this reservoir is attributable, in part, to the proliferation of infected cells (10–19). This clonal expansion can result from antigen stimulation (15, 20, 21), responses to homeostatic signals (12), or effects related to proviral integration into host genes involved in cell survival or proliferation (13, 14). If cells comprising the reservoir become activated, viral genes can be expressed, and virions can be released (1–3). However, ART prevents additional rounds of infection. Upon cessation of ART, there is exponential viral growth and rebound viremia (22, 23). These properties of the latent reservoir make it the major barrier to an HIV-1 cure.
Substantial efforts have been made to target this reservoir to allow ART-free remission (24). In the “shock and kill” strategy (25, 26), HIV-1 gene expression is induced pharmacologically, rendering infected cells susceptible to viral cytopathic effects or lysis by HIV-1–specific cytotoxic T lymphocytes (CTLs) (27–30). Another approach involves the use of broadly neutralizing antibodies (bNAbs) to prevent or control viral rebound after interruption of ART (31–37). These antibodies recognize the HIV-1 envelope (Env) protein, which is expressed both on productively infected cells and free virions (37). bNAbs are unique in their ability to recognize diverse HIV-1 isolates from many different individuals. During the normal course of infection, HIV-1 evolves rapidly to escape from antibody responses, largely through point mutations, recombination, and in-frame insertions and deletions in the env gene (34–42). The Env protein has five regions of extensive variability, known as the hypervariable loops. The ability of the Env protein to incorporate large genetic variability in these regions without loss of function facilitates progressive evolution of the env gene and allows viral escape mutants to arise (34, 37, 41). Pioneering studies by Shaw and coworkers (41) and Richman et al. (42) showed that, during periods of ongoing viral replication, the constant evolution of the Env protein leads to continual escape from the evolving neutralizing antibody response. In addition, the HIV-1 Env protein is highly glycosylated (43). N-linked oligosaccharides comprise up to 50% of the mass of the Env protein and produce a glycan shield which can inhibit antibody access to neutralizing viral epitopes. This glycan shield is also frequently altered by mutations leading to escape from neutralization (41).
Potent bNAb responses arise in only a small percentage of HIV-1–infected individuals (31, 34, 36, 37, 44). In these individuals, B cells producing bNAbs are rare and comprise <1% of the Env-specific memory B cell repertoire (36). bNAbs arise by somatic mutation and selection of the immunoglobulin variable regions during B cell affinity maturation in germinal centers (34, 36). bNAbs have been developed as antiviral agents and have certain advantages over classical ART, such as lower toxicity and the potential for less frequent administration. Most significantly, they can promote the elimination of HIV-1–infected cells through Fc receptor-dependent mechanisms, potentially enabling the clearance of long-lived CD4+ T cells from the latent reservoir following latency reversal (45). Infusions of HIV-1–specific bNAbs have been shown to prevent transmission of HIV-1 or simian HIV (SHIV) in animal models (46–49) and to suppress viral replication, both in infected individuals and in mouse and nonhuman primate models (50–56). These findings collectively led to clinical studies examining whether viral rebound following analytical treatment interruption (ATI) could be delayed or prevented by passive bNAb infusions (57, 58) and whether such infusions could affect reservoir size and diversity (59, 60). These clinical studies have shown that, in some cases, bNAbs delay viral rebound after interruption of ART, likely due to their long in vivo half-life (55–58). However, no clinical trial has demonstrated a substantial decrease in the size or diversity of the latent reservoir following bNAb infusions (59, 60). Interestingly, a recent study has shown decreases in viral DNA following bNAb and TLR7 agonist administration in ART-treated, SHIV-infected macaques (61). These data suggest that the administration of bNAbs in the presence of innate immune stimulation could reduce viral reservoir size. However, most proviral DNA is defective (62, 63), and bNAb-mediated reductions in the amount of replication-competent virus have not yet been demonstrated with assays selective for intact proviruses, such as the quantitative viral outgrowth assay (QVOA) (1, 59, 60, 64) or the intact proviral DNA assay (IPDA) (65).
Clinical studies investigating changes in the latent reservoir following bNAb infusion have shown that HIV-1 variants detected in the plasma during a subsequent ATI are often genetically distinct from the viruses found in the latent reservoir during ART using the QVOA (59, 60). Potential explanations include sampling issues, differences between in vitro stimulation and in vivo reactivation, anatomic compartmentalization of rebound viruses, and innate and adaptive immune system pressures in vivo. Recent studies have also suggested that viral recombination may be involved in the emergence of distinct variants during rebound (59, 66, 67).
Understanding the discrepancy between viruses detected in QVOAs and rebound virus seen following ATI requires a consideration of the clonal nature of the latent reservoir. Clonal expansion of latently infected cells is prevalent in HIV-1–infected individuals (10–19). Clonal populations of infected cells predominantly carry defective proviruses (62, 63). However, recent reports have shown that expanded clones carrying replication-competent proviruses can dominate the latent reservoir, accounting for greater than 50% of the latently infected cells detected in QVOAs (15–18). These clonal populations are capable of producing infectious virus and are conceivably the source of low-level viremia in individuals on ART, which is often oligoclonal (10, 11, 15). Although individual clones harboring replication-competent proviruses wax and wane over time, at any given time, the reservoir is dominated by expanded clones (68). These clones would therefore be expected to contribute to viral rebound following ATI. To explore why the initial rebound viruses are often distinct from these clonal sequences, we asked whether immune system pressures in vivo could play a role in the selection of viruses that cause rebound during an ATI, specifically focusing on the autologous antibody response to the HIV-1 Env protein.
Results
Autologous Neutralizing Antibodies Limit In Vitro Outgrowth from the HIV-1 Reservoir.
To explain why the rebound viruses appearing in the plasma following ATI are often genetically distinct from viruses detected in the reservoir using the QVOA (59, 60), we asked whether the autologous neutralizing antibody response could inhibit a subset of viruses in the reservoir from rebounding following ATI. It has been unclear to what extent viruses deposited in the latent reservoir are susceptible to autologous neutralizing antibodies and to what degree these antibodies can block viral rebound.
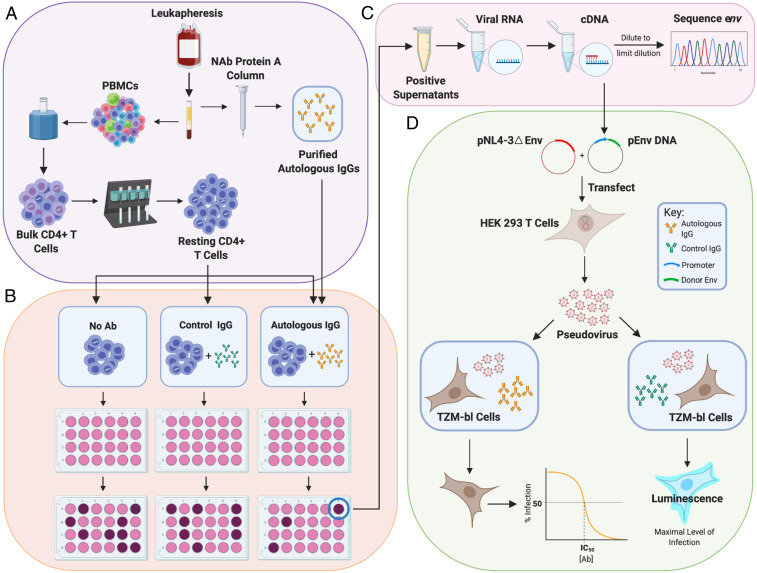
To address this question, we used the QVOA to model viral rebound following treatment interruption. In the QVOA, T cell activation is used to reverse latency in limiting dilution cultures of resting CD4+ T cells from infected individuals. Viruses released following latency reversal replicate in CD4+ T cells present in the culture, with exponential growth similar to that seen following ATI in vivo (22). In previous studies, it has been difficult to determine whether immune responses can block viral outgrowth in this system. Because the frequency of latently infected cells detected in the QVOA is low (∼1 in 106), only a small number of isolates are generally obtained from blood samples. It is thus difficult to test factors affecting outgrowth in a statistically meaningful way. Therefore, we obtained leukapheresis samples from 15 HIV-1–infected individuals on long-term ART (see SI Appendix, Table S1 for characteristics of study participants) and purified large numbers of resting CD4+ T cells (60,000,000 to 120,000,000) for use in the QVOA. The frequency of cells with intact proviruses was initially measured using the IPDA (65) to identify limiting dilutions to use in the QVOA. These limiting dilution cultures allow us to determine whether individual latently infected cells can give rise to a spreading infection in the presence of autologous antibodies largely without confounding effects of recombination between different viral variants in the same culture well. QVOAs were carried out in the absence of antibodies, in the presence of autologous contemporaneous leukapheresis plasma at a dilution of 1:50, in the presence of immunoglobulin G (IgG) purified from autologous contemporaneous leukapheresis plasma using protein A affinity chromatography (50 μg/mL), or in the presence of control IgG from HIV-negative donors at the same concentration (Fig. 1 A and B). IgGs were supplemented into the media during culture splits to ensure that the concentration remained constant over the course of the 14- to 21-d cultures. Western blot analysis of IgG preparations from all participants showed HIV-1–specific antibodies, including antibodies to gp120 (SI Appendix, Fig. S1).
Fig. 1.
Evaluation of suppression of viral outgrowth by autologous neutralizing antibodies. (A) HIV-1–infected volunteers on long-term ART underwent leukapheresis, and resting CD4+ T cells and autologous IgG were purified from the same leukapheresis sample as described in Methods. (B) After an initial assessment of the frequency in intact proviruses by IPDA (65), resting CD4+ T cells were plated in limiting dilutions with no antibody, with control IgG from an HIV-negative donor (50 μg/mL), or with purified autologous IgG (50 μg/mL). Cells were then activated with phytohemagglutinin (PHA) and irradiated allogeneic PBMC according to the standard QVOA protocol (64). MOLT4/CCR5 cells were added to expand virus released following latency reversal. Enzyme-linked immunosorbent assays (ELISAs) for HIV-1 p24 on days 14 and 21 detected wells with viral outgrowth. (C) Viral RNA was isolated from p24+ supernatants, converted to cDNA, and diluted for SGS of the env gene. Sequences from isolates obtained in the presence of autologous antibodies were compared with sequences from isolates in control wells using phylogenetic analysis and neutralization assays. (D) For neutralization assays, cDNA was used to generate env-expression vectors for selected isolates. These were cotransfected into HEK 293 cells along with an env-deficient proviral construct to generate pseudoviruses. Pseudoviruses were incubated with autologous or control antibodies and then used to infect the TZM-bl reporter cell line (70, 71). Luminescence was measured at 72 h. Dose–response curves were analyzed as described in Methods.
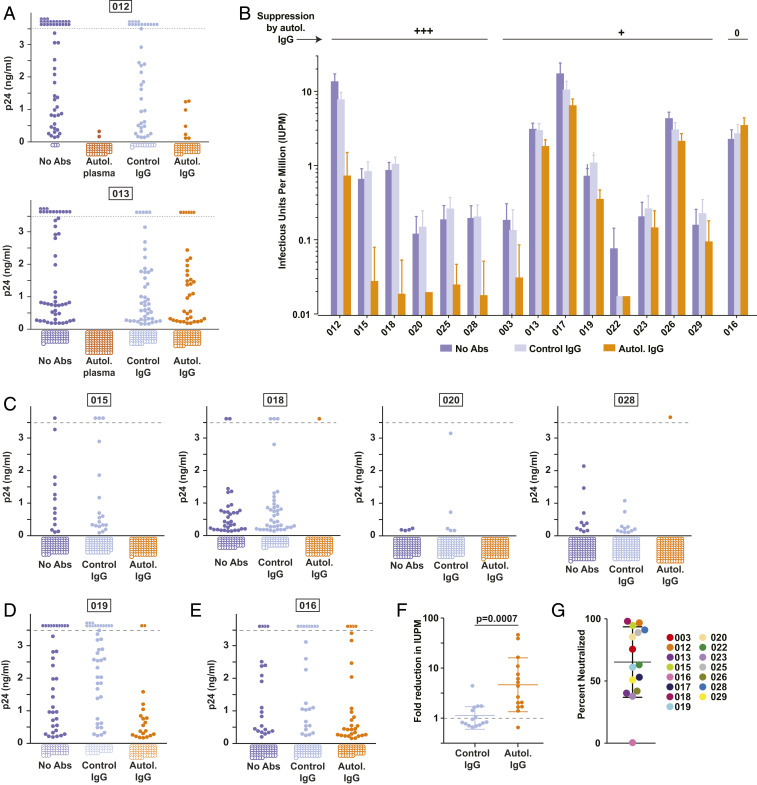
In initial experiments with cells from two donors, we found complete or near complete suppression of viral outgrowth by a 1:50 dilution of dialyzed, heat-inactivated autologous plasma in cultures set up near the limit dilution (Fig. 2 and SI Appendix, Table S2). However, suppression by plasma could reflect the presence of other inhibitory factors, and therefore we used purified IgG in this and all subsequent experiments. For study participant 012, viral outgrowth was largely suppressed by autologous IgG at a concentration of 50 μg/mL while control IgG from HIV-negative donors had no effect. Suppression was evident both as a reduction in the number of wells with outgrowth and a decreased level of p24 in wells with outgrowth (Fig. 2A). Interestingly, in the second participant (013) (Fig. 2A), autologous IgG had only a small suppressive effect at 50 μg/mL, reducing the fraction of wells with outgrowth (36 of 100 vs. 45 of 100). Given that normal IgG concentrations in plasma are ∼104 μg/mL, the 50 μg/mL concentration used in these experiments represents only ∼0.5% of the normal concentration and thus likely underestimates the suppressive effects of autologous antibodies.
Fig. 2.
Autologous plasma and purified autologous IgGs inhibit viral rebound in QVOAs. (A) QVOAs for two participants (012 and 013) set up under four conditions: no antibodies added, autologous plasma (1:50 dilution), control IgG from HIV-negative donors (50 μg/mL), and purified autologous IgG (50 μg/mL). Graphs show p24 values in the culture supernatants from individual wells at the end of the 14- to 21-d culture and include data from all dilutions except those at which most (>60%) wells were positive under all conditions. The dotted line indicates the upper limit of quantification for the p24 ELISA. Off-scale positive wells are shown above the dotted line. Negative wells are shown as open circles below the graph. (B) Frequency of CD4+ T cells giving rise to viral outgrowth in cultures without antibodies, with control IgG (50 μg/mL), or with autologous IgG (50 μg/mL). Results of QVOAs are expressed as estimated infectious units per million (IUPM) cells ± SD calculated using maximum likelihood (see Methods). (C) Supernatant p24 levels for representative participants showing strong suppression (>80%) of viral outgrowth by autologous IgG. (D) Representative culture data for a donor (019) showing weak suppression of outgrowth by autologous IgG. (E) Culture data for donor 016, who showed no suppression of outgrowth by autologous IgG. (F) Fold change in IUPM relative to cultures without antibody. Each symbol represents a different donor. Bars indicate geometric mean ± SD. (G) Estimated fraction of latent proviruses that fail to grow out in the presence of autologous IgG for each study participant calculated on the basis of the reduction in IUPM. Bars indicate mean ± SD.
To further explore the suppressive potential of autologous antibodies, we tested additional donors to determine whether low concentrations of autologous IgG antibodies could suppress outgrowth from the latent reservoir (Fig. 2B). In total, cultures from 14 of 15 individuals showed a decrease in viral outgrowth in the presence of autologous contemporaneous IgG, as evidenced by a decrease in infectious units per million (IUPM) resting CD4+ T cells relative to the control QVOAs set up either without antibodies or with control IgG from uninfected donors (Fig. 2B). However, there was considerable variability in the extent of this decrease (Fig. 2 and SI Appendix, Fig. S2 and Table S2). For 6 of 15 participants, the decreases were substantial (>80%). Examples are shown in Fig. 2C. For 8 of the remaining participants, autologous IgG caused only a minor reduction in the number of positive wells or in the amount of p24 produced in positive wells (Fig. 2D and SI Appendix, Fig. S2). Only participant 016 showed no apparent decrease in viral outgrowth (Fig. 2E).
To further quantitate the suppressive effect of autologous IgG on viral outgrowth, we calculated the fold reduction in IUPM values caused by control IgG from HIV-negative donors and by autologous IgG from study participants (Fig. 2F). Paired analysis revealed a significant reduction in the frequency of latently infected cells giving rise to viral outgrowth in the presence of autologous IgG (P = 0.0007). On average, there was a 4.6-fold reduction in IUPM. Autologous IgG blocked outgrowth of an average of 65% of viruses in the latent reservoir (range 0 to 98%) (Fig. 2G). For 11 of 15 participants, there was a reduction of >50%, and, for 6 of 15 participants, the reduction was >80% (Fig. 2G).
Phylogenetic Analysis of Reservoir Viruses Neutralized by Autologous IgG.
We next investigated whether the viruses growing out in the presence of autologous IgGs were genetically distinct from other replication-competent HIV-1 variants persisting in the latent reservoir of the same individual. Viral RNA was isolated from supernatants of p24+ QVOA wells, and the env gene was amplified using RT-PCR. The entire env gene (nt 6225 to 8795 of the HXB2 sequence) was then analyzed by single genome sequencing (SGS) (Fig. 1C). The env amplicon had a clonal prediction score of 94, suggesting that 94% of sequences that are identical in this region are also identical throughout the whole genome (69). A total of 699 independent QVOA isolate sequences from 12 participants were examined. Four of 15 participants (015, 016, 017, and 019) had previously participated in the AIDS Clinical Trials Group (ACTG) trial A5340 (57, 60). Sequences obtained during that trial 3 to 4 y prior to this study were also included in the corresponding phylogenetic trees. Phylogenetic analysis demonstrated the expected donor-specific clustering of sequences (SI Appendix, Fig. S3). The average pairwise distance within each participant varied considerably among participants (SI Appendix, Fig. S3).
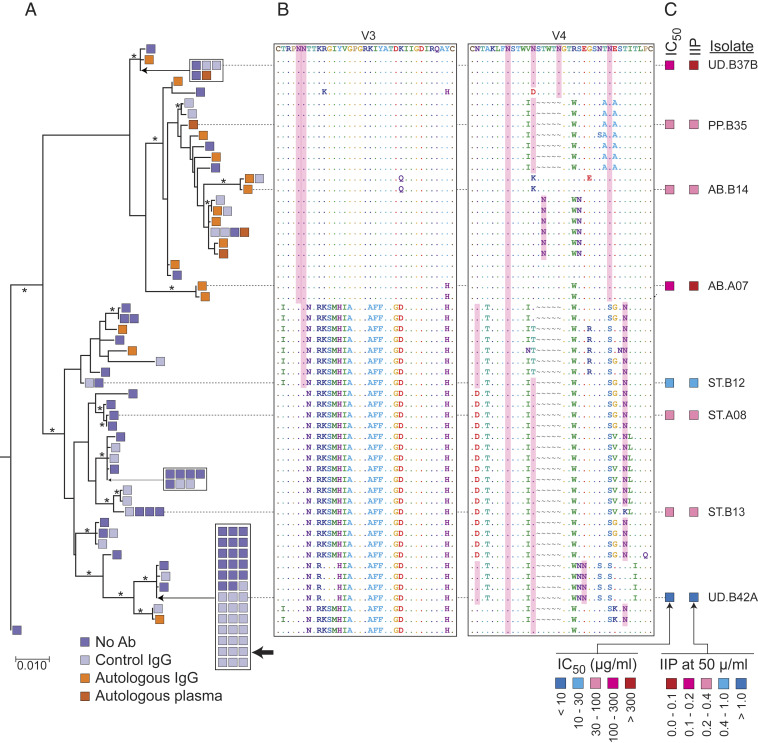
We first investigated env sequences from participants with a large decrease in viral outgrowth in the presence of autologous IgG. Participant 012 fit this criterion and had sufficient sequence data to infer relationships among sequences collected from the different treatment conditions. There was distinct clustering of isolates growing out in the presence of autologous antibodies (Fig. 3A). The overall composition of the reservoir is best appreciated by examining sequences of viruses growing out in the absence of antibody (blue and gray sequences in Fig. 3A). There are two main branches of this phylogenetic tree that appear with high bootstrap values (99.9%). One branch contains only 18% of the sequences obtained from wells without antibody but 84% of the isolates growing out in the presence of autologous IgG. This branch is distinguished by sequence changes and altered glycosylation sites in the V3 and V4 loops, as well as additional changes elsewhere in the env gene (Fig. 3B). These changes are consistent with viral evolution to escape from autologous neutralizing antibodies (41). Importantly, the other branch contained a large set of identical sequences accounting for 39 of 94 isolates (41%) growing out in the absence of antibody. This sequence was not found among 19 isolates growing out in the presence of antibody (P = 0.00375). Neutralization assays confirmed that this clone was antibody-sensitive (see Fig. 4 below). These results demonstrate that the latent reservoir includes viral variants, including expanded clones, that are sensitive to neutralization by autologous antibodies. The presence of expanded cellular clones carrying neutralization-sensitive variants may explain why viruses rebounding upon ART interruption can differ from those detected in standard viral outgrowth assays conducted in the absence of antibodies.
Fig. 3.
Phylogenetic relationship between antibody-sensitive and antibody-resistant QVOA isolates. (A) Maximum likelihood phylogenic tree of full-length env sequences of participant 012 QVOA isolates obtained under the four indicated conditions. Bar indicates substitutions per site. Bootstrap values greater than 70% are indicated by an asterisk. The large black arrow indicates a large set of independent, identical, antibody-sensitive isolates. (B) Sequences of the V3 and V4 regions of Env for the each isolate in A. Identity with isolate at the top of the tree is indicated by a period (.). Gaps are indicated by a “∼.” Asparagine residues conforming to the consensus sequence for N-linked glycosylation are shaded in pink. (C) Results of neutralization assays on pseudoviruses generated with the env sequences of the indicated isolates. Neutralization is quantified as the concentration of autologous IgG required for 50% neutralization (IC50); and the instantaneous inhibitory potential (IIP), the number of logs of inhibition produced by autologous IgG at 50 μg/mL (Fig. 4 and Methods).
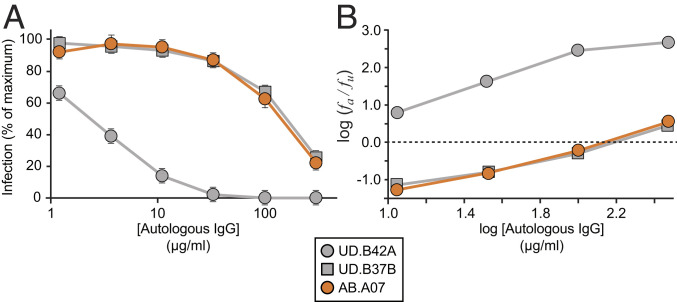
Fig. 4.
Representative dose–response curves for the inhibition of infectivity of pseudoviruses generated from participant 012 by autologous IgG. (A) Standard dose–response curves. Pseudoviruses were generated from representative QVOA isolates obtained in the presence of control IgG from uninfected donors (UD.B37B and UD.B42A) or of autologous IgG antibodies (AB.A07). Pseudoviruses were used to infect Tzm-bl cells (70, 71) in the presence of the indicated concentrations of autologous IgG. IC50 and IIP values were calculated using the median effect plot in B as previously described (72). The mean + SD of triplicate measurements are shown. Results for all isolates tested are in SI Appendix, Table S3. (B) Median effect plot of the data from A. fa is the fraction of infection events affected (inhibited) by antibody, and fu is the fraction unaffected. IIP is the number of logs of inhibition at a given antibody concentration and is determined from the slope and y intercept (IC50, dotted line) of regression lines for each isolate as previously described (72).
Direct Measurement of Neutralization of Latent Reservoir Isolates by Autologous IgG.
The observed reduction in viral outgrowth in the presence of autologous IgG could reflect neutralization of cell-free virus particles or inhibition of cell-to-cell spread. To provide direct evidence that the autologous IgG does have neutralizing activity against some Env variants, we pseudotyped an env-defective HIV-1 reporter virus with participant-derived Env variants (Fig. 1D), as described in Methods. Pseudoviruses were subsequently titrated on TZM-bl cells (70) to determine a linear range of infection for each pseudovirus. The pseudoviruses were then tested in a TZM-bl–based neutralization assay (70, 71) in the presence of a range of autologous IgG concentrations. Raw data from the TZM-bl–based neutralization assays were plotted in standard dose–response curves and analyzed using a median effect model to estimate the concentration that inhibits response by 50% (IC50) and instantaneous inhibitory potential (IIP). IIP is a pharmacodynamic measure that uses the IC50 and the slope of the dose–response curve to estimate the number of logs of suppression of infectivity produced by antiretroviral drugs (72) or neutralizing antibodies (73). Given that replication defective pseudoviruses are used in this assay, any suppression observed can be attributed to neutralization rather than inhibition of cell-to-cell spread.
The large clone found in participant 012 (represented by isolate UD.B42A) was sensitive to neutralization by autologous IgG, with an IC50 value of 2.4 μg/mL (Figs. 3 A and C and 4 and SI Appendix, Table S3). For monoclonal bNAbs, IC50 values below 2 μg/mL are considered to indicate significant antiviral activity (74). For autologous IgG, antibodies specific for the HIV-1 Env protein likely constitute a minute fraction of all of the antibodies present in the plasma. Therefore, the actual IC50 for the relevant autologous neutralizing antibodies in the plasma is likely much lower than the measured IC50 values for polyclonal IgG preparations. Another measure of antiviral activity is the IIP, which, for autologous IgG at 50 μg/mL, was 1.8 against isolate UD.B42A pseudovirus. This indicates almost 100-fold suppression of infectivity at this dose and likely much greater suppression at plasma IgG concentrations. Isolates with high sensitivity to neutralization, including those present in some expanded cellular clones, may not contribute to viral rebound following ART interruption.
Other isolates from the latent reservoir of participant 012 showed considerable resistance to neutralization by autologous IgG (Figs. 3C and 4 and SI Appendix, Table S3). For example, isolate AB.A07, which clustered with most of the isolates growing out in the presence of antibody, had an IC50 of 128 μg/mL and an IIP of 0.11 (Figs. 3 B and C and 4). As expected, some of the isolates from QVOAs set up in the absence of autologous IgG were also antibody-resistant. For example, isolate UD.B37B, which also clustered with most of the isolates growing out in the presence of autologous IgG, had an IC50 of 146 μg/mL and an IIP of 0.12 (Figs. 3 B and C and 4). Most of the other isolates tested from this participant showed some degree of resistance to autologous IgG (Fig. 3C and SI Appendix, Table S3). Similar results were obtained for other participants (Figs. 5–7). Together, these results demonstrate that the latent reservoir contains viruses that are sensitive to neutralization by autologous antibodies as well as viruses that are resistant. Moreover, the neutralization susceptibility of some expanded clones provides one explanation for the observed differences between viruses obtained in standard viral outgrowth assays and viruses growing out following treatment interruption.
Fig. 5.
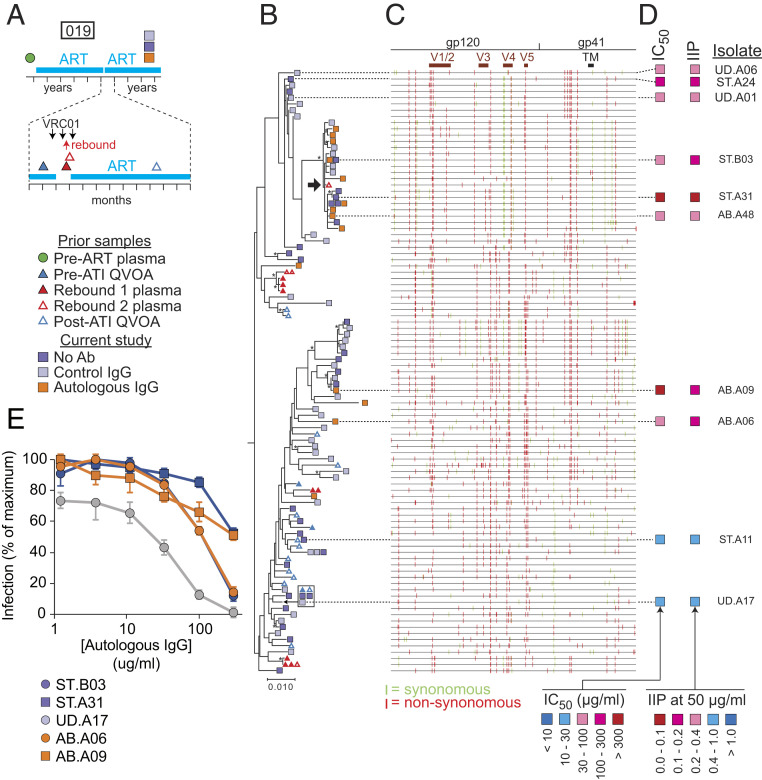
Phylogenetic analysis and neutralization sensitivity of isolates from participant 019. (A) Sampling time line. Participant 019 was part of a previous trial of bNAb VRCO1 that included a treatment interruption (57, 60). Times of VRC01 infusion and viral rebound are indicated by black and red arrows, respectively. Symbols indicate times of pre- and post-ATI QVOA samples, rebound plasma samples, and QVOA samples for the current study. (B) Maximum likelihood phylogenetic tree with midpoint rooting of full-length env sequences from the current study and the VRC01 study. One rebound sequence (black arrow) is closely related to several sequences growing out in the presence of autologous IgG in the current study. The bar indicates substitutions per site. Bootstrap values greater than 70% are indicated by an asterisk. (C) Highlighter plot indicating synonymous (green) and nonsynonymous (red) nucleotide differences relative to a participant consensus sequence. Positions of the variable loops (V1 to V5) in gp120 and the transmembrane (TM) domain in gp41 are indicated. (D) Results of neutralization assays on pseudoviruses generated with the env sequences of the indicated isolates. Neutralization is quantified as the concentration of autologous IgG required for 50% neutralization (IC50); and the instantaneous inhibitory potential (IIP), the number of logs of inhibition produced by autologous IgG at 50 μg/mL. See also SI Appendix, Table S3. (E) Representative dose–response curves for the inhibition of infectivity of participant 019 pseudoviruses by autologous IgG. Pseudoviruses were generated from representative isolates from a standard (ST) QVOA set up without antibodies, a QVOA set up with control IgG from uninfected donors (UD), or a QVOA set up with autologous IgG antibodies (AB). Pseudoviruses were used to infect Tzm-bl cells (70, 71) in the presence of the indicated concentrations of autologous IgG. IC50 and IIP values were calculated using the median effect model (72). Data are plotted as mean ± SD. See also SI Appendix, Table S3.
Fig. 7.
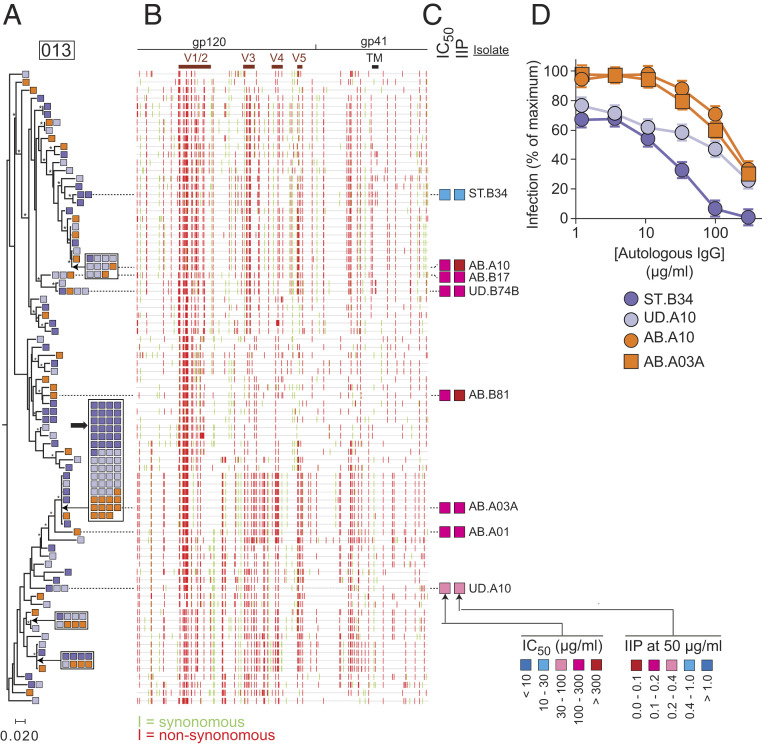
Phylogenetic analysis and neutralization sensitivity of isolates from participant 013. (A) Maximum likelihood phylogenetic tree with midpoint rooting of full-length env sequences. (B) Highlighter plot indicated synonymous (green) and nonsynonymous (red) nucleotide differences relative to the participant consensus sequence. Positions of the variable loops (V1 to V5) in gp120 and the transmembrane (TM) domain in gp41 are indicated. The large black arrow indicates one large potential clone including 12 isolates that grew out in the presence of autologous IgG. The bar indicates substitutions per site. Bootstrap values greater than 70% are indicated by an asterisk. (C) Results of neutralization assays on pseudoviruses generated with the env sequences of the indicated isolates. Neutralization is quantified as the concentration of autologous IgG required for 50% neutralization (IC50); and the instantaneous inhibitory potential (IIP), the number of logs of inhibition produced by autologous IgG at 50 μg/mL. See also SI Appendix, Table S3. (D) Representative dose–response curves for the inhibition of infectivity of pseudoviruses generated from participant 013 by autologous IgG. Pseudoviruses were generated from representative isolates from a standard (ST) QVOA set up without antibodies, a QVOA set up with control IgG from uninfected donors (UD), or a QVOA set up with autologous IgG antibodies (AB). Pseudoviruses were used to infect Tzm-bl cells (70, 71) in the presence of the indicated concentrations of autologous IgG. IC50 and IIP values were calculated using the median effect model (72). Data are plotted as mean ± SD.
In general, viruses sensitive to neutralization by autologous antibodies did not appear any less fit than resistant viruses. For example, the p24 levels achieved in outgrowth cultures carried out in the absence of autologous IgG were not correlated with neutralization sensitivity, as measured by IC50 (correlation coefficient = 0.154).
Relationship Between Viral Outgrowth and Rebound Viruses.
To explore the relationship between ex vivo outgrowth of viruses in the presence of autologous antibody and in vivo viral rebound following treatment interruption, we analyzed samples from participants who had previously participated in a clinical trial of the bNAb VRC01 (57, 60). In this trial, ART was interrupted during administration of VRC01 (Fig. 5A). ART was reinitiated after rebound to >1,000 copies per milliliter was detected and continued throughout sampling for the present study (∼3 y later, after infused VRC01 antibodies were no longer present). Thus, we can compare viruses growing out in the presence of autologous antibodies with viruses growing out during this previous treatment interruption. Results from representative participants (019 and 015) are shown in Figs. 5 and 6. For participant 019, rebound occurred 3 wk after ART interruption. We found moderate suppression (∼60%) of viral outgrowth in the QVOA by autologous IgG (Fig. 2 D and G). Phylogenetic analysis (Fig. 5B) showed a prominent branch containing 9 of 14 isolates (64.2%) growing out in the presence of autologous IgG but only 11 of 69 isolates (15.9%) growing out in the absence of autologous IgG (P = 0.0139). This branch showed sequence differences throughout the env gene, including differences in the variable loops and in the extracellular domain of gp41 (Fig. 5C). Importantly, this branch also contained a sequence detected upon viral rebound 3 y earlier (Fig. 5B, black arrow). Neutralization assays confirmed the relative resistance of isolates from this branch to autologous IgG (Fig. 5D and SI Appendix, Table S3). These results indicate a phylogenetic relationship between the ability of a viral variant to cause rebound upon treatment interruption in vivo and its ability to grow out in vitro in the presence of autologous IgG.
Fig. 6.
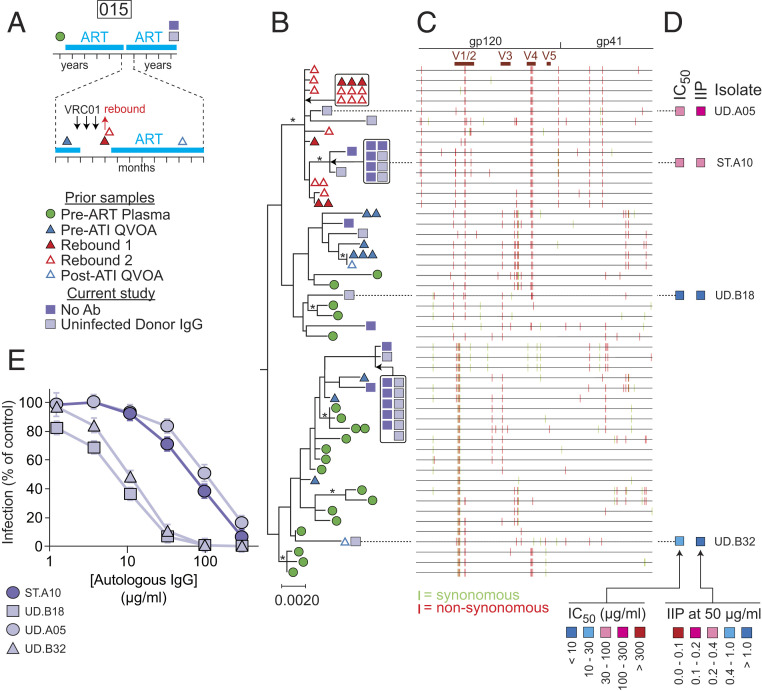
Phylogenetic analysis and neutralization sensitivity of isolates from participant 015. (A) Sampling time line. Participant 015 was part of a previous trial of bNAb VRCO1 that included a treatment interruption (57, 60). Times of VRC01 infusion and viral rebound are indicated by black and red arrows, respectively. Symbols indicate times of pre- and post-ATI QVOA samples, rebound plasma samples, and QVOA samples for the current study. (B) Maximum likelihood phylogenetic tree with midpoint rooting of full-length env sequences from the current study and the VRC01 study. The bar indicates substitutions per site. Bootstrap values greater than 70% are indicated by an asterisk. (C) Highlighter plot indicating synonymous (green) and nonsynonymous (red) nucleotide differences relative to the participant consensus sequence. Positions of the variable loops (V1 to V5) in gp120 and the transmembrane (TM) domain in gp41 are indicated. (D) Results of neutralization assays on pseudoviruses generated with the env sequences of the indicated isolates. Neutralization is quantified as the concentration of autologous IgG required for 50% neutralization (IC50); and the instantaneous inhibitory potential (IIP), the number of logs of inhibition produced by autologous IgG at 50 μg/mL. See also SI Appendix, Table S3. (E) Representative dose–response curves for the inhibition of infectivity of pseudoviruses generated from participant 015 by autologous IgG. Pseudoviruses were generated from representative isolates from a standard (ST) QVOA set up without antibodies and a QVOA set up with control IgG from uninfected donors (UD). No isolates were obtained in a QVOA set up with autologous IgG antibodies. Pseudoviruses were used to infect Tzm-bl cells (70, 71) in the presence of the indicated concentrations of autologous IgG. IC50 and IIP values were calculated using the median effect model (72). Data are plotted as mean ± SD.
Other regions of the 019 tree contained isolates sensitive to neutralization by autologous IgG (ST.A11 and UD.A17) (Fig. 5 B, D, and E). Antibody-sensitive isolate UD.A17 was identical in sequence throughout the env gene to two other isolates obtained in the absence of autologous antibodies and to isolates obtained before and after (but not during) ATI. Sensitive isolates lacked mutations in V1/2 and the extracellular domain of gp41 seen in resistant and rebound isolates (Fig. 5C). These results support the conclusions that latent reservoir viruses growing out in the presence of autologous IgG have Env proteins that are resistant to neutralization by autologous IgG and that other reservoir isolates from the same individual, including those that have undergone clonal expansion, may be more readily neutralized. These isolates can be detected in QVOAs set up without autologous antibodies but may not contribute to viral rebound in vivo.
Similar results were obtained with samples from other participants in the VRC01 trial. For participant 015, QVOAs performed ∼3 y after the VRC01 trial (Fig. 6A) showed a high level of suppression by autologous IgG (Fig. 2C), and no outgrowth was observed in the presence of autologous IgG, due in part to the low frequency of latently infected cells. The plasma rebound sequences obtained following ATI were likely from a single lineage that clustered with a group of env sequences obtained in the current study that demonstrated significant resistance to autologous IgG (isolates UD.A05 and ST.A10) (Fig. 6 B–E). Other isolates from separate branches of the tree showed sensitivity to neutralization by autologous IgG (UD.B18 and UD.B32). In participant 016, some rebound sequences from the ATI were identical to a large set of identical QVOA isolates obtained 3 y later, including isolates growing out in the presence of autologous IgG (SI Appendix, Fig. S4). These results highlight the relationship between viral rebound in vivo and in vitro outgrowth in the presence of autologous antibodies. Due to limited sampling prior to ATI, our results do not exclude reservoir reseeding during ATI with antibody-resistant variants.
Variable Patterns in the Autologous Neutralizing Antibody Response.
For several participants, the presence of autologous IgG in the QVOA caused only a small decrease in IUPM, and, for one participant (016), there was no decrease (Fig. 2 E and G). Phylogenetic trees for some of these participants (013, 016, and 017) did not demonstrate clustering of antibody-resistant env sequences (Fig. 7 and SI Appendix, Figs. S4 and S5). For example, isolates for participant 013 that grew out in the presence of autologous IgG were present throughout the phylogenetic tree (Fig. 7A). Tree branches characterized by differences in V3, V4, and V5 all included sequences that were resistant to autologous IgG, as confirmed by neutralization assays (Fig. 7 A–D). However, antibody-sensitive isolates were also found (for example, ST.B34). Importantly, several potential clones were detected. One very large potential clone, represented by isolate AB.A03A, showed relative resistance to neutralization by autologous IgG (IC50 = 134 μg/mL) and included several isolates that grew in the presence of 50 μg/mL autologous IgG.
Similar patterns were seen in other participants. Participant 016 also had a large set of identical isolates, including several that grew out in the presence of autologous IgG (SI Appendix, Fig. S4). Participant 017 showed overall low genetic diversity (SI Appendix, Fig. S3) but had numerous potential clones (SI Appendix, Fig. S5). Consistent with previous results (64), some of these appeared to have expanded in the 3 y after the A5340 trial. Two rebound sequences were closely related to QVOA isolates. Some potential clones included isolates growing out in the presence of autologous IgG. These results confirm that the latent reservoir contains viruses with different degrees of sensitivity to neutralization by autologous antibodies and suggest that, while some expanded clones of infected CD4+ T cells carry isolates sensitive to autologous neutralizing antibodies, others carry resistant variants. Due to low reservoir size, fewer isolates were obtained from the remaining participants, but, in general, there was a tendency for a reduction in outgrowth in the presence of autologous IgG (Fig. 2 and SI Appendix, Fig. S6).
Discussion
The latent reservoir in resting memory CD4+ T cells is the major barrier to curing HIV-1 infection (1–3). We modified the QVOA to reflect immune pressure from low concentrations of autologous IgG antibodies and showed that a substantial but variable fraction of latent reservoir viruses are susceptible to neutralization and unable to grow out in the presence of autologous contemporaneous IgG. A reduction in outgrowth was seen in 14 of 15 individuals, and, in 11 of 15, the reduction was greater than 50%. Using sequences obtained in a previous treatment interruption study (57, 60), we established a phylogenetic relationship between viruses growing out in a QVOA set up with autologous antibodies and viruses causing rebound in vivo. Together, these results indicate that the size of the reservoir likely to cause viral rebound may be considerably smaller than estimates based on standard viral outgrowth assays carried out in the absence of autologous antibodies or estimates based on analysis of genetically intact proviruses.
Our results are fully consistent with elegant studies from the laboratories of Shaw and coworkers (41) and Richman et al. (42) showing that, in untreated infection, the evolving antibody response neutralizes previous but not contemporaneous viral variants. Prior to initiation of ART, archiving of viral variants in the latent reservoir at different time points during chronic infection should result in a reservoir that includes both antibody-sensitive and antibody-resistant variants, as proven here. Recent studies have shown many replication-competent proviruses comprising the latent reservoir are incorporated near the time of ART initiation (75, 76). If these variants are less susceptible to antibody neutralization, then variation in the susceptibility of reservoir viruses to the repertoire of antibodies circulating during ART is expected.
Our study also provides one simple and direct explanation for the puzzling recent observation that viruses that rebound in vivo after treatment interruption and the viruses that grow out in the QVOA prior to interruption are often genetically distinct (59, 60). Given the large number of viral variants that arise in individuals who begin ART during chronic infection, sampling issues provide one explanation. However, in many individuals, large clones of infected CD4+ T cells carrying identical proviruses dominate the latent reservoir (10–19). For this reason, it is unlikely that the disparity between these two viral populations is solely caused by sampling error. Recombination of viruses in vivo has also been suggested as a factor affecting viral rebound following ATI (59, 66, 67). However, it is unclear when this recombination occurs. Immune system pressures have also been implicated in the selection of rebound viruses during ATIs. This would provide a much simpler explanation for the differences between rebound viruses and those obtained in the absence of immune system pressure in standard outgrowth assays. In a trial of the bNAb VRC01, Bar et al. (57) and Bar and coworkers (60) explored the possibility that antibodies could act as a sieve determining which viruses in the reservoir contribute to rebound viremia. They noted a trend toward greater susceptibility of QVOA viruses to neutralization by autologous plasma relative to rebound viruses although VRC01 was also present in the plasma.
In light of these studies (57, 59, 60, 66, 67), we explored the effect of immune pressure in determining which latent reservoir viruses can contribute to rebound. Here, we directly demonstrate that the autologous neutralizing antibody response can block outgrowth of a significant fraction of viruses in the reservoir. If the reservoir is dominated by large clones carrying antibody-sensitive proviruses (for example, participant 012, isolate UD.B42A) (Figs. 3 and 4), then a difference between rebound viruses and those obtained in standard QVOA assays would clearly be expected. Compared to other isolates from participant 012, this clone was readily neutralized by autologous IgG. The fraction of reservoir viruses neutralized by autologous IgG varied in different individuals, and, in some participants, we found large clones carrying antibody-resistant viruses. Nevertheless, our results establish that immune pressure can affect which viruses in the reservoir are capable of causing viral rebound and suggest that QVOAs carried out in the presence of autologous antibodies might provide more accurate prediction of time to viral rebound than current approaches. Importantly, recombination is unlikely to be a factor in viral outgrowth under the limiting dilution conditions used here. During periods of active viral replication, recombination clearly contributes to the generation of viral variants which can be selected under immune pressure and deposited in the latent reservoir. Our results demonstrate that immune pressure can regulate viral outgrowth from the latent reservoir in conditions where further recombination is not a factor.
Our results also suggest that other forms of immune pressure be considered in assessing what fraction of latent reservoir viruses can cause rebound. For example, Hahn and coworkers have shown that resistance to inhibition by type I interferons is a critical determinant of transmission (77), and the same effects may be operative during viral rebound following treatment interruption. The ability of cytolytic T lymphocyte and natural killer (NK) cell responses to control rebound remains unclear. bNAbs show variable activity against viruses comprising the reservoir (78, 79), and infusions of bNAbs can delay viral rebound (57, 58). Continuous production of bNAbs delivered by adeno-associated virus vectors has suppressed viral replication and prevented rebound in one SHIV-infected macaque (80).
Overall, these results support the hypothesis that autologous neutralizing antibody responses can prevent viral rebound from at least a fraction of viruses in the reservoir. This is important as eradication strategies have previously focused on completely eliminating the reservoir of replication-competent viruses. Given that a subset of these viruses are unable to rebound due to their susceptibility to antibody neutralization, developing therapeutic immunization strategies to induce antibodies to the nonneutralized fraction is an aspirational goal that might allow treatment-free remission. Our initial analysis of antibody-resistant viruses did not reveal shared features that could be readily targeted, but additional analysis is needed. A recently described comprehensive atlas of bNAb escape mutations (35) may be useful in this effort. A more distant possibility is a personalized therapeutic vaccine approach targeting viral variants resistant to existing immune responses. Another important unknown is the stability of the autologous neutralizing antibody response. It is also unclear whether the antibody response can continue to evolve in the setting of ART, perhaps in response to residual viremia (10, 11). In any event, our results support the optimistic conclusion that autologous antibody responses control rebound from some of the viruses in the latent reservoir. Finding a way to induce antibodies or other immune responses to block outgrowth of the remaining viruses could lead to a functional cure.
Methods
Study Participants.
HIV-1–infected adults on suppressive ART were enrolled at the University of Pennsylvania. Selection criteria included viral suppression for >6 mo and generally undetectable plasma HIV-1 RNA levels (<50 copies per mL). Participants underwent leukapheresis under a protocol approved by the University of Pennsylvania Institutional Review Board (IRB). All participants provided written informed consent. Coded, deidentified leukapheresis products were shipped by same-day courier to the Johns Hopkins School of Medicine for processing and analysis under a protocol approved by a Johns Hopkins University School of Medicine IRB. Participant characteristics are given in SI Appendix, Table S1.
Resting CD4+ T Cell and Plasma Isolation.
Peripheral blood mononuclear cells (PBMCs) and autologous plasma were isolated from the leukapheresis product using density centrifugation on a Ficoll-Hypaque gradient. CD4+ T cells were enriched using the EasySep Human CD4+ T Cell Negative Depletion Kit (Stemcell Technologies). Resting CD4+ T cells were then subsequently enriched from total CD4+ T cells using negative depletion via anti-CD69, anti-CD25, and anti-HLA-DR MicroBeads (Miltenyi Biotech). Collected resting CD4+ T cell populations are greater than 95% pure (81).
Isolation of IgG Antibodies.
Autologous IgG antibodies were isolated from heat-inactivated leukapheresis plasma using protein A spin purification columns (Thermo Scientific Pierce Protein Biology). IgG antibodies were eluted from protein A columns, neutralized, and dialyzed using multiple exchanges of phosphate-buffered saline (PBS), pH 7.2, at 4 °C. Final antibody concentrations were in the range of 3 to 5 mg/mL The antibody preparations had no nonspecific neutralizing activity against murine leukemia virus (MuLV) in Tzm.bl assays (IC50 > 500 μg/mL).
Western Blot Analysis.
Qualitative detection of HIV-1 antigen reactivity of purified participant IgG was analyzed using a GS HIV-1 diagnostic Western blot kit (Bio-Rad) according to the manufacturer’s instructions.
QVOAs.
After initial assessment of intact proviral frequencies using the IPDA (65), purified resting CD4+ T cells were seeded into QVOA culture wells at 1 × 106, 2 × 105, or 4 × 104 cells per well with up to 100 replicate wells per dilution. Cultures were set up without antibody, with 50 μg/mL IgG from uninfected donors (Sigma Aldrich), or with purified autologous IgG at 50 μg/mL Cultures were carried out as described (64, 81). With each split and media change over the course of 21 d, the appropriate IgG was added to maintain a final concentration of 50 μg/mL. Culture wells were assayed for HIV-1 p24 on day 14. If no wells were positive, cultures were continued until day 21.
RNA Isolation and Complementary DNA Synthesis.
Viral RNA was isolated from supernatants of p24+ wells using a 96-Well Spin Plate RNA Isolation Kit according to the manufacturer’s instructions (Zymo Research). RNA was subsequently subjected to env-specific complementary DNA (cDNA) synthesis using the primer envB3out (5′ TTGCTACTTGTG-ATTGCTCCATGT 3′; nt 8913 to 8936 of the HXB2 sequence) (82) and the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific). Remaining RNA was digested with RNAseH at 37 °C for 20 min.
Single Genome Envelope Sequencing.
Env-specific cDNA was amplified with nested PCR. The outer PCR was completed in a volume of 10 μL containing 1× High Fidelity PCR Buffer, 2 mM MgCl2, 0.2 mM 2′-deoxynucleoside 5′-triphosphate (dNTP), and 0.4 U of Platinum Taq DNA Polymerase High Fidelity with a 0.2 μM final concentration of primers envB5out (5′ TAGAGCCCTGGAAGCATCCAGGAAG 3′; nt 5853 to 5877 of the HXB2 sequence) (82) and envB3out. PCR conditions were 94 °C for 2 min (94 °C for 30 s; 50 °C for 30 s; 72 °C for 2.5 min) times 44, and 72 °C for 3 min. The nested, inner PCR was completed in a total volume of 20 μL using 2 μL of the first round PCR product that had been diluted 1:1 with Tris⋅HCl, pH 8.0, and 0.2 μM of the primers envB5in (5′ TTAGGCATCTCCTATGGCAGGAAGAAG 3′; nt 5957 to 5983 of the HXB2 sequence) (82) and envB3in (5′ GTCTCGAGATACTGCTCCCACCC 3′; nt 8882 to 8904 of the HXB2 sequence) (82). PCR conditions were 94 °C for 2 min (94 °C for 30 s; 55 °C for 30 s; 72 °C for 2.5 min) times 41, and 72 °C for 3 min. PCR products were subsequently run on a 1% agarose gel for analysis. Positive QVOA supernatant PCR products that had a lower than 40% amplification efficiency were subsequently cleaned using the Monarch PCR and DNA Cleanup Kit according to the manufacturer’s instructions (New England Biolabs). Sanger sequencing was completed using the primers YH5EnvOut (5′ ATGGCAGGAAGAAGCGGAGACAG 3′; nt 5970 to 5992 of the HXB2 sequence) (62), BKRev16 (5′ ATGGGAGGGGCATACATTGCT 3′; nt 7520 to 7540 of the HXB2 sequence) (18), BKFor16 (5′ TTTAATTGTGGAGGAGAATTTTTCTA 3′; nt 7350 to 7375 of the HXB2 sequence) (18), and envB3in. Consensus sequences of individual env variants were assembled using multiple SGS-derived env sequences for each culture well. GenBank accession numbers for QVOA env sequences are MW077921–MW078304.
Pseudovirus Generation.
For selected isolates, env expression cassettes were generated using the pcDNA 3.4 TOPO TA Cloning Kit (Thermo Fisher Scientific) as per the manufacturer’s instructions. Then 25 μg of cytomegalovirus (CMV)-driven env-expressing plasmids were cotransfected into HEK293T cells with 30 μg of pNL4-3 ΔEnv GFP (72) using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific) in the presence of 5 μg of pAdvantage (Promega) to enhance protein expression. HEK293T cells were incubated at 37 °C for 72 h, and supernatants containing isolate-specific env-expressing pseudoviruses were subsequently harvested, centrifuged to remove debris, and snap frozen.
Neutralization Assay.
Pseudoviruses were initially titrated on Tzm-bl cells (70, 71) to determine a linear range of infection for each pseudovirus. Infections of Tzm-bl cells were then performed in triplicate with a concentration of virus in the linear range. The virus was preincubated with autologous IgG at 37 °C for 90 min starting at a concentration of 300 μg/mL and threefold serially diluting to a concentration of 1.23 μg/mL. Virus–antibody mixtures were then added to Tzm-bl cells and incubated for 48 h at 37 °C. Control wells with cells only, virus only, and cells with virus and uninfected donor IgG were included. All infections were performed in the presence of 10 μg/mL DEA-Dextran. Infection was measured by luciferase production after 48 h using Bright-Glo Reagent (Promega). For each viral isolate, the maximum degree of infection was determined using wells with no antibody or very low concentrations of antibody. Inhibition was expressed as a fraction of maximum infection. IC50 and IIP values were determined as previously described (72) using the linear part of the median-effect curve.
Sequence and Phylogenetic Analysis.
Reads from SGS experiments were aligned to HXB2 using Geneious Prime software. Nucleotide assemblies were codon aligned by hand, and unique sequences were obtained using ElimDupes (Los Alamos National Laboratories, https://www.hiv.lanl.gov/content/index) with a 100% threshold for identical sequences. Maximum likelihood trees were constructed from these alignments with PhyML v3.0 using a general time reversible (GTR) substitution model, incorporating invariant and gamma-distributed sites with four rate categories. Bootstrapping with 1,000 replicates was implemented, and tree improvement was optimized by the nearest neighbor interchange branch-swapping algorithm. Midpoint rooting was determined by Tree Rate (Los Alamos National Laboratories, https://www.hiv.lanl.gov/content/index). Trees were visualized in MEGA7, and annotations with data were performed in Adobe Illustrator.
To exclude cross-contamination and determine average pairwise distances, env sequence sets from all participants were compiled with Clade B reference sequences and codon aligned using MUSCLE 3.8. A neighbor-joining tree was constructed with Treemaker from Los Alamos National Laboratories (https://www.hiv.lanl.gov/content/index) under a GTR substitution model.
Supplementary Material
Acknowledgments
This work was supported by the NIH Martin Delaney Immunotherapy for HIV Cure (UM1 AI126603), BEAT-HIV (UM1 AI126620), and Delaney AIDS Research Enterprise (UM1 AI12661) Collaboratories, by the Johns Hopkins Center for AIDS Research (P30AI094189), by the Howard Hughes Medical Institute, the Bill and Melinda Gates Foundation (OPP1115715), the Kean Family Professorship, and the Robert I. Jacobs Fund of the Philadelphia Foundation.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020617117/-/DCSupplemental.
Data Availability.
Viral sequences have been deposited in the GenBank database (accession nos. MW077921–MW078304).
References
- 1.Finzi D., et al. , Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Chun T.-W., et al. , Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 94, 13193–13197 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong J. K., et al. , Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Mbonye U., Karn J., The molecular basis for human immunodeficiency virus latency. Annu. Rev. Virol. 4, 261–285 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Chun T.-W., et al. , Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. U.S.A. 95, 8869–8873 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitney J. B., et al. , Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512, 74–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siliciano J. D., et al. , Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Strain M. C., et al. , Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: Intrinsic stability predicts lifelong persistence. Proc. Natl. Acad. Sci. U.S.A. 100, 4819–4824 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crooks A. M., et al. , Precise quantitation of the latent HIV-1 reservoir: Implications for eradication strategies. J. Infect. Dis. 212, 1361–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobin N. H., et al. , Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: Expression of archival virus and replication of virus. J. Virol. 79, 9625–9634 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey J. R., et al. , Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80, 6441–6457 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomont N., et al. , HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15, 893–900 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maldarelli F., et al. , HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner T. A., et al. , HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonetti F. R., et al. , Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, 1883–1888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenzi J. C. C., et al. , Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc. Natl. Acad. Sci. U.S.A. 113, E7908–E7916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosmane N. N., et al. , Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J. Exp. Med. 214, 959–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bui J. K., et al. , Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 13, e1006283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee G. Q., et al. , Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J. Clin. Invest. 127, 2689–2696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrich T. J., et al. , Human immunodeficiency virus type 1 persistence following systemic chemotherapy for malignancy. J. Infect. Dis. 216, 254–262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendoza P., et al. , Antigen-responsive CD4+ T cell clones contribute to the HIV-1 latent reservoir. J. Exp. Med. 217, e20200051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davey R. T., Jr, et al. , HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc. Natl. Acad. Sci. U.S.A. 96, 15109–15114 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothenberger M. K., et al. , Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc. Natl. Acad. Sci. U.S.A. 112, E1126–E1134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sengupta S., Siliciano R. F., Targeting the latent reservoir for HIV-1. Immunity 48, 872–895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deeks S. G., HIV: Shock and kill. Nature 487, 439–440 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Archin N. M., et al. , Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487, 482–485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker B. D., et al. , HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature 328, 345–348 (1987). [DOI] [PubMed] [Google Scholar]
- 28.Borrow P., Lewicki H., Hahn B. H., Shaw G. M., Oldstone M. B., Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68, 6103–6110 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koup R. A., et al. , Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68, 4650–4655 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz J. E., et al. , Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283, 857–860 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Burton D. R., et al. , Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Saphire E. O., et al. , Crystal structure of a neutralizing human IGG against HIV-1: A template for vaccine design. Science 293, 1155–1159 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Calarese D. A., et al. , Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300, 2065–2071 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Scheid J. F., et al. , Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Dingens A. S., Arenz D., Weight H., Overbaugh J., Bloom J. D., An antigenic atlas of HIV-1 escape from broadly neutralizing antibodies distinguishes functional and structural epitopes. Immunity 50, 520–532.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwong P. D., Mascola J. R., Human antibodies that neutralize HIV-1: Identification, structures, and B cell ontogenies. Immunity 37, 412–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burton D. R., Mascola J. R., Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol. 16, 571–576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albert J., et al. , Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4, 107–112 (1990). [DOI] [PubMed] [Google Scholar]
- 39.Montefiori D. C., et al. , Homotypic antibody responses to fresh clinical isolates of human immunodeficiency virus. Virology 182, 635–643 (1991). [DOI] [PubMed] [Google Scholar]
- 40.Burns D. P., Collignon C., Desrosiers R. C., Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J. Virol. 67, 4104–4113 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei X., et al. , Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Richman D. D., Wrin T., Little S. J., Petropoulos C. J., Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U.S.A. 100, 4144–4149 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doores K. J., et al. , Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. U.S.A. 107, 13800–13805 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simek M. D., et al. , Human immunodeficiency virus type 1 elite neutralizers: Individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83, 7337–7348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C.-L., et al. , Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science 352, 1001–1004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mascola J. R., et al. , Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6, 207–210 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Ko S.-Y., et al. , Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature 514, 642–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pegu A., et al. , Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci. Transl. Med. 6, 243ra88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gautam R., et al. , A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature 533, 105–109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein F., et al. , HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492, 118–122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barouch D. H., et al. , Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503, 224–228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horwitz J. A., et al. , HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc. Natl. Acad. Sci. U.S.A. 110, 16538–16543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shingai M., et al. , Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503, 277–280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halper-Stromberg A., et al. , Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 158, 989–999 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lynch R. M. et al.; VRC 601 Study Team , Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 7, 319ra206 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Caskey M., et al. , Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 23, 185–191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bar K. J., et al. , Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N. Engl. J. Med. 375, 2037–2050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheid J. F., et al. , HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535, 556–560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen Y. Z., et al. , Relationship between latent and rebound viruses in a clinical trial of anti-HIV-1 antibody 3BNC117. J. Exp. Med. 215, 2311–2324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salantes D. B., et al. , HIV-1 latent reservoir size and diversity are stable following brief treatment interruption. J. Clin. Invest. 128, 3102–3115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borducchi E. N., et al. , Antibody and TLR7 agonist delay viral rebound in SHIV-infected monkeys. Nature 563, 360–364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ho Y.-C., et al. , Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bruner K. M., et al. , Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 22, 1043–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laird G. M., Rosenbloom D. I., Lai J., Siliciano R. F., Siliciano J. D., Measuring the frequency of latent HIV-1 in resting CD4+ T cells using a limiting dilution coculture assay. Methods Mol. Biol. 1354, 239–253 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Bruner K. M., et al. , A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vibholm L. K., et al. , Characterization of intact proviruses in blood and lymph node from HIV-infected individuals undergoing analytical treatment interruption. J. Virol. 93, e01920-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu C. L., et al. , Relationship between intact HIV-1 proviruses in circulating CD4+ T cells and rebound viruses emerging during treatment interruption. Proc. Natl. Acad. Sci. U.S.A. 115, E11341–E11348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z., et al. , Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc. Natl. Acad. Sci. U.S.A. 115, E2575–E2584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laskey S. B., Pohlmeyer C. W., Bruner K. M., Siliciano R. F., Evaluating clonal expansion of HIV-infected cells: Optimization of PCR strategies to predict clonality. PLoS Pathog. 12, e1005689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Montefiori D. C., Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 485, 395–405 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Sarzotti-Kelsoe M., et al. , Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 409, 131–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen L., et al. , Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat. Med. 14, 762–766 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Webb N. E., Montefiori D. C., Lee B., Dose-response curve slope helps predict therapeutic potency and breadth of HIV broadly neutralizing antibodies. Nat. Commun. 6, 8443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bar-On Y., et al. , Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat. Med. 24, 1701–1707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abrahams M. R., et al. , The replication-competent HIV-1 latent reservoir is primarily established near the time of therapy initiation. Sci. Transl. Med. 11, eaaw5589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pankau M. D., et al. , Dynamics of HIV DNA reservoir seeding in a cohort of superinfected Kenyan women. PLoS Pathog. 16, e1008286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iyer S. S., et al. , Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness. Proc. Natl. Acad. Sci. U.S.A. 114, E590–E599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chun T.-W., et al. , Broadly neutralizing antibodies suppress HIV in the persistent viral reservoir. Proc. Natl. Acad. Sci. U.S.A. 111, 13151–13156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ren Y., et al. , Susceptibility to neutralization by broadly neutralizing antibodies generally correlates with infected cell binding for a panel of clade B HIV reactivated from latent reservoirs. J. Virol. 92, e00895-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martinez-Navio J. M., et al. , Adeno-associated virus delivery of anti-HIV monoclonal antibodies can drive long-term virologic suppression. Immunity 50, 567–575.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Siliciano J. D., Siliciano R. F., Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol. Biol. 304, 3–15 (2005). [DOI] [PubMed] [Google Scholar]
- 82.Evering T. H., et al. , Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog. 8, e1002506(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Viral sequences have been deposited in the GenBank database (accession nos. MW077921–MW078304).