Abstract
Oncolytic virus is an effective therapeutic strategy for cancer treatment, which exploits natural or manipulated viruses to selectively target and kill cancer cells. However, the innate antiviral system of cancer cells may resistant to the treatment of oncolytic virus. M1 virus is a newly identified oncolytic virus belonging to alphavirus species, but the molecular mechanisms underlying its anticancer activity are largely unknown. Cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assays. RNA seq analysis was used to analyze the gene alternation after M1 virus infection. Small interfering RNAs transfection for gene knockdown was used for gene functional tests. Caspase-3/7 activity was detected by Caspase-Glo Assay Systems. A mice model of orthotopic bladder tumor was established to determine the oncolytic effectiveness of the M1 virus. The expression of cleaved-Caspase 3 as well as Ki-67 in tumor cells were detected by immunohistochemical analysis. To further define the molecular factors involved in M1 virus-mediated biological function, we knocked down genes related to alphavirus’ activity and found that CCDC6 plays an important role in the oncolytic activity of M1 virus. Moreover, knocked down of CCDC6 augments the reproduction of M1 virus and resulted in endoplasmic reticulum (ER) stress-induced cell apoptosis in vitro as well as in vivo orthotopic bladder cancer model. Our research provides a rational new target for developing new compounds to promote the efficacy of oncolytic virus therapy.
Keywords: Oncolytic virus, M1 virus, CCDC6, Interferon pathway
Abbreviations: ATM, ataxia telangiectasia-mutated; CCDC6, Coiled-coil-domain containing 6; DAUCs, area under the curve; DNA-PK, DNA-dependent protein kinase; DNA-PKI, DNA-PK inhibitor; HRP, horseradish peroxidase; IFNs, Interferons; ISG, interferon-stimulated genes; PVDF, polyvinylidene fluoride; RT, room temperature; siRNA, Small interfering RNAs; VCP, valosin-containing protein; XBP1, X-box binding protein 1; ZAP, zinc finger antiviral protein
Introduction
Oncolytic virus is developing as an effective anticancer therapy for its dual effect to directly kill cancer cells and induce anticancer immunity [1], [2], [3], [4], [5]. After selectively targeting tumor cells, the oncolytic virus replicates and lyse the malignant cells that lead to the expression of abundant tumor antigen and subsequently accelerate the process of antitumor memory. Nonetheless, the therapeutic effectiveness of oncolytic virus as a single agent in patients is inadequate [6]. Hence, efforts to pursue a different strategy to augment the prospective of oncolytic therapy are still needed.
One of the most effective methods to enhance oncolytic therapy is to find out the candidates that facilitate virus replication in the cancer cells. Many viruses have been engineered to exploit the aberrantly expressed surface proteins or have taken benefit of the atypical signaling pathway in tumor cells to enhanced the survival rate and increased the infective efficiency. The virus either kills cancer cells or creates a favorable environment for adaptive immunity via replication inside the cancer cells. The mechanism for selectively infect cancer cells is needed to be illuminated to guarantee the therapeutic effect. Alphavirus M1, a natural oncolytic virus found in the 1960s from the Hainan province of China and belongs to getah-like virus species [7]. It has been identified as a probable antitumor agent that specifically targets numerous cancer cells in vitro, in vivo, as well as ex vivo [7,8]. M1 virus induces sustained and severe ER stress and leads to apoptosis of tumor cells with Zinc-finger antiviral protein deficiency [9,10]. However, the underlying mechanisms for the M1 virus specifically recognize and replicate in cancer cells has not been fully explained.
Interferons (IFNs) are pleiotropic cytokines and involved in the development of cancer [11,12]. IFNs play an important role in various biological processes, such as cell differentiation, apoptosis, immune responses, antipathogens, and anticancers [12]. Type I interferon (type I IFN) is critical for immune responses to cancer by promoting memory T-cell survival and then control the immunosurveillance of cancer cells [13,14]. Furthermore, type I IFN showed inhibitory effects in several types of tumor cells via limits cell proliferation, drive senescence, and apoptosis. Type I IFN may have contrary effects to promote tumor progression [15], therefore, understanding the associated signaling pathways of type I IFN is critical for therapeutic intervention. Coiled-coil-domain containing 6 (CCDC6) is the substrate of ataxia telangiectasia-mutated (ATM) and involved in ATM-mediated cellular response, such as cell apoptosis, DNA damage response, and DNA repair [16], [17], [18]. Loss of the DNA repair function of CCDC6 has been found in many human cancers [19,20]. Moreover, some studies have also shown that CCDC6 is associated with drug resistance of lung cancer [21]. But the role of CCDC6 in oncolytic virus-mediated function has not been clarified yet.
Here, we identify that the antitumor potency of M1 is related to its replication in the cancer cells, which is blocked by the antiviral interferon system. Small interfering RNAs (siRNA) screening indicates CCDC6 has an antiviral influence against the M1 virus via regulating interferon-stimulated genes (ISG). Knockdown of CCDC6 (siCCDC6) enhanced oncolytic effects of M1 virus through ER stress-mediated apoptosis. The current study explains the vital role of CCDC6 as a potential biomarker in the prediction of the efficacy of antitumor activity of oncolytic M1 virus.
Materials and methods
Cell lines and viruses
The cells were cultured at 37°C, 5% CO2, and complemented with 10% (v/v) fetal bovine serum plus 1% penicillin/streptomycin (Life Technologies, China). Cell lines were purchased from the American Type Culture Collection (Maryland, USA) and the Shanghai Institute of Cell Biology. All cell lines used in this study were listed in Supplementary Table. M1 virus was developed in Vero cells (OPTI-SFM, 12309-019, Thermo Fisher, USA) and collected for experiments. M1 virus titers were detected by TCID50 assay by utilizing BHK-21 cells. The data were converted to plaque-forming units (PFU) according to the converting equation from ATCC (https://www.atcc.org/en/Global/FAQs/4/8/Converting_TCID50_to_plaque_forming_units_PFU-124.aspx) that was TCID50 titer (per mL) by 0.7 to predict the mean number of PFU.
Cell viability assays
Exponential-phase cells were sowed in 24-well plates at 30,000 cells/well. After 12 h, M1 virus (MOI = 10 PFU/cell) was added to the wells. After 48 h, cell viability was measured by MTT assays. The detailed process was described as a previous study [22]. Differences in the area under the curve (DAUC) were calculated by Graphpad Prism 6 (USA). DAUC specifies (area (a)-area (b)) /area (b).
Flow cytometry analysis
Apoptosis analysis: For flow cytometry analysis, the cells were suspended in 400 μL binding buffer and incubated with 5 μL of Annexin V-FITC at 37°C for 15 min and 10 μL of PI (PIECD) at 37°C for 5 min in the dark. The stained cells were immediately analyzed by flow cytometry (Becton–Dickinson, USA). This assay discriminates intact (FITC−/PI−) from early apoptotic (FITC+/PI−).
The infection rate of M1 virus: After wash with phosphate-buffered saline (PBS), all samples treated by GFP-M1 were analyzed by flow cytometry within 30 min. GFP percentage was detected by flow cytometry.
Quantitative reverse transcription-PCR
Total RNA was extracted by Trizol (Life Technologies) reagent according to the manufacturer's protocol. The acquired RNAs were treated with RNase-free DNase. Next, these RNAs were reverse transcribed to cDNA with oligo (dT). Precise gene manifestation was computed using SuperReal PreMix SYBR Green (FP204-02, TIANGEN, China). β-actin was used as the internal control for normalization. Following primers (Thermo Fisher) were utilized: IFIH1 sense (TCACAAGTTGATGGTCCTCAAGT), IFIH1 antisense (CTGATGAGTTATTCTCCATGCCC); IRF3 sense (AGAGGCTCGTGATGGTCAAG), IRF3 antisense (AGGTCCACAGTATTCTCCAGG); IRF7 sense (CCCACGCTATACCATCTACCT), IRF7 antisense (GATGTCGTCATAGAGGCTGTTG); IFIT1 sense (TTGATGACGATGAAATGCCTGA), IFIT1 antisense (CAGGTCACCAGACTCCTCAC); IFNB sense (GCTTGGATTCCTACAAAGAAGCA), IFNB antisense (ATAGATGGTCAATGCGGCGTC); M1 NS3 sense (GGGGAGGGCTTTCTTTGTCA), M1 NS 3 antisense (CACCCTGTCTTGTCTTTGCTG); β-actin sense (GATCATTGCTCCTCCTGAGC); β-actin antisense (ACTCCTGCTTGCTGATCCAC).
RNA interference
SiRNAs were purchased from Ribobio (Guangzhou, China). Cells were transfected with indicated siRNAs by Lipofectamine RNAiMAX (Thermo Fisher, USA) and OPTI-MEM (Thermo Fisher) according to recommended concentrations. The concentrations of siRNAs are mentioned below: CCDC6-1 (25 nM), CCDC6-2 (25 nM), CCDC6-3 (25 nM), ACYP1 (20 nM), ATF1 (25 nM), E2F3 (20 nM), GP1BB (25 nM), GUCY1B3 (20 nM), NDUFA7 (20 nM), NPAT (20 nM), PTBP1 (20 nM), TRAF2 (20 nM), CUL4A (20 nM), ZNF261 (15 nM), CRSP7 (25 nM), CRSP8 (20 nM), ULK2 (20 nM), LRIG2 (10 nM), TADA3L (10 nM), PITRM1 (50 nM), UBE2C (50 nM), TREX2 (20 nM), SULF1 (20 nM), PSD3 (20 nM), CIZ1 (25 nM), WDR40A (20 nM), ZZZ3 (25 nM), APPL (20 nM), RPS6KC1 (50 nM), SDFR1 (20 nM), APEX2 (50 nM), CGI-12 (20 nM), EXOSC1 (20 nM), PCQAP (20 nM), RARSL (20 nM), C14orf160 (10 nM), THRAP6 (20 nM), MGC13138 (20 nM), ADPRHL1 (20 nM), SIMP (25 nM), DOK6 (20 nM), MGC21830 (25 nM).
Caspase activity analyses
Cells treated with siCCDC6 and negative control siRNA (siNC) were cultivated in 96-well plates and infected with or without M1 virus (MOI = 10 PFU/cell). Caspase-3/7 activity was detected by Caspase-Glo Assay Systems (Promega, USA) according to the manufacturer's protocol, and the data were standardized to the results of cell viability.
RNAseq
Oligo(dT)-attached magnetic beads were used to purified mRNA. Purified mRNA was fragmented into small pieces with fragment buffer at appropriate temperature. Then First-strand cDNA was generated using random hexamer-primed reverse transcription, followed by second-strand cDNA synthesis. Afterward, A-Tailing Mix and RNA Index Adapters were added by incubating to end repair. The cDNA fragments obtained from the previous step were amplified by PCR, and products were purified by Ampure XP Beads, then dissolved in EB solution. The product was validated on the Agilent Technologies 2100 bioanalyzer for quality control. The double-stranded PCR products from the previous step were heated denatured and circularized by the splint oligo sequence to get the final library. The single-strand circle DNA (ssCir DNA) was formatted as the final library. The final library was amplified with phi29 to make DNA nanoball (DNB) which had more than 300 copies of one molecule. DNBs were loaded into the patterned nanoarray and pair-end 150 base reads were generated on the BGISEQ platform (BGI-Shenzhen, China).
Western blot analysis
Proteins were extracted using extraction reagent (M-PER; Thermo Scientific). An equivalent quantity of protein samples (20 μg) were separated by 10% SDS-PAGE and transferred to the polyvinylidene fluoride (PVDF) membrane (Millipore). The membranes were blocked by 5% nonfat milk for 1 h at room temperature (RT). Membranes were first incubated with primary antibodies overnight at 4°C and then incubated with suitable horseradish peroxidase (HRP) conjugated secondary antibodies. Proteins were envisaged by Chemi Doc XRS System (Bio-Rad) with Immobilon Western Chemiluminescent HRP substrate (Millipore). Following antibodies were used: CCDC6 (Abcam), Bip (Cell Signaling Technology [CST]), IRE1α (Abcam), PERK (CST), Ki-67 (CST), Cleaved-Caspase-3 (CST), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Bioworld), M1 E1, and NS3 (manufactured by Beijing Protein Innovation, China).
Virus preparation and infection
Lentiviral plasmids were co-transfected with VSVG and CCDC6 vectors into 293T cells. The CCDC6 lentivirus vector is GL407 and this vector is purchased from Obio Technology (Shanghai, China). Virus containing supernatant was harvested and aliquoted after 72 h, and stored at −80°C. About 1 μg/mL Polybrene (Sigma, Cat.No TR-1003) was added into virus-containing medium and incubated with target cells for 24h. Puromycin (Thermo Fisher, Cat.No A1113802) was used to select the infected cells for 72 h. The M1-GFP virus was derived from the M1 virus by our research team. We have used the M1-GFP virus as the experimental material for several years [10,[22], [23], [24], [25], [26], [27], [28], [29]].
Animal models
The experimental protocols of animal studies were approved by the Animal Ethics and Welfare Committee of Sun Yat-Sen University. Female BALB/c-nu/nu mice, 4 to 5 weeks old, were obtained and kept in cages with water as well as food. General anesthesia was given in mice by providing isoflurane (3% for induction, 1%–2% for maintenance) via inhalation and sustained by the nose cone. A sterilized ophthalmic cream was smeared on the eyes of the animal and a heating pad was utilized to preserve body heat. In the supine position, the abdominal wall of the animal was sterilized by chlorhexidine, a low longitudinal cut was made. Suspension of 50 μL of freshly harvested sh NC UM-UC-3 cells or sh CCDC6 UM-UC-3 cells (2.5 × 106 cells) was straightforwardly injected into the empty bladder wall by 30 G needle. The cut was then sealed with absorbable sutures M1 virus were injected into the mice via tail vein injection. The establishment of in situ animal models has been reported in a previous study [23]. The mice were arbitrarily separated into four indicated groups. The treatment plan is described in the legend. Orthotopic bladder cancer-bearing mice were treated with the M1 virus for 3 days. Mice were sacrificed on day 23rd and key tissues, such as heart, muscles, liver, spleen, lung, kidney, brain, along with tumor were separated to identify M1 virus gene expression by qRT-PCR.
Immunohistochemical analysis
The expression of cleaved-Caspase 3 as well as Ki-67 in tumor cells were detected by immunohistochemical analysis utilizing precise antibodies. Tumor-bearing bladder sections (4 μm) were first dewaxed by xylene, hydrated in downward concentrations of ethanol, submerged in 0.3% H2O2-methanol for 30 min, rinsed with phosphate-buffered saline and treated with cleaved-Caspase 3 antibody (1:100) or Ki-67 antibody (1:100) or isotype control overnight at 4°C. After washing, the segments were treated with biocatalytic goat anti-rabbit or anti-mouse IgG for 2 h at room temperature. Immunostaining was envisaged through streptavidin/peroxidase complex as well as diaminobenzidine, and segments were then counterstained using hematoxylin. Image-Pro Plus 6.0 software (MediaCybernetics) was utilized for quantification of relative protein expression.
Statistical analysis
All data analyses were accomplished by SPSS 20.0 (IBM, USA). Student's t test or 1-way ANOVA, followed by Dunnett's multiple post hoc tests were used. The in situ tumor volume was examined by repeated measures 1-way ANOVA. P < 0.05 suggested statistical significance.
Results
Interferon pathway is regulated by the M1 virus in cancer cells with differential oncolytic sensitivities
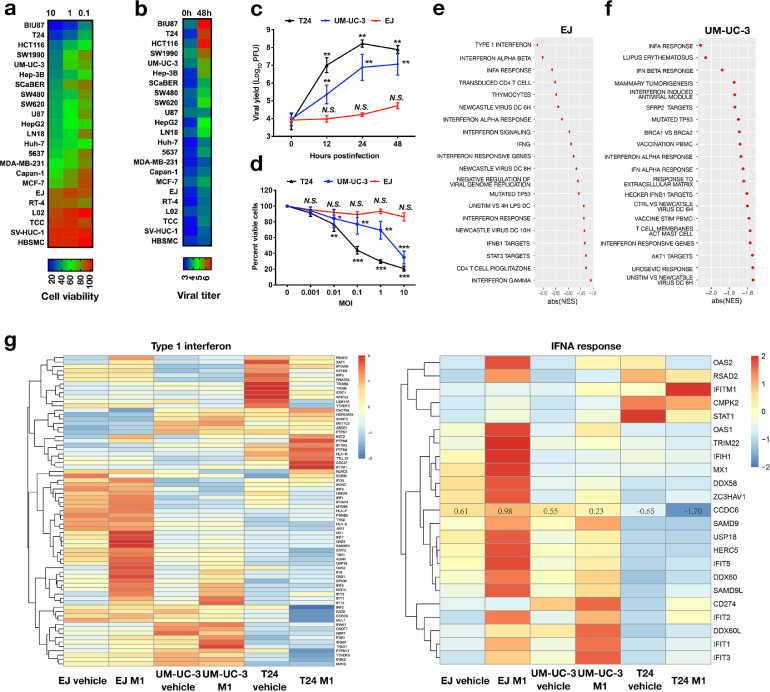
To evaluate the oncolytic effectiveness of M1 virus, we treated a panel of tumor cell lines with M1 virus for 48 h and determined the cell viability. We discovered that M1 virus reduced cell viability in tested cell lines with different abilities, some tumor cell lines and normal cell lines were resistant to M1 virus treatment (Figure 1a). We also found that the viral replication was much higher in M1 virus sensitive cell line than in the refractory cell line (Figure 1b). These outcomes indicate that the reproduction of M1 is greatly associated with virus oncolysis. To illustrate the factors involved in the replication of M1 virus, we used 3 bladder cancer cell lines with different sensitivities to M1 virus, including hypersensitive cell line T24, less sensitive cell line UM-UC-3 and refractory cell line EJ, to analyzed the cell response and gene alternation resulted from M1 virus. We found the cell viability was dose-dependently decreased by treatment with M1 virus in T24 cells and UM-UC-3 cells, but not EJ cells (Figure 1c). Consistently, the efficiency of viral production was much higher in T24 cells and UM-UC-3 cells than in EJ cells (Figure 1d). Cells were infected with the M1 virus or vehicle control for 24 h and performed the RNAseq analysis. Gene ontology analysis showed substantial enhancement of genes related to Interferon-α/β signaling cascade in the resistant and less sensitive cell lines after M1 infection (Figure 1e, f and Supplementary Figure 1a). Among these differentially expressed genes, more than 60% are interferon antiviral cascade genes (Figure 1g). Furthermore, we also found that expression of these interferon antiviral cascade-related genes was upregulated in both nonsensitive UM-UC-3 and EJ cell lines, while there was a minor change of these genes in sensitive cancer cell line T24 after M1 infection (Figure 1g). These results indicated that type I interferon pathway has an imperative role in combating duplication of M1 virus and ensuing cell death.
Fig. 1.
Oncolytic impacts of M1 in different tumor cell lines. (a) 23 cell lines, including tumor cells and normal cells, were infected with M1 virus (MOI = 10, 1, or 0.1 PFU/cell) and cell viability was detected by MTT assay after infection for 48 h. (b) Heat map of M1 virus reproduction levels in the corresponding cells (mean ± SD) (MOI = 0.01 PFU/cell), virus titer was determined at 48 h after infection. (c) The survival rate of bladder tumor cell lines EJ, UM-UC-3, as well as T24 after treatment with M1 virus were determined by MTT assay. (d) M1 virus titer overtime after infection of 3 representative bladder cancer cells (MOI = 10). ** indicates the significant difference compared to the control group and the P value <0.01. For EJ cells, NS is the data from 12 h, 24 h, and 48 h groups compared to the 0 h group. For T24 cells, ** is the data from 12 h, 24 h, and 48 h groups compared to the 0 h group. (e-g) EJ, UM-UC-3, and T24 bladder cancer cell lines were treated with vehicle or M1 virus (MOI = 0.1 PFU)/cell for 24 h, and then the RNA was acquired and examined. RNAseq analysis was done on 6 samples. The clustering of the transcriptome profiles of the most prominent genes expressed in the 3 cell lines when treated with M1 are listed (e, f). Heat map of type 1 interferon gene expression and INFA reaction after M1 treatment (g). *P < 0.05; **P < 0.01; ***P < 0.001; N.S., not significant.
CCDC6 is critical for the oncolytic function of M1 virus
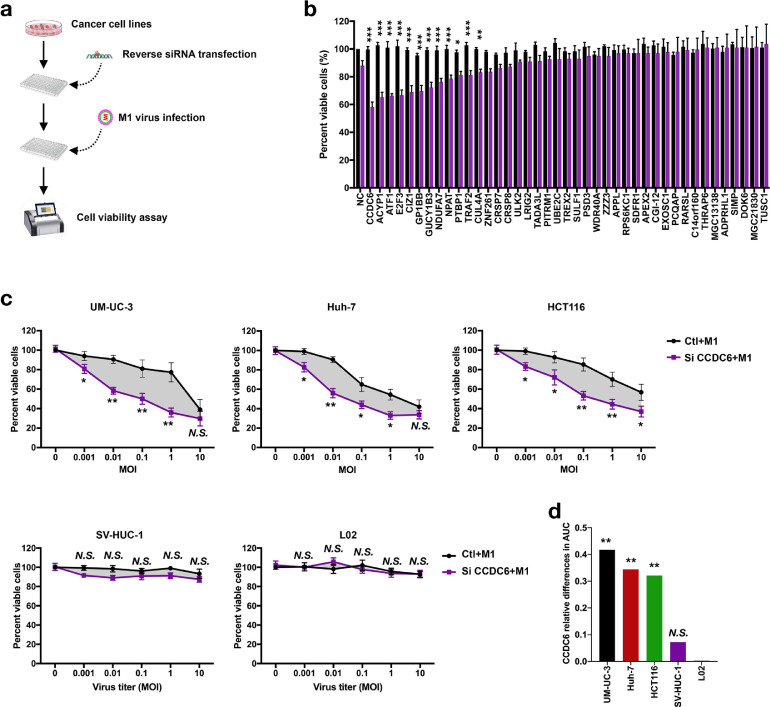
Type I interferons are well-known innate antiviral system which triggered by pattern recognition receptors to block the replication and spread of a broad range of viruses through inducing multiple ISGs [30]. To further determine the underlying mechanisms for induce ISGs after M1 virus infection and enhance virus-induced cell death, we performed siRNA screen of 40 genes [31], which has been reported to inhibit the replication of alphavirus (Figure 2a). After knocked down 40 genes separately, we found that 12 genes augmented M1 virus-induced cell death after siRNA treatment (Figure 2b). Among these candidate genes, CCDC6, ACYP1, ATF1, E2F3, and CIZ1 were the top 5 genes with dramatic ability to the alternation of M1 virus-induced cell death (P < 0.001; Figure 2b). Among the top 5 selected genes, knockdown of CCDC6 showed the most significant reduction of cell viability.
Fig. 2.
Small interfering RNA screen CCDC6 renders tumor cells; however, not normal cells, sensitive to oncolytic virus M1. (a) Schematic of screening strategy to find out CCDC6. (b) UM-UC-3 cells were treated with negative control (NC) or siRNAs for 24 h and then infected with M1 virus (MOI = 1). Cell viability was measured 72 h after the M1 virus infection (n = 3). (c) Tumor cells (UM-UC-3 Huh-7 and HCT116) and normal cells (SV-HUC-1 and L-02) were infected with M1 virus and treated with negative control (NC) or siCCDC6. After 72 h, cell viability was measured (mean ± SD). (d) Bar graphs describe the relative changes in AUC (area under the curve). Ctl, control groups. Data shown in b, c, and d were the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; N.S., not significant.
It has been reported that loss of function or inhibition of CCDC6 affects the DNA repair function as well as reduces the apoptosis in cancer cells [21,32]. To confirm the role of CCDC6 in M1 virus-induced cell death, 3 human cancer cell lines from different organs, comprising bladder tumor cell line UM-UC-3, liver tumor cell line HUH7 and colorectal cancer cell line HCT116, and 2 human normal immortalized cell lines SV-HUC-1 and L-02, were used to detect the cell viability after knocked down of CCDC6 and treatment with M1 virus. As shown in Figure 2c, siCCDC6 significantly increased the oncolytic impacts of the M1 virus in all tested tumor cells, but not in SV-HUC-1 and L-02 normal cells (Figures 2c). DAUC represent the effect of siCCDC6 on M1 virus-induced cell death, also showed that knockdown of CCDC6 specifically promoted M1 virus-induced cell death of cancer cells rather than normal cells (Figure 2c and d).
Knockdown of CCDC6 promotes M1 virus-mediated ISGs expression and the oncolytic effect
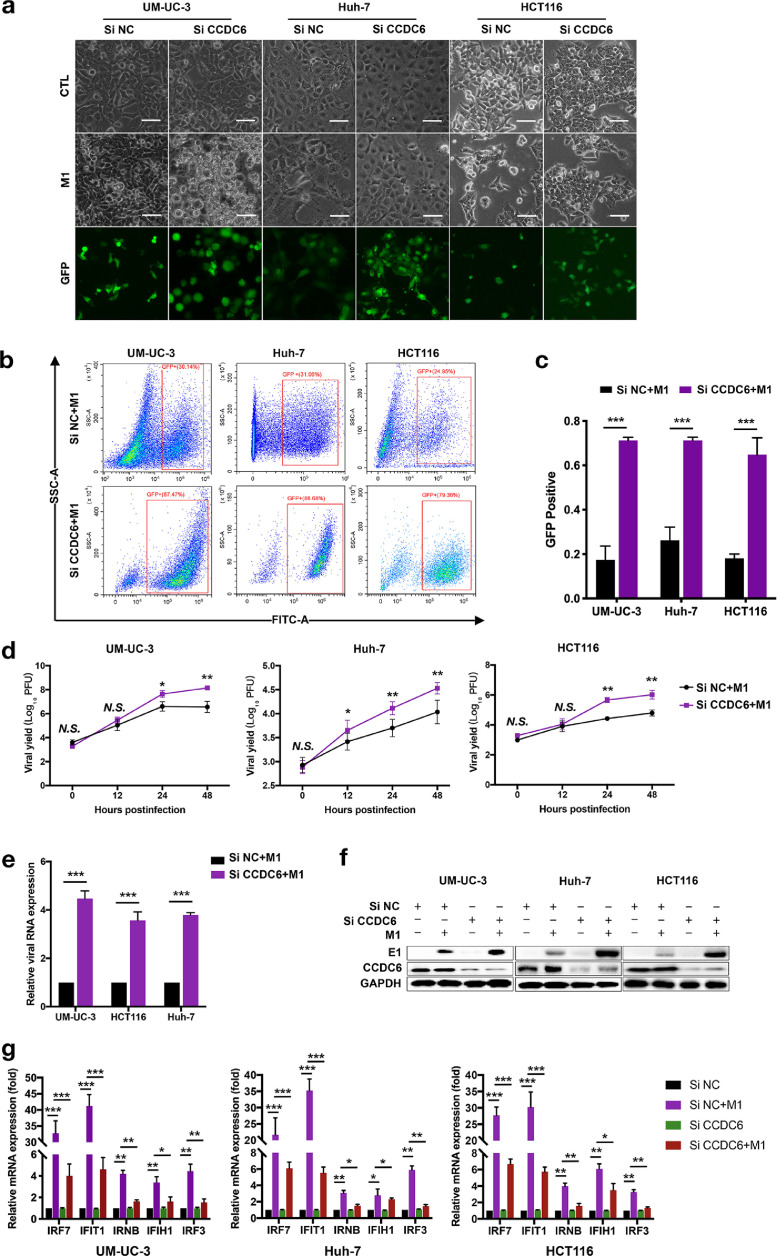
To further define whether siCCDC6 promotes M1 virus-induced cell death by accelerating virus replication, we used multiple methods to analyze the replication of the M1 virus after treatment with various siCCDC6 siRNAs. We used 3 different siRNAs specific target to CCDC6 and detected the expression of CCDC6 by Western blot assay. As shown in Supplementary Figure 2d, 3 siRNAs (siCCDC6-1, siCCDC6-2, and siCCDC6-3) significantly decreased the expression of CCDC6 (Supplementary Figure 2d). Phase-contrast and fluorescence microscopy indicated that the M1 virus caused the apparent cytopathic impact after siCCDC6 treatment, and the virus replication was also enhanced (Figure 3a and Supplementary Figure 2a). Furthermore, we also determined the viral infection rate, viral titer, the expression of viral protein E1 as well as NS3 after knocked down of CCDC6 by 3 siCCDC6 siRNAs. Our data showed that siCCDC6 significantly increased the amplification of M1 virus in tumor cells in a time-reliant manner (Figure 3b and d and Supplementary Figure 2b). Consistently, expressions of M1 virus marker E1 were also significantly increased by siCCDC6 in mRNA and protein levels (Figure 3e and f and Supplementary Figure 2c). We further found that overexpression of CCDC6 significantly suppressed the oncolytic effects of M1 virus in T24 cells (Supplemental Figure 1b). These results indicated that siCCDC6 enhanced oncolysis of M1 due to the elevated replication of M1 virus and suggest CCDC6 may have antiviral function.
Fig. 3.
SiRNA CCDC6 prevents M1 virus-induced expression of antiviral indicators and activates replication of M1 virus. (a) Pretreatment with CCDC6 siRNA 24 h after UM-UC-3 Huh-7 and HCT116 cells are infected with M1 virus (MOI = 1). Phase contrast microscope image of 48 h. Scale bar: 100 μm. (b and c) M1-GFP virus (MOI = 1) was infected for 24 h, the M1 virus infection rate was measured by flow cytometry (b), and the comparison of the GFP positive proportion of total cell number between CCDC6 siRNA group and negative control (NC) siRNA group for UM-UC-3, Huh-7 and HCT116 cells (c). (d) After 48 h of M1 virus infection, the M1 virus titer (MOI = 1) was measured by the TCID50 assay. (e) Expression of E1 mRNA of the M1 virus was performed by qRT-PCR 4 h after infection with M1 virus (MOI = 10). Relative gene manifestation levels were standardized to β-actin. (f) M1 viral protein levels were distinguished by Western blot 24 h after M1 virus infection (MOI = 1). (g) After 12 h of treatment with M1 virus (MOI = 10), mRNA levels of IRF7, IFIT1, IRNB, IFIH1, and IRF3 were detected by qRT-PCR (mean ± SD). The expression of genes was normalized to β-actin. The data are shown in mean ± SEM. * P < 0.05; ** P < 0.01; ***P < 0.001; N.S., not significant. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To further characterize the antiviral effect of CCDC6, we next want to find out whether CCDC6 regulates the interferon pathway. By detecting the mRNA expression of 5 reported ISGs (IFNB, IFIH1, IRF3, IRF7, and IFIT1) in UM-UC-3, HUH7, and HCT116 cells, we found that siCCDC6 abrogated M1 virus-mediated transcriptional upregulation of ISGs (Figure 3g). We also found that siCCDC6 did not affect the inhibitory effect of IFN-α on the M1 virus (Supplementary Figure 1c and d). The above results indicated that CCDC6 is a critical antiviral factor to suppress the replication and oncolytic effect of the M1 virus.
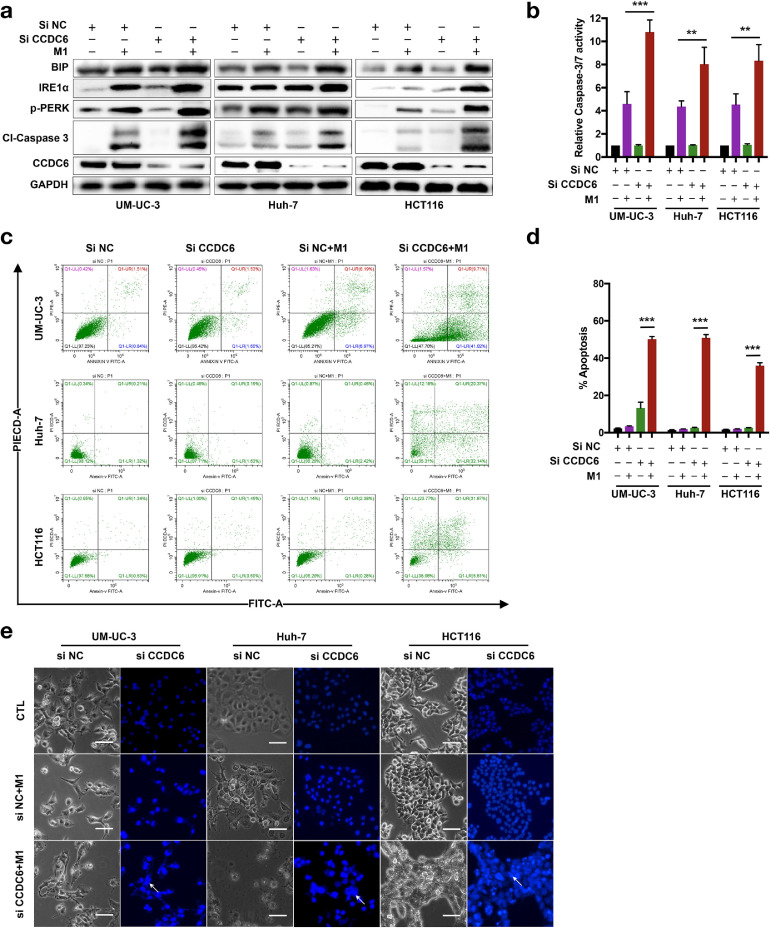
Knocked down of CCDC6 enhanced M1 virus-induced apoptosis via induction of ER stress
Since knockdown of CCDC6 promotes the replication of M1 virus, but how the increased M1 virus resulted in cancer cell death needs further investigation. It has been reported that increased viral replication might lead to the accumulation of great quantity of viral proteins in the ER lumen and subsequently induced severe ER stress [33]. We further asked whether siCCDC6-mediated viral protein accumulation stimulates ER stress and induces devastation of ER as well as cell apoptosis. We found that M1/siCCDC6 combinational treatment increased the expression of ER stress indicators, including Bip, IRE1α, and p-PERK in UM-UC-3, Huh-7, and HCT116 cancer cells (Figure 4a). These data indicated that M1/siCCDC6 combinational treatment induced severe ER stress in cancer cells. Combination of siCCDC6 and M1 virus treatment significantly increased the expression of cleavage (active) form of caspase 3 (Figure 4a) and the activity of caspase-3/7 (Figure 4b), recommending that downstream apoptotic cascades are stimulated. Next, we observed that apoptotic cells were dramatically elevated in M1/siCCDC6 combinational treatment group than in M1 treatment alone group (Figure 4c and d). We can also found more apoptotic bodies in M1/siCCDC6 combinational treatment group than in M1 treatment alone group (Figure 4e). These data suggested that the M1/siCCDC6 combinational therapy induces cell death may due to the induction of ER stress. Although M1 treatment alone can cause mild ER stress, inadequate to stimulate apoptosis efficiently. When this process is promoted by siCCDC6, ER stress becomes unresolvable and then turn on apoptosis.
Fig. 4.
siCCDC6 increases ER stress prompted by M1 virus and stimulates ER stress-related apoptosis. Cells were treated with negative control (NC) or siCCDC6 for 24 h and then infected with M1 virus (MOI = 0.01) for 48 h. (a) The expression of ER stress markers was evaluated by Western blot assay. (b) Relative Caspase-3/7 activity assays were performed in indicated conditions. (c) Flow cytometry and annexing V staining analyses. (d) Apoptotic cells were measured by flow cytometry analysis. (e) Chromatin condensation was demonstrated via Hoechst 33342 staining. Arrows indicate the apoptotic cells. Data are means ± SD (n = 3). **P < 0.01; ***P < 0.001.
CCDC6 knockdown augments the oncolytic effectiveness of M1 virus in vivo
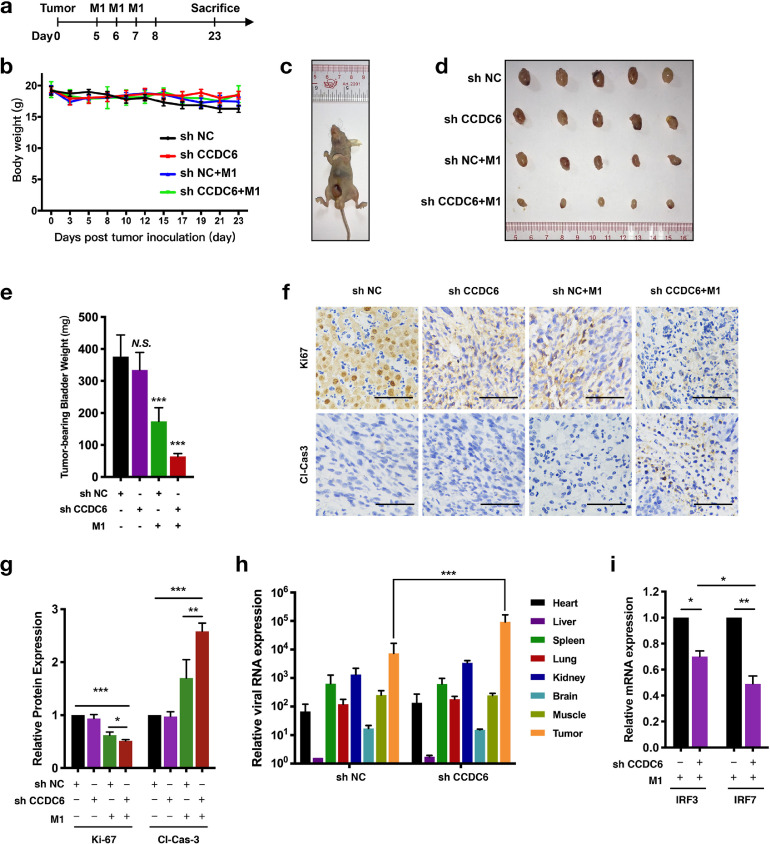
To evaluate the effects of CCDC6 on the oncolytic ability of M1 virus in vivo, we stably knocked down CCDC6 expression in UM-UC-3 cells (shCCDC6) to establish the orthotopic bladder tumor models in female BALB/c-nu/nu mice. The treatment schedule is presented in Figure 5a. Intravenous injection of M1 virus did not affect the bodyweight of the tumor-bearing mice (Figure 5b) and the tumors were observed 5 to 7 days after orthotopic injection (Figure 5c). More importantly, we found that shCCDC6 alone did not affect the volume of tumor and the weight of tumor-bearing bladder, but M1 virus alone group showed a significant reduction of tumor volume and bladder weight compared with shNC group; however, shCCDC6 combined with the treatment of M1 virus further dramatically decreased the tumor volume and the weight of tumor-bearing bladder (Figure 5d and e).
Fig. 5.
Knocked down of CCDC6 potentiates the oncolytic effectiveness of M1 virus in vivo. Laparotomy and bladder wall injections were performed to the establishment of the orthotopic bladder tumor mouse model. (a) Timeline of the experimental arrangement. (b) The weight of the tumor-bearing mice. (c) Tumors were observed 5 to 7 days after orthotopic injection. (d) Tumors were successively separated and images were captured. (e) Tumor-bearing bladder weight was measured. (f) Immunohistochemistry was accomplished to identify the expression of Ki-67 as well as cleaved-caspase-3. (g) Quantitation of Ki-67 and cleaved-caspase-3 expressing levels in indicated groups. (h and i) Orthotopic bladder cancer-bearing mice were treated with the M1 virus for 3 days (n = 3). (h) Mice were sacrificed and key tissues such as heart, muscles, liver, spleen, lung, kidney, brain, along with tumor were separated to identify M1 virus gene expression by qRT-PCR. (i) Tumors are examined to identify interferon-stimulated genes IRF7 and IRF3 manifestation by qRT-PCR. Data shown in b, e, g, h, and i were the mean ± SEM.*P < 0.05; **P < 0.01; ***P < 0.001.
Immunohistochemistry staining showed that tumors from shCCDC6 combined with M1 virus group expressed lower Ki-67 and higher cleaved caspase-3/7 (Figure 5f and g). These results showed consistency with the in vitro study, demonstrating that combinational treatment of M1 virus and shCCDC6 inhibited tumor development via suppressed tumor cell proliferation and stimulated apoptosis in vivo. Moreover, we assessed whether the reproduction of the M1 virus is specifically augmented in cancer tissue by identifying the virus genome. We found that shCCDC6 prompted an induction of the M1 virus in cancer tissues, besides viral replication was extremely enhanced (Figure 5h). As shown in Figure 5h, IRF3 and IRF7 were repressed in the shCCDC6 group (Figure 5i), which is consistent with the previous results that knocked down of CCDC6 promotes M1 virus reproduction through inhibiting antiviral factors. The above results showed that CCDC6 plays an antiviral role and inhibition of CCDC6 increases the replication of M1 virus and the subsequent oncolytic effect.
Discussion
Through analysis of a variety of cell lines, we found these cells showed differential sensitivity to Mi virus. We further defined the mechanisms involved in the induction of M1 viral replication to improve M1 efficiency in refractory cancers. As the antitumor capability of the oncolytic virus often lags during the clinical trial compared with inspiring in vitro results, further research focused on improving oncolytic effects became a tendency [34]. The main course of these researches is the development of small molecules, which enhance tumor cell death by enhancing virus-induced cancer cell stress or inducing bystander killing, like second mitochondria-derived activator of caspases (Smac) mimetic compounds [24] or tumor enhancement agent that promotes virus replication, such as dbcAMP, tipifarnib, and H89 [27,35,36].
The interferon pathway plays an important role in the anticancer effects of oncolytic virus M1. Moreover, we found a factor CCDC6 that exerts the antiviral effect by regulating the interferon pathway, thereby inhibiting viral replication. Inhibition of CCDC6 promoted viral replication and then induced irreversible endoplasmic reticulum stress to enhance the oncolytic impact of M1 virus in vitro as well as in vivo. Our data showed that the infective efficiency and oncolytic activity of M1 virus were higher in cancer cells than in normal cells, this phenomenon might due to the difference in the immune response of cancer cells and normal cells. In refractory tumor cells, M1 virus infection triggers the antiviral elements and limits reproduction and oncolytic effect of M1 virus. Knocked down of CCDC6 inhibited the expression of ISGs and enhanced M1 virus-mediated antitumor effect. In previous reports, it was found that siCCDC6 reduced the replication of alphavirus Chikungunya virus [31], but the underlying mechanism needs to be further explored. Because of the limited viral protein production in refractory cancer cells, the amount of M1 virus is not enough to stimulate severe ER stress. This study defined that knocked down of CCDC6 increases the reproduction of the M1 virus, induces severe ER stress, and leading to apoptosis of cancer cells in vitro and in vivo.
CCDC6 is a coiled-coil domain-containing protein and concurrent loss or inhibition of CCDC6 affects the DNA damage repair and reduces the apoptosis [21]. CCDC6 is decreased in various types of cancer due to somatic mutations, chromosomal rearrangements, as well as abridged manifestation [21,32,37,38]. CCDC6 has been reported to regulate the cellular checkpoints for DNA damage recovering and preservation of the stability of cell cycle genome; alternation of CCDC6 involves in cancer development and progression [20,39,40].
Oncolytic virotherapy is an attractive novel therapeutic strategy selectively infect and kill tumor cells. A recent study has shown that through drug screening, DNA-dependent protein kinase (DNA-PK) inhibition sensitizes cancer cells to the M1 virus and improves therapeutic effects in refractory cancer models and patient tumor samples [29]. M1 virus-mediated induction of interferons and antiviral response could be abolished by DNA-PK inhibitor (DNA-PKI), resulting in increased replication of M1 virus in tumor cells [29]. It has also been reported that the inhibitor of valosin-containing protein (VCP) acts as the sensitizer to selectively increasing oncolytic efficacy up to 3600-fold by modulated M1 virus-suppressed inositol-requiring enzyme 1α (IRE1α)/X-box binding protein 1 (XBP1) pathway and triggered (ER) stress-mediated apoptosis in HCC [10].
In our study, we have defined that CCDC6 plays a critical role in determining the efficacy of oncolytic virotherapy via regulating ER stress; these results suggested CCDC6 may be a potential molecular target to design the selective inhibitor to combined with oncolytic virotherapy. Figure out the critical regulator of CCDC6 to enhance the oncolytic effect of M1 would be an urgent need. CCDC6 is an antiviral factor; therefore, in future research and clinical applications, it is necessary to further explore its possibility as a marker for oncolytic virus therapy. Further clinical studies to investigate whether CCDC6 could be a prognostic biomarker of M1 virus anticancer therapies are an urgent need.
Conclusion
The current study demonstrates that targeted inhibition of CCDC6 enhances the sensitivity of tumor cells to the oncolytic virus and increases the antitumor activity of the M1 virus in vitro and in vivo.
Author contributions
LK, HC, and LJK conceived and directed the project. HC wrote the manuscript. ZWB edited the manuscript. LY, HC, LY, HWT, CJ, ZH, QJG, LXC, and PL performed and analyzed experiments. YGM provided key services, materials and guidance, and commented on the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (No. 81802536, 81570539, 81672701, and 81370535); the Science and Technology Planning Project of Guangdong Province, China (No. 20160909, 411308023039, and 2016A020215221); the Natural Science Foundation of Guangdong Province (No. 2017A030313620); Guangdong Medical Science and Technology Research Fund (No. A2020545); the Research and Development Project of Applied Science and Technology of Guangdong Province, China (No. 2016B020237004); Open project of Key Lab of Tropical Disease Control (Sun Yat-sen University 2020kfkt09), Ministry of Education (No. 2020kfkt09); Fundamental Research Funds for the Central Universities (No. 19ykpy36, and 20ykpy22); the Special Funding Project of National Natural Science Foundation of the Third Affiliated Hospital of Sun Yat-sen University (No. 2020GZRPYQN01); and the Guangzhou Science and Technology Project (No. 1561000155). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
Conflict of interests relevant to this article was not reported.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2020.12.003.
Contributor Information
Jian-kai Liang, Email: liangjk5@mail.sysu.edu.cn.
Cheng Hu, Email: hucheng2@mail.sysu.edu.cn.
Appendix. Supplementary materials
References
- 1.Au GG, Lindberg AM, Barry RD, Shafren DR. Oncolysis of vascular malignant human melanoma tumors by Coxsackievirus A21. Int J Oncol. 2005;26:1471–1476. doi: 10.3892/ijo.26.6.1471. [DOI] [PubMed] [Google Scholar]
- 2.Kim MK, Breitbach CJ, Moon A, Heo J, Lee YK, Cho M, Lee JW, Kim S-G, Kang DH, Bell JC. Oncolytic and immunotherapeutic vaccinia induces antibody-mediated complement-dependent cancer cell lysis in humans. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3005361. 185ra63. [DOI] [PubMed] [Google Scholar]
- 3.Rudin CM, Poirier JT, Senzer NN, Stephenson J, Loesch D, Burroughs KD, Reddy PS, Hann CL, Hallenbeck PL. Phase I clinical study of Seneca Valley Virus (SVV-001), a replication-competent picornavirus, in advanced solid tumors with neuroendocrine features. Clin Cancer Res. 2011;17:888–895. doi: 10.1158/1078-0432.CCR-10-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nat Biotechnol. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tai CK, Kasahara N. Replication-competent retrovirus vectors for cancer gene therapy. Front Biosci. 2008;13:3083–3095. doi: 10.2741/2910. [DOI] [PubMed] [Google Scholar]
- 6.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 7.Hu J, Cai XF, Yan G. Alphavirus M1 induces apoptosis of malignant glioma cells via downregulation and nucleolar translocation of p21WAF1/CIP1 protein. Cell Cycle. 2009;8:3328–3339. doi: 10.4161/cc.8.20.9832. [DOI] [PubMed] [Google Scholar]
- 8.Lin Y, Zhang H, Liang J, Li K, Zhu W, Fu L, Wang F, Zheng X, Shi H, Wu S. Identification and characterization of alphavirus M1 as a selective oncolytic virus targeting ZAP-defective human cancers. Proc Natl Acad Sci U S A. 2014;111:E4504–E4512. doi: 10.1073/pnas.1408759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Li K, Lin Y, Xing F, Xiao X, Cai J, Zhu W, Liang J, Tan Y, Fu L. Targeting VCP enhances anticancer activity of oncolytic virus M1 in hepatocellular carcinoma. Sci Transl Med. 2017;9:eaam7996. doi: 10.1126/scitranslmed.aam7996. [DOI] [PubMed] [Google Scholar]
- 11.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 12.Medrano RFV, Hunger A, Mendonça SA, Barbuto JAM, Strauss BE. Immunomodulatory and antitumor effects of type I interferons and their application in cancer therapy. Oncotarget. 2017;8:71249–71284. doi: 10.18632/oncotarget.19531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budhwani M, Mazzieri R, Dolcetti R. Plasticity of type I interferon-mediated responses in cancer therapy: from anti-tumor immunity to resistance. Front Oncol. 2018;8:322. doi: 10.3389/fonc.2018.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15:405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 15.Snell LM, McGaha TL, Brooks DG. Type I interferon in chronic virus infection and cancer. Trends Immunol. 2017;38:542–557. doi: 10.1016/j.it.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merolla F, Pentimalli F, Pacelli R, Vecchio G, Fusco A, Grieco M, Celetti A. Involvement of H4(D10S170) protein in ATM-dependent response to DNA damage. Oncogene. 2007;26:6167–6175. doi: 10.1038/sj.onc.1210446. [DOI] [PubMed] [Google Scholar]
- 17.Jungang Z, Jun T, Wanfu M, Kaiming R. FBXW7-mediated degradation of CCDC6 is impaired by ATM during DNA damage response in lung cancer cells. FEBS Lett. 2012;586:4257–4263. doi: 10.1016/j.febslet.2012.10.029. [DOI] [PubMed] [Google Scholar]
- 18.Merolla F, Luise C, Muller MT, Pacelli R, Fusco A, Celetti A. Loss of CCDC6, the first identified RET partner gene, affects pH2AX S139 levels and accelerates mitotic entry upon DNA damage. PLoS One. 2012;7:e36177. doi: 10.1371/journal.pone.0036177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malapelle U, Morra F, Ilardi G, Visconti R, Merolla F, Cerrato A, Napolitano V, Monaco R, Guggino G, Monaco G. USP7 inhibitors, downregulating CCDC6, sensitize lung neuroendocrine cancer cells to PARP-inhibitor drugs. Lung Cancer. 2017;107:41–49. doi: 10.1016/j.lungcan.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Morra F, Luise C, Visconti R, Staibano S, Merolla F, Ilardi G, Guggino G, Paladino S, Sarnataro D, Franco R. New therapeutic perspectives in CCDC6 deficient lung cancer cells. Int J Cancer. 2015;136:2146–2157. doi: 10.1002/ijc.29263. [DOI] [PubMed] [Google Scholar]
- 21.Leone V, Mansueto G, Pierantoni GM, Tornincasa M, Merolla F, Cerrato A, Santoro M, Grieco M, Scaloni A, Celetti A. CCDC6 represses CREB1 activity by recruiting histone deacetylase 1 and protein phosphatase 1. Oncogene. 2010;29:4341–4351. doi: 10.1038/onc.2010.179. [DOI] [PubMed] [Google Scholar]
- 22.Ying L, Cheng H, Xiong XW, Yuan L, Peng ZH, Wen ZW, Ka LJ, Xiao X, Jing C, Qian TY. Interferon alpha antagonizes the anti-hepatoma activity of the oncolytic virus M1 by stimulating anti-viral immunity. Oncotarget. 2017;8:24694–24705. doi: 10.18632/oncotarget.15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu C, Liu Y, Lin Y, Liang J-K, Zhong W-W, Li K, Huang W-T, Wang D-J, Yan G-M, Zhu W-B. Intravenous injections of the oncolytic virus M1 as a novel therapy for muscle-invasive bladder cancer article. Cell Death Dis. 2018;9:1–10. doi: 10.1038/s41419-018-0325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai J, Lin Y, Zhang H, Liang J, Tan Y, Cavenee WK, Yan G. Selective replication of oncolytic virus M1 results in a bystander killing effect that is potentiated by Smac mimetics. Proc Natl Acad Sci U S A. 2017;114:6812–6817. doi: 10.1073/pnas.1701002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li K, Hu C, Xing F, Gao M, Liang J, Xiao X, Cai J, Tan Y, Hu J, Zhu W. Deficiency of the IRE1α-autophagy axis enhances the antitumor effects of the oncolytic virus M1. J Virol. 2017;92:e01331. doi: 10.1128/jvi.01331-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan Y, Lin Y, Li K, Xiao X, Liang J, Cai J, Guo L, Li C, Zhu W, Xing F. Selective antagonism of Bcl-xL potentiates M1 oncolysis by enhancing mitochondrial apoptosis. Hum Gene Ther. 2018;29:950–961. doi: 10.1089/hum.2017.055. [DOI] [PubMed] [Google Scholar]
- 27.Liang J, Guo L, Li K, Xiao X, Zhu W, Zheng X, Hu J, Zhang H, Cai J, Yu Y. Inhibition of the mevalonate pathway enhances cancer cell oncolysis mediated by M1 virus. Nat Commun. 2018;9:1–12. doi: 10.1038/s41467-018-03913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Hu C, Zhu WB, Xu WX, Li ZY, Lin Y, Cai J, Liang JK, Zhu X, Gao ZL. Association of low zinc finger antiviral protein expression with progression and poor survival of patients with hepatocellular carcinoma. Cell Physiol Biochem. 2018;49:1048–1059. doi: 10.1159/000493285. [DOI] [PubMed] [Google Scholar]
- 29.Xiao X, Liang J, Huang C, Li K, Xing F, Zhu W, Lin Z, Xu W, Wu G, Zhang J. DNA-PK inhibition synergizes with oncolytic virus M1 by inhibiting antiviral response and potentiating DNA damage. Nat Commun. 2018;9:1–15. doi: 10.1038/s41467-018-06771-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Goraya MU, Zaighum F, Sajjad N, Anjum FR, Sakhawat I, Rahman S. Web of interferon stimulated antiviral factors to control the influenza A viruses replication. Microb Pathog. 2020;139 doi: 10.1016/j.micpath.2019.103919. [DOI] [PubMed] [Google Scholar]
- 31.Karlas A, Berre S, Couderc T, Varjak M, Braun P, Meyer M, Gangneux N, Karo-Astover L, Weege F, Raftery M. A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs. Nat Commun. 2016;7:1–14. doi: 10.1038/ncomms11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerrato A, Merolla F, Morra F, Celetti A. CCDC6: the identity of a protein known to be partner in fusion. Int J Cancer. 2018;142:1300–1308. doi: 10.1002/ijc.31106. [DOI] [PubMed] [Google Scholar]
- 33.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zemp FJ, Corredor JC, Lun X, Muruve DA, Forsyth PA. Oncolytic viruses as experimental treatments for malignant gliomas: using a scourge to treat a devil. Cytokine Growth Factor Rev. 2010;21:103–117. doi: 10.1016/j.cytogfr.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Li K, Liang J, Lin Y, Zhang H, Xiao X, Tan Y, Cai J, Zhu W, Xing F, Hu J. A classical PKA inhibitor increases the oncolytic effect of M1 virus via activation of exchange protein directly activated by cAMP 1. Oncotarget. 2016;7:48443–48455. doi: 10.18632/oncotarget.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li K, Zhang H, Qiu J, Lin Y, Liang J, Xiao X, Fu L, Wang F, Cai J, Tan Y. Activation of cyclic adenosine monophosphate pathway increases the sensitivity of cancer cells to the oncolytic Virus M1. Mol Ther. 2016;24:156–165. doi: 10.1038/mt.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Ding X, Wang S, Moser CD, Shaleh HM, Mohamed EA, Chaiteerakij R, Allotey LK, Chen G, Miyabe K. Antitumor effect of FGFR inhibitors on a novel cholangiocarcinoma patient derived xenograft mouse model endogenously expressing an FGFR2-CCDC6 fusion protein. Cancer Lett. 2016;380:163–173. doi: 10.1016/j.canlet.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y-M, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin S-F. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerrato A, Morra F, Celetti A. Use of poly ADP-ribose polymerase [PARP] inhibitors in cancer cells bearing DDR defects: the rationale for their inclusion in the clinic. J Exp Clin Cancer Res. 2016;35:179. doi: 10.1186/s13046-016-0456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lord CJ, Tutt ANJ, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu Rev Med. 2015;66:455–470. doi: 10.1146/annurev-med-050913-022545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.