Significance
The molecular origin of basal activity in G protein coupled receptors and its suppression by inverse agonists are poorly understood. Here we employ pressure-resolved DEER spectroscopy to explore the structural origin of basal activity and the mechanism of inverse agonism in the β2 adrenergic receptor. Collectively, the results support a conformational selection model wherein basal activity is due to a preexisting equilibrium between inactive and active conformations in the unliganded receptor, and inverse agonists stabilize the inactive conformation. Furthermore, the results illustrate the importance of rare states in molecular mechanisms of protein function and the capability of pressure-resolved DEER for revealing structural features of sparsely populated conformations of membrane proteins and for determining their thermodynamic properties.
Keywords: double electron–electron resonance, β2 adrenergic receptor, high pressure, conformational selection, basal activity
Abstract
The β2 adrenergic receptor (β2AR) is an archetypal G protein coupled receptor (GPCR). One structural signature of GPCR activation is a large-scale movement (ca. 6 to 14 Å) of transmembrane helix 6 (TM6) to a conformation which binds and activates a cognate G protein. The β2AR exhibits a low level of agonist-independent G protein activation. The structural origin of this basal activity and its suppression by inverse agonists is unknown but could involve a unique receptor conformation that promotes G protein activation. Alternatively, a conformational selection model proposes that a minor population of the canonical active receptor conformation exists in equilibrium with inactive forms, thus giving rise to basal activity of the ligand-free receptor. Previous spin-labeling and fluorescence resonance energy transfer experiments designed to monitor the positional distribution of TM6 did not detect the presence of the active conformation of ligand-free β2AR. Here we employ spin-labeling and pressure-resolved double electron–electron resonance spectroscopy to reveal the presence of a minor population of unliganded receptor, with the signature outward TM6 displacement, in equilibrium with inactive conformations. Binding of inverse agonists suppresses this population. These results provide direct structural evidence in favor of a conformational selection model for basal activity in β2AR and provide a mechanism for inverse agonism. In addition, they emphasize 1) the importance of minor populations in GPCR catalytic function; 2) the use of spin-labeling and variable-pressure electron paramagnetic resonance to reveal them in a membrane protein; and 3) the quantitative evaluation of their thermodynamic properties relative to the inactive forms, including free energy, partial molar volume, and compressibility.
Many aspects of physiology in health and disease are regulated by signal transduction through G protein coupled receptors (GPCRs). Among these, the β2 adrenergic receptor (β2AR) is an archetype for the subset of family A GPCRs activated by hormones and neurotransmitters, as well as a pharmaceutical target for asthma and chronic obstructive pulmonary disease. The activity of β2AR and other ligand-binding GPCRs can be finely tuned by ligands of varying efficacy, with agonists stimulating an increase in activation of cognate G proteins and inverse agonists decreasing G protein activation below a basal level.
β2AR crystal structures (1) have defined the outward movement of transmembrane helix 6 (TM6) as the largest structural rearrangement associated with activation of the receptor, as was originally found for rhodopsin (2) and subsequently for other receptors. This movement of TM6 is required for β2AR to productively couple to its signaling partners (3).
Ligand-independent or basal activity has been observed in many GPCRs. The level of basal receptor activity is highly receptor-specific and is important in maintaining homeostasis in physiologic systems independent of agonist stimulation (4). Basal activity in the β2AR (5–7) and other receptors (4, 8) could arise from distinct receptor conformations that promote weak activation of the G protein, possibly involving an induced fit mechanism for receptor–G protein interaction. Alternatively, the basal activity could arise from a preexisting equilibrium (9, 10) between inactive and active conformations, where a small fraction of active receptors could account for baseline levels of signaling in physiologic systems. In support of this mechanism, sparse NMR data on the unliganded adenosine receptor provides evidence for the existence of an equilibrium between the inactive and active conformations (11).
Although data from single-molecule fluorescence on β2AR (12) have been interpreted in terms of the presence of the active form of the unliganded receptor, direct structural evidence for the existence of an active-like conformation in equilibrium with the unliganded, inactive conformation is lacking for this pharmacologically important GPCR. Indeed, structural studies of equilibrium populations by single-molecule fluorescence resonance energy transfer (FRET) (13) and double electron–electron resonance (DEER) spectroscopy (5) did not detect the presence of the active conformation in the ensemble of the unliganded β2AR. This leaves the mechanism of basal activity for β2AR unresolved. If true conformational selection plays a role, then a population of active form must exist in the equilibrium manifold of the unliganded receptor. The goal of the present work is to investigate the possibility that an active conformation in fact exists but is too sparsely populated to detect with the methods employed.
Detection and characterization of sparsely populated (rare or excited) conformations is a general problem in elucidating the molecular mechanisms of protein function. Such low-lying excited states may play important functional roles despite being sparsely populated (14). Extensive empirical evidence indicates that the application of pressure may provide a solution to this problem by reversibly increasing the population of excited states (15, 16), allowing the use of standard experimental techniques to characterize these states and elucidate their functional roles. The mechanisms underlying the pressure-dependent conformational shifts are discussed in recent literature (17–20). It is important to note that as long as the pressure-induced shift in population is reversible, the rare state must also exist at atmospheric pressure and is not an artifact of pressure itself.
Recently, site-directed spin labeling (SDSL) together with variable pressure electron paramagnetic resonance (EPR) was developed to monitor protein conformational shifts due to applied hydrostatic pressure (21). Of particular interest for the present study is SDSL with DEER (22). DEER provides a probability distribution of the distances between site-specifically introduced spin labels. Thus, each conformation in an ensemble generates a different distance in the distribution with a probability proportional to the population of the associated conformation, provided that the spin labels are properly placed to monitor the conformational equilibrium. In this respect, DEER is well suited to provide a structural definition to excited states revealed by pressure because it does not require a single conformation to be fully populated but instead reveals each conformation in the ensemble that is sufficiently populated (above ∼5 to 10%). Here we employ pressure-resolved DEER (23) to identify and characterize sparsely populated states (below 1%) in the conformational ensemble of unliganded β2AR. The data provide direct structural evidence for the presence of an active-like conformation in the equilibrium ensemble of the unliganded receptor. The addition of agonists and inverse agonists increase and decrease the equilibrium population of the active form, respectively. The results indicate a mechanism for basal activity as well as that for inverse agonists in the β2AR. Equally important, the variable pressure data provide a thermodynamic characterization of the active state including the partial molar volume, free energy, and compressibility relative to the ground (inactive) state.
Results and Discussion
Basal Activity of β2AR.
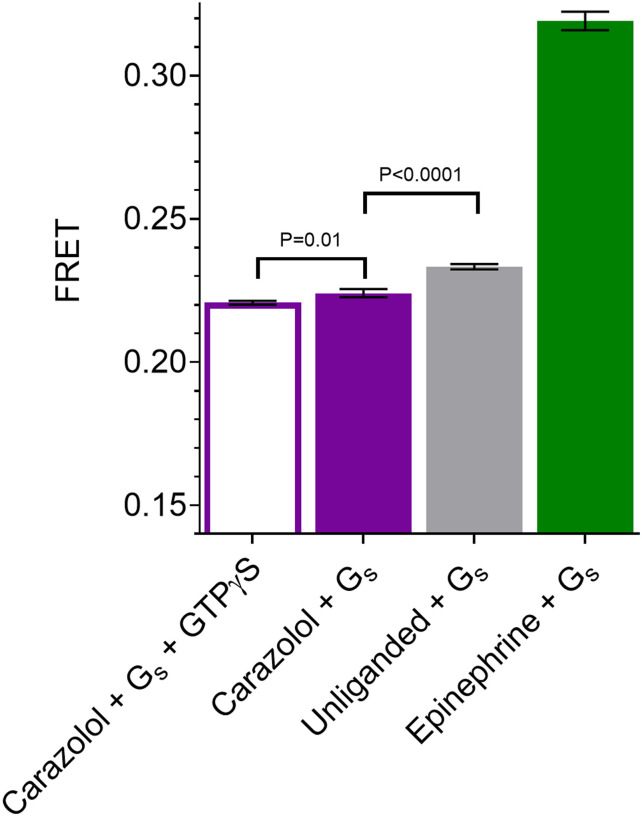
Since purified β2AR in detergent is not a physiologic environment, we wanted to confirm that β2AR in fact has basal activity under conditions similar to those used for DEER studies. Given the small fraction of unliganded β2AR anticipated to be in the active state (∼0.5%; see Pressure-Resolved DEER Reveals a Preexisting Equilibrium in the Unliganded Receptor), we performed an assay to detect constitutive activity under conditions that would trap the active β2AR in a β2AR-Gs complex, thereby amplifying the response. For this purpose, FRET experiments were designed to monitor β2AR-Gs complex formation in the presence of a low concentration of GDP (0.24 µM) and in the absence of GTP to capture and accumulate the β2AR-GsEMPTY state. For these experiments the β2AR was labeled with Cy3B (donor fluorophore), and Gs was labeled with Cy5 (acceptor fluorophore). FRET was measured for receptor either without ligand or in the presence of saturating concentrations of the agonist epinephrine or inverse agonist carazolol (Fig. 1). The level of FRET observed for carazolol likely represents the nonspecific interactions between the lipidated Gs and the detergent micelle of the receptor. This is supported by the observation that a similar level of FRET was obtained when an excess of the nonhydrolyzable GTP analog GTPγS was added to dissociate any nucleotide-free β2AR-Gs complexes. While small, there is a statistically significant increase in FRET for the unliganded receptor compared with the carazolol-bound receptor, verifying that the unliganded receptor exhibits a measurable level of basal activity under the in vitro conditions used in the DEER experiments. In the presence of epinephrine there is an ∼10-fold greater increase in FRET compared with unliganded receptor.
Fig. 1.
FRET measurement of basal activity of the β2AR. β2AR-Gs complex formation was monitored under buffer conditions similar to DEER sample conditions, either without ligand or in the presence of saturating concentrations of epinephrine or carazolol. FRET experiments on β2AR labeled with Cy3B (donor fluorophore) and Gs labeled with Cy5 (acceptor fluorophore) reveal the FRET intensity due to β2AR-Gs complex formation in the presence or absence of an excess of the nonhydrolyzable GTP analog GTPγS (Materials and Methods). Error bars denote SD, four replicates. Statistical significance is tested with Welch’s t test.
Monitoring the Conformational Manifold of β2AR with DEER.
The above FRET assay identifies the existence of basal activity in the preparation under study but provides no information on possible mechanisms. For example, the unliganded receptor may exist in a conformation with weak affinity for the G protein, and this weak interaction leads to a low level of nucleotide exchange. Alternatively, the weak interaction could lead to a fully active β2AR-Gs complex by an induced fit mechanism. In such models, the canonical active conformation of the receptor does not exist in the absence of G protein. On the other hand, if a conformational selection mechanism pertains, then the active conformation of receptor must exist prior to the interaction with G protein. We note that both mechanisms could in principle coexist, but if the active conformation is present at equilibrium in the unliganded receptor, then conformational selection must be an active pathway for activation. In the present experiments, we employ pressure-resolved DEER spectroscopy to look for the hallmark TM6 movement that identifies the presence of the active conformation in the absence of ligand and absence of G protein.
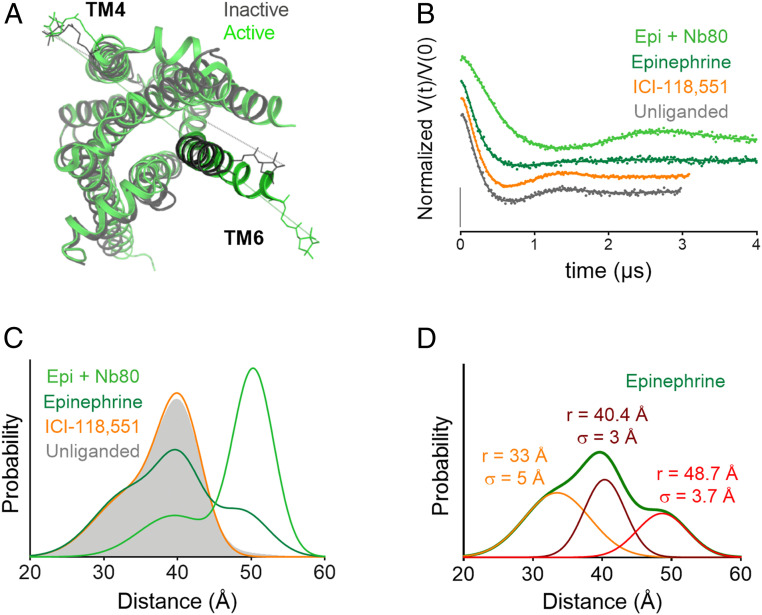
For a conformational readout of the β2AR equilibrium, nitroxide spin labels were introduced at the cytoplasmic ends of transmembrane helices 4 and 6 (TM4 and TM6) at positions 148 and 266 (Ballesteros–Weinstein 4.40 and 6.28) (24), respectively. Based on crystallographic coordinates of β2AR (25), the TM4 position is expected to remain relatively fixed and therefore serves as a useful reference to monitor outward movement of TM6 (Fig. 2A). Previous DEER (26) and single-molecule FRET studies (13) have demonstrated the utility of this distance pair in monitoring receptor activation. As mentioned above, outward movement of TM6 is a hallmark of receptor activation by ligand binding and is a key element of the structure of the receptor in the β2AR-Gs complex.
Fig. 2.
Monitoring the β2AR TM6 conformational equilibrium with DEER. (A) Crystal structures of inactive (gray; Protein Data Bank [PDB] ID 2RH1) and active (green; PDB ID 3SN6) β2AR with a PROXYL side chain shown at labeled sites in TM4 (148) and TM6 (266). (B) Background-corrected DEFs and fits for the unliganded receptor as well as receptor bound to an inverse agonist (ICI-118,551), agonist (epinephrine), and agonist + G protein mimic (Nb80) at atmospheric pressure. The data are displayed as dots and the fits as solid lines. (C) Distance distributions obtained from fits of the data to a Gaussian model. (D) Individual Gaussian components of the distance distribution for the receptor bound to epinephrine. All pressure-dependent and ligand-bound DEER distance distributions are composed of these three components with varying amplitude.
In the present experiments, distance distributions between nitroxides at 148 and 266 were determined as a function of pressure for the unliganded state, in the presence of an agonist, in the presence of an inverse agonist, and in the presence of a G protein mimic nanobody + agonist. In all cases, the distance distributions derived from the DEER dipolar evolution functions (DEFs) were multimodal and could be reasonably well fit to a sum of Gaussian distributions (17, 27, 28). The goodness of fit of the data to the Gaussian model was similar to that for a model-free Tikhonov analysis (SI Appendix, Fig. S1).
Fig. 2B shows the background-corrected DEFs for the unliganded receptor as well as the receptor bound to an agonist (epinephrine), an inverse agonist (ICI-118,551), and a G protein mimic nanobody (Nb80) + epinephrine at atmospheric pressure. The data collection times were longer for the epinephrine and epinephrine + Nb80 bound states to improve the resolution of longer distances detected in these distributions. Fig. 2C shows the corresponding distance distributions derived from the DEFs assuming a Gaussian model. The unliganded receptor generates a relatively broad distance distribution with significant probability density in the range of ∼25 to 45 Å and a most-probable distance of 40.4 Å (solid gray). Binding of the inverse agonist ICI-118,551 has negligible impact on the distance distribution (orange trace), whereas the agonist epinephrine increases a population at 48.7 Å, consistent with the range of accessible distances for this spin pair with TM6 in an active outward conformation (Fig. 2) (26). In agreement with previous studies, epinephrine alone does not fully stabilize an active state (26). Epinephrine + Nb80 binding to the receptor dramatically shifts the population to an outward tilted position of TM6, although of a slightly longer distance (1.6 Å) than epinephrine alone, perhaps reflecting different positions of TM6. Alternatively, this shift in distance could result from direct contact of Nb80 with the spin labels at the cytoplasmic surface or minor secondary structure changes local to the labeling sites due to contact of Nb80 with TM4 and TM6. The influence of Nb80 is only included to illustrate the large shift in population to the outward tilted position of TM6 but was not studied as a function of pressure; the present study is focused on the unliganded receptor and its interaction with small molecule ligands.
As illustrated in Fig. 2D for epinephrine bound to the receptor, three populations (conformational states) are needed to fit the distribution; the peak positions and widths are noted in the figure. Importantly, the entire set of ligand- and pressure-dependent DEER data presented below are well fit globally using this set of three Gaussian distributions of varying amplitude and fixed position and width (Fig. 2D and SI Appendix, Table S1). It is apparent in Fig. 2C and SI Appendix, Fig. S2 that the population at 48.7 Å, corresponding to the outward tilted conformation, is too small to be reliably detected in the unliganded state.
The Gaussian peak centered at 48.7 Å is assigned to the active conformation based on the prominence of a similar peak in the distribution of the receptor bound to epinephrine plus Nb80, although in principle, there could be conformations with the outward position of TM6 that are not fully active but are on the pathway to activation (26). Similarly, the peaks centered at 33 and 40.4 Å are taken to represent inactive conformations based on the distance distribution of the receptor bound to the inverse agonist ICI-188,551. The ∼20-Å width of the distribution characteristic of the inactive state is taken to reflect significant structural heterogeneity, perhaps including conformations with the ionic lock in place or broken as previously suggested based on earlier DEER and NMR measurements (26) and molecular dynamics simulations (29).
Pressure-Resolved DEER Reveals a Preexisting Equilibrium in the Unliganded Receptor.
The distance distributions generated by global fitting of the unliganded and ligand-bound dataset in Fig. 2 recapitulate the major features of the model-free fits to the individual DEFs (SI Appendix, Fig. S1). An underlying assumption of this global analysis of the DEER data is that the different ligand and pressure conditions shift the equilibrium between a shared set, or ensemble, of conformations, and the high quality of the global fit supports the validity of this model. For the unliganded receptor at atmospheric pressure, the population of the active conformation in an unbiased global fit was 2 to 4% (SI Appendix, Table S1), although the quality of the fits was negligibly affected by fixing the population of this conformation at 0% (SI Appendix, Fig. S2). This is consistent with previous DEER experiments that could not conclusively identify the presence of the active conformation in the ensemble of the unliganded receptor (26), due to a combination of low active conformation population and the difficulty of accurately quantifying long distances, which require extended dipolar evolution times and extremely high signal-to-noise.
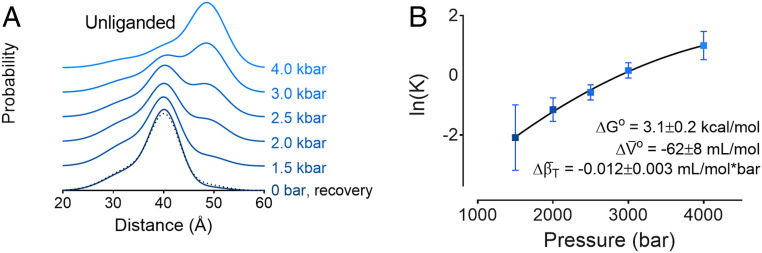
Upon pressurization, the population of the active conformation increases continuously to a population of 73% at 4 kbar, the highest pressure tested here. Importantly, the changes observed at 4 kbar are completely reversible (black dashed line, Fig. 3A and SI Appendix, Fig. S3), demonstrating that the population shift caused by pressure is not due to an irreversible physical process such as aggregation but rather is due to a preexisting equilibrium between inactive and active conformations at atmospheric pressure. The pressure dependence of the equilibrium between inactive and active states is well fit to a two-state model (Fig. 3B), yielding a ΔGo for activation of 3.1 ± 0.2 kcal/mol. This corresponds to 0.5% of unliganded receptors in an active conformation under ambient conditions, consistent with this conformation being below the detection limit at atmospheric pressure.
Fig. 3.
Pressure increases the population of the active conformation in unliganded β2AR. (A) Distance distributions of unliganded receptor as a function of pressure, as well as the recovery distance distribution (dotted black line) collected for the 4-kbar sample after allowing it to reequilibrate at atmospheric pressure. (B) Plot of ln(K) vs. pressure and fit to a two-state model to yield the difference in free energy (), partial molar volume changes (), and compressibility () for the inactive-to-active transition. DEFs for these samples are shown in SI Appendix, Fig. S3.
Can such a small population be responsible for the observed levels of basal activity as viewed in the context of the conformational selection mechanism? A study of β2AR basal activity in cardiac myocytes found that the unliganded receptor overexpressed to 69 times the endogenous level produces 66% of the G protein steady-state signaling generated by isoproterenol-bound receptor at the endogenous level (5). Assuming that ligand binding and conformational equilibria are fast compared to G protein activation (30) and that about 32% of the receptor is in the active conformation with isoproterenol bound (SI Appendix, Fig. S6 and Table S1), one can estimate that the apparent concentration of activated conformation generating signaling in the unliganded receptor is only 0.3% of the receptor population. The similarity of this value to the active conformation population determined by pressure-resolved DEER is striking and further substantiates the assumption of a direct correlation between TM6 conformation and G protein activation under the in vitro conditions employed herein.
It should be emphasized that the assay for constitutive activity presented in Fig. 1 is not a steady-state activation but is designed to trap the active nucleotide-free β2AR-Gs complex and amplify the response and cannot provide an estimate of the actual amount of active species in equilibrium with the inactive state. The assay only confirms that there is constitutive activity, while the variable-pressure DEER experiment directly determines the true equilibrium amount of activated species, which is rather like that estimated from data from an in vivo cellular response in the context of the conformational selection model.
Mechanism of the Pressure Response.
Proteins respond to pressure both by compression and by shifting conformational equilibria toward structures with lower partial molar volume. For soluble proteins, it has been found that a lower partial molar volume can be achieved by filling internal packing defects with water, along with any concomitant structural changes, or by repacking the protein interior to fill packing defects with neighboring side chains (17, 18). Far less is known about pressure effects on membrane proteins, but one expects similar responses, perhaps modulated by the nature of the lipidic or micellar environment. The outward motion of TM6 at the intracellular surface in the active state opens the TM bundle and therefore is unlikely to lead to increased packing in the protein interior. On the other hand, the opening of the cytoplasmic surface by the helical motion suggests that any internal packing defects (cavities) are now accessible to water. To test whether cavity hydration contributes to the mechanism of the β2AR response to pressure, we examined the behavior of the receptor in the presence of a strong protecting osmolyte, sucrose, which stabilizes more compact and less solvated protein conformations (31, 32) (SI Appendix, Fig. S4). At 2 kbar, the presence of 25% (wt/wt) sucrose reduced the active state population from 24 to 16% (SI Appendix, Table S1). The counteracting effects of pressure and the protecting osmolyte indicate that cavity hydration does contribute to the mechanism of the pressure response in β2AR, although other factors such as protein–detergent interactions likely also play a role. A reduction in the active state population of rhodopsin (TM6 outward tilt) was similarly observed upon addition of sucrose (32), consistent with earlier data indicating that rhodopsin internal hydration increases upon activation (33).
Interestingly, the slight curvature in the plot of Fig. 3B is accounted for by a lower compressibility of the active relative to the inactive conformations. It is often assumed that the compressibility difference between conformations is negligible, but at the relatively high pressures used here (4 kbar) this assumption is invalid (34). The small negative value of the compressibility difference (−0.012 ± 0.003 mL/mol*bar) and the relatively large errors in the data preclude a high degree of precision, but the fitted value verifies the algebraic sign of the parameter. This result is consistent with numerous observations that partially unfolded and hydrated excited states of soluble proteins (i.e., the molten globule) have lower compressibility than the ground state (35, 36). Collectively, the results suggest that the activated conformation of the receptor may be viewed as a partially unfolded and internally hydrated structure.
Pressure Provides Insight into the Mechanism of Ligand-Mediated Signaling.
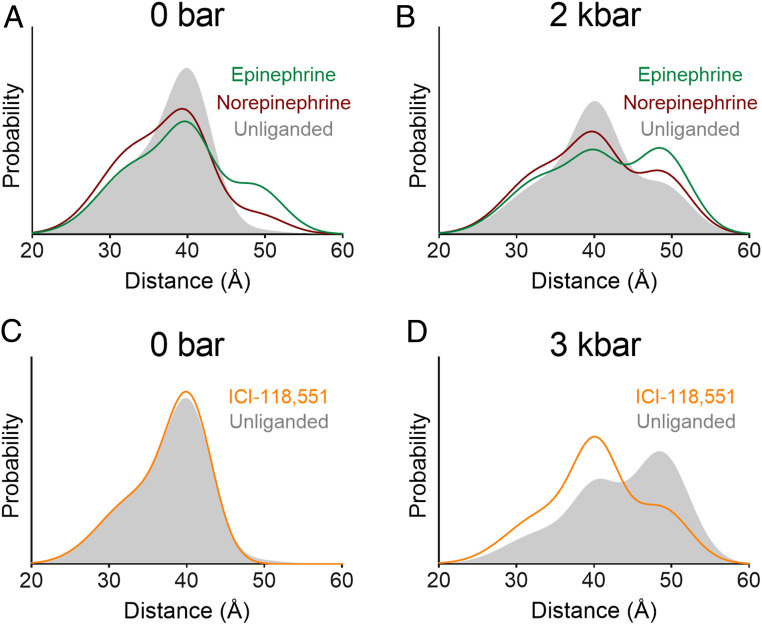
To examine the combined effects of pressure and ligand binding, we compared the inactive–active equilibrium at high pressure in the unliganded vs. ligand-bound receptor. The endogenous agonists epinephrine and norepinephrine increased the population of the active conformation at 2 kbar compared with the unliganded receptor (Fig. 4 and SI Appendix, Fig. S5 and Table S1). Importantly, the additive effect of pressure and agonist is consistent with stabilization of the same conformation by both agonist binding and pressurization. These results are also consistent with recent high-pressure NMR experiments on the β1AR that revealed a pressure-populated shift toward the active conformation in receptor bound to the commonly used βAR-selective catecholamine agonist isoproterenol (37). However, the NMR experiments were restricted to the agonist bound receptor and could not demonstrate the presence of the active form in the unliganded receptor, the key focus of the present work focused on the mechanism for basal activity. Moreover, the NMR study only examines pressure-dependent changes near the orthosteric ligand site at the extracellular surface. On the other hand, the DEER data presented here were obtained at the intracellular surface, where the large structural changes in TM6 take place that allow binding of Gs. While it is clear that changes near the ligand binding site and at the cytoplasmic end of TM6 are allosterically coupled in the presence of a ligand (38), it is not obvious that the coupling is strong in the more flexible apo-protein. The volume changes observed by EPR may be unrelated to those at the other surface, and the mechanism of the volume change may be different.
Fig. 4.
β2AR pressure response is ligand-dependent. Distance distributions for the unliganded receptor overlaid with receptor bound to endogenous agonists at (A) atmospheric pressure and (B) 2 kbar, as well as inverse agonist (ICI-118,551) at (C) atmospheric pressure and (D) 3 kbar. Ligands are present at saturating concentrations in each sample. DEFs for these samples are shown in SI Appendix, Fig. S5.
Here the pressure response was also measured for the isoproterenol-bound receptor (SI Appendix, Fig. S6), and the results are consistent with epinephrine and norepinephrine. The relative populations of active conformation for the agonists at 2 kbar, epinephrine > norepinephrine, were the same as those at atmospheric pressure. Interestingly, these agonists stimulate Gs signaling to a similar degree in cell-based assays (39, 40) and in neonatal cardiac myocytes (41). However, differences between epinephrine and norepinephrine in bulk fluorescence (39) and FRET-based (40) biophysical assays suggest that norepinephrine may stabilize a distinct state, which is consistent with the observation that norepinephrine is less efficacious than epinephrine at promoting receptor coupling to β-arrestin 2 (40) and Gi (41). The DEER results show that norepinephrine stabilizes the same outward displacement of TM6 as epinephrine but to a smaller extent; however, they do not exclude the possibility that norepinephrine stabilizes a distinct state in other TM segments.
The DEER data from the TM4–TM6 spin pair at atmospheric pressure reveal no discernable effect of the inverse agonist ICI-118,551 on the distance distribution (Fig. 4 and SI Appendix, Fig. S5), but they do reveal a bimodal distribution in each case with a difference of ∼7 Å between the most probable positions. In principle, this could arise from different rotamers of the spin label. However, previous DEER and NMR measurements (26) suggest that the heterogeneity of TM6 within this inactive state is due to fluctuations between conformations with the ionic lock intact or broken, as mentioned above. In that earlier study, Carr–Purcell–Meiboom–Gill (CPMG)–NMR experiments using a 19F probe on TM6 showed that the inverse agonists ICI-118,551 and carazolol increase the rate of fluctuations within the inactive conformation without shifting the equilibrium populations (26), suggesting that the bimodal distribution observed here is due to distinct conformational substates involving TM6, the inverse agonist-dependent-exchange rate of which is measured by the 19F CPMG-NMR experiment.
Importantly, upon pressurization a clear difference is observed in the distance distributions of the unliganded and inverse agonist-bound receptor. Both ICI-118,551 and carazolol suppress the population of the active conformation relative to the unliganded receptor at the same pressure (by 27 and 18%, respectively) (Fig. 4 and SI Appendix, Fig. S6 and Table S1). Assuming that ligand binding does not differentially influence the partial molar volumes of the inactive and active conformations, this indicates that inverse agonists suppress the equilibrium population of the active conformation below the 0.5% level of the unliganded receptor at atmospheric pressure, suggesting a structural basis for the mechanism of inverse agonism.
This structural effect may be related to the kinetic effect of inverse agonists revealed by CPMG-NMR, namely, a decrease in the lifetimes of inactive states on the pathway to activation (26). Reducing the lifetime of on-pathway inactive states below the characteristic timescale for conversion to active states was speculated to result in a decreased active state population, which is fully consistent with our results.
Summary and Conclusion.
The pressure-resolved DEER results presented here constitute direct structural evidence for the presence of the active conformation in the equilibrium ensemble of the unliganded β2AR at levels undetectable at atmospheric pressure. By populating this excited state with pressure, it was shown that inverse agonists reduce the population of the active conformation. This effect was not seen in earlier DEER studies, since the active state was too sparsely populated to detect. Taken together, these results reveal an expanded role for conformational selection in GPCRs to include not only agonism but inverse agonism and basal activity as well. Basal activity assay results highlight how subtle changes in conformational equilibria involving rare states may translate to large physiologic effects.
Furthermore, the present study highlights the importance of detecting rare states in order to elucidate molecular mechanisms of catalytic protein function and illustrate the utility of SDSL in combination with variable pressure EPR for defining structural and thermodynamic features of sparsely populated conformations of membrane proteins. This capability will be important in future studies of allosteric and biased agonism, where the delicate interplay of different ligands may have subtle effects on conformational equilibria.
Materials and Methods
β2AR Expression and Purification.
The β2AR construct employed in DEER experiments in this study contains N148C and L266C mutations introduced into a minimal cysteine version of the full-length human β2AR (β2Δ6: C77V, C265A, C327S, C341L, C378A, and C406A) with an N-terminal FLAG tag and C-terminal hexahistidine tag. FRET experiments were performed using a similar β2Δ4 construct (C77V, C327S, C378A, and C406A) without the cysteine mutations at N148 and L266. These constructs, β2Δ6–148C/266C and β2Δ4, have been used previously in biophysical studies and have similar Gs activation in cell-based assays and ligand binding affinities compared with unmodified β2AR (13, 26, 42). Here these constructs were expressed in Sf9 insect cells using a baculovirus expression system (Expressions Systems BestBac) and purified using a combination of anti-FLAG antibody chromatography and functional purification through alprenolol-Sepharose ligand affinity chromatography, similar to previous studies (3, 26, 43). Baculovirus encoding β2Δ6–148C/266C or β2Δ4 was used to infect Sf9 insect cells, which were harvested 60 h postinfection. Membranes were prepared by hypotonic lysis and centrifugation, and the receptor was solubilized from membranes with douncing and stirring in a buffer composed of 1% (wt/vol) n-dodecyl-β-d-maltopyranoside (DDM), 0.01% (wt/vol) cholesteryl hemisuccinate (CHS), 20 mM Hepes, pH 7.4, 350 mM NaCl, 2 mM MgCl2, 2 mM benzamidine, 5.25 μM leupeptin, 1 μM alprenolol, and 1.25 units/mL benzonase in H2O. The soluble fraction was supplemented with 2 mM CaCl2, loaded onto an anti-FLAG M1 antibody affinity column, and washed with 0.1% DDM, 0.01% CHS, 20 mM Hepes, pH 7.4, 350 mM NaCl, and 2 mM CaCl2 in H2O. The receptor was eluted at a concentration of ∼5 μM in 0.1% DDM, 0.01% CHS, 20 mM Hepes, pH 7.4, 350 mM sodium chloride, 200 μg/mL FLAG peptide, and 5 mM EDTA in H2O.
For DEER experiments, FLAG-pure β2Δ6–148C/266C was spin-labeled overnight on ice using 20-fold molar excess of the spin label reagent iodoacetamido-PROXYL in the presence of 100 μM TCEP. This label was selected to facilitate comparison of the data presented herein with those published previously (26). The spin-labeled receptor was loaded onto an alprenolol affinity column and washed with 4 column volumes (CV) of wash buffer consisting of 0.1% DDM, 0.01% CHS, 20 mM Hepes, pH 7.4, and 350 mM NaCl in H2O. Functional receptor was eluted with wash buffer supplemented with 300 μM alprenolol and 2 mM CaCl2 and captured by direct elution onto an anti-FLAG M1 column. M1-bound receptor was rinsed with buffer containing a saturating concentration of the antagonist atenolol, which has low affinity and fast dissociation kinetics. The M1-bound receptor was then rinsed with ligand-free wash buffer for 1 h and eluted as above. The receptor was exchanged into DEER buffer (0.1% DDM, 0.01% CHS, 20 mM Hepes, pH 7.4, 100 mM NaCl in D2O) via size exclusion chromatography (EnRich 650 or Superdex 200 10/300 GL column). The monomeric fraction was collected and concentrated to 100 to 150 μM using a 100-kDa spin concentrator. Finally, 12-μL aliquots of unliganded receptor were flash frozen in liquid nitrogen in the presence of 20% (vol/vol) deuterated glycerol as cryoprotectant.
For FRET experiments, FLAG-pure β2Δ4 was subjected to alprenolol affinity purification and ligand removal as described above. Samples of β2Δ4 at 10 μM were incubated with fivefold molar excess of Cy3 at room temperature for 45 min to label the native C265 residue. After quenching with cysteine, size exclusion chromatography on a Superdex 200 10/300 Increase column in 20 mM Hepes, pH 7.5, 100 mM sodium chloride, 0.01% MNG/0.001% CHS was performed to remove excess label.
Nanobody 80 (Nb80) Expression and Purification.
Nb80 was produced as previously described (25). In brief, Nb80 containing an N-terminal signal sequence and a carboxyl-terminal His6 tag was cloned into the periplasmic expression vector pMES4 and transformed into the BL21(DE3) Escherichia coli cell line (Novagen). Cells were induced in Terrific Broth at an OD600 of 0.8 with 1 mM IPTG and incubated with shaking at 22 °C for 24 h. Periplasmic protein was obtained by osmotic shock, and the nanobody was purified using Ni-chelating Sepharose chromatography followed by size-exclusion chromatography (Sephadex S200 column).
Gs Expression and Purification.
Heterotrimeric Gs was expressed and purified as previously described (13). Briefly, Gα and Gβγ were coexpressed in baculovirus-infected Tni insect cells (Expression Systems BestBac). After 48 h of infection at 27 °C, cells were harvested by centrifugation and lysed in 10 mM Tris, pH 7.5, 100 μM MgCl2, 5 mM β-mercaptoethanol (βME), 50 μM GDP, and protease inhibitors. The membrane fraction was solubilized by dounce homogenization in 20 mM Hepes, pH 7.5, 100 mM NaCl, 1% sodium cholate, 0.1% DDM, 5 mM MgCl2, 5 mM βME, 5 mM imidazole, 50 μM GDP, and protease inhibitors. The protein was loaded on a nickel column, washed, and exchanged into buffer containing 20 mM Hepes, pH 7.5, 100 mM NaCl, 0.05% DDM, 1 mM MgCl2, 20 μM GDP, and 5 mM βME. Protein was eluted with 200 mM imidazole, and the Gβ hexahistidine tag cleaved with 3C protease (1:500 by weight), followed by dialysis overnight in buffer lacking imidazole. The cleaved protein was reverse-purified by flowing over nickel resin in 20 mM imidazole and dephosphorylated by adding 1 mM MnCl2, 5 µL lambda phosphatase, 5 µL antarctic phosphatase, and 5 µL calf intestinal alkaline phosphatase and incubating on ice for 1 h. G protein was then purified on a MonoQ 10/100 GL column (GE Healthcare) as previously described (13). The protein was loaded onto the column and washed with 5 CV 20 mM Hepes, pH 7.5, 0.05% DDM, 1 mM MgCl2, 100 μM TCEP, 20 μM GDP containing 50 mM NaCl, followed by gradient elution using 50 to 335 mM NaCl in the same buffer over 7.5 CV to separate the heterotrimer from free Gβγ. Purified protein was dialyzed into 20 mM Hepes, pH 7.5, 100 mM NaCl, 0.1% DDM, 1 mM MgCl2, 100 µM TCEP, and 20 µM GDP.
Purified Gs samples at 10 µM were incubated with fivefold molar excess of Cy5 at room temperature for 45 min before quenching with cysteine, resulting in three Cy5 labels attached to each Gs heterotrimer. To remove excess label, size exclusion chromatography was performed on a Superdex 200 10/300 Increase column in 20 mM Hepes, pH 7.5, 100 mM sodium chloride, 0.05% DDM/0.001% CHS, 10 µM GDP, 1 mM MgCl2, and 100 µM TCEP.
FRET Assay.
Cy3B-labeled β2Δ4 and Cy5-labeled Gs were used to measure receptor–Gs association using FRET. Cy3B-labeled β2Δ4 was incubated with no ligand (unliganded) or a fivefold molar excess epinephrine or carazolol for 1 h at room temperature. Afterward, either Gs alone or Gs and GTPγS were added to the receptor samples and incubated for 1 h at room temperature. Final concentrations were 100 nM receptor, 250 nM Gs, and 1 mM GTPγS (where applicable). Fluorescence spectra were recorded on a Fluorolog instrument (Horiba) in photon-counting mode. Spectra were collected with emission from 550 to 700 nm and excitation at 520 nm with bandpass of 2 nm and integration time of 0.4 s*nm−1. FRET was calculated as maximum acceptor intensity (IA) divided by maximum acceptor intensity plus maximum donor intensity (IA + ID). Error analysis and plots were done in GraphPad Prism 7 or later.
Sample Preparation for DEER.
Ligands were prepared in 100 mM stocks of DMSO or water, according to their solubility. Working stocks of 13× were prepared in DEER buffer or 26% (vol/vol) DMSO in DEER buffer to aid in ligand solubility. One μL of ligand working stock was added to a thawed receptor aliquot (12 μL) and incubated for 1 h at room temperature. Final ligand concentrations were set fivefold higher than receptor concentration for all samples to ensure saturation. Sucrose samples were prepared by diluting receptor aliquots 1:1 with a solution of 50% (wt/wt) sucrose in DEER buffer. For ambient pressure DEER measurements, samples were transferred to borosilicate capillary tubes (1.4 mm ID × 1.7 mm OD; VitroCom) and flash-frozen in liquid nitrogen. Existing evidence indicates that flash-freezing faithfully captures the room-temperature conformational equilibrium of the protein (44–52) and rotameric equilibrium of the spin label side chain (53).
Pressure-resolved DEER samples were prepared as described elsewhere (17, 23), with the following modifications: a TE Technologies CP-031 Peltier temperature control unit was used to maintain the pressurized sample at 298 K prior to freezing, and all measurements were performed using a Barocycler HUB880 pressure intensifier (Pressure Biosciences, Inc.) and high-pressure tubing and connectors rated to 6.9 kbar (Maxpro Technologies). Briefly, samples were loaded into a standard capillary cell, and a silicone piston was inserted to separate the sample from the pressurization fluid. A magnetic collar was glued to the outside of the upper portion of the cell to allow its position inside a pressure chamber to be controlled externally using a magnet. Samples were pressurized at 298 K, held for 30 s to equilibrate, then rapidly cooled to 200 K while still under pressure. Freezing the pressurized sample kinetically traps the high-pressure conformational ensemble. The sample was then depressurized, removed from the pressure chamber, and immediately transferred to a liquid nitrogen bath in preparation for DEER data acquisition at 80 K. To test reversibility of the pressure response, pressurized samples were brought to ambient temperature and pressure and allowed to equilibrate for 2 to 5 min before flash-freezing the sample to 200 K in a dry ice/ethanol bath for subsequent DEER data collection. All pressures listed in this article are gauge pressure (i.e., 0 bar is equal to atmospheric pressure).
DEER Experiments.
DEER data were acquired on a Bruker ELEXSYS E580 spectrometer equipped with a SuperQ-FT bridge, 10 W AmpQ amplifier, and SpinJetAWG operated at Q-band frequency (∼33.5 GHz) with an ER 5107D2 Q-Band pulse resonator (Bruker Biospin). Sample temperature was maintained at 80 K during data collection using an Oxford ITC503S temperature controller (Oxford Instruments). A dead-time free four-pulse DEER sequence (54) was used with an eight-step suppression of the deuterium ESEEM signal (55) and 16-step phase cycle. The pump pulse (linear frequency-swept [chirp] (56), 100 ns, 100 MHz bandwidth) was applied at the minimum between the low- and center-field maxima of the absorbance spectrum. The observe pulses (rectangular, ∼20 ns π/2, ∼40 ns π) were applied at a −70 MHz offset from the pump pulse. Data were acquired with dipolar evolution times of 3 to 4.5 μs, using >4 μs for all pressure samples.
Background correction and model-free fitting analysis of DEFs were performed with the program LongDistances590 (developed by Christian Altenbach and available at www.biochemistry.ucla.edu/biochem/Faculty/Hubbell/). The dimensionality was allowed to vary during optimization of the background correction; typical optimized values were in the range of 2.5 to 3.5. The program DeerA (57) (developed and kindly provided by Richard A. Stein, Vanderbilt University, Nashville, TN, and available online via https://www.mathworks.com/) was used for Gaussian model-based analysis of background-corrected DEFs. DeerA was used for global analysis of the unliganded and ligand-bound dataset, in which a minimal set of Gaussians required to fit the background-corrected DEFs was identified. The mean and width of each Gaussian distribution were kept constant across all samples, and only the relative amplitudes were allowed to vary. The fractional population of each of the three individual Gaussians is listed in SI Appendix, Table S1, for each sample, calculated by dividing the area for the individual Gaussian by the total area for all Gaussians. The comparison in SI Appendix, Fig. S2, was performed using the model-based function in LongDistances. Means and widths of individual Gaussians were taken from the global analysis, and amplitudes were allowed to vary. The amplitude of the 48.7-Å peak was fixed at 0 for the two-Gaussian fit. All distributions were area-normalized and plotted in GraphPad Prism 7.
Thermodynamic Analysis of the Inactive–Active Equilibrium.
The pressure dependence of the unliganded receptor distribution (Fig. 2A) is well-fit to a two-state equilibrium between the active and inactive conformations, where the Gaussians centered at 33 Å and 40.4 Å are attributed to the inactive state and the Gaussian centered at 48.7 Å is attributed to the active state (see Results). The equilibrium constant (K) is given by Eq. 1:
| [1] |
where fa and fi are the fractional populations of active and inactive conformations, respectively. The pressure dependence of the Gibbs free energy () of the system is described by Eqs. 2 and 3:
| [2] |
| [3] |
Combining Eqs. 2 and 3 yields Eq. 4, where R is the gas constant and T is the temperature. A temperature of 298 K was used in this analysis because it was the holding temperature for all samples prior to freezing for DEER data acquisition.
| [4] |
A plot of ln(K) vs. pressure for the unliganded receptor was fit to Eq. 4 using GraphPad Prism 7 to solve for the change in standard Gibbs free energy (), partial molar volume (), and isothermal compressibility () for the transition from inactive to active state (Fig. 3B). Values of fa and fi at atmospheric pressure were calculated using with Eqs. 1 and 2.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01GM135581 (M.T.L.), NSF Graduate Research Fellowships Program (R.A.M.), American Heart Association Postdoctoral Fellowship 17POST33410958 (M.M.), German Research Foundation Research Fellowship (M.E.), the German Academic Exchange Service (D.H.), NIH Grant R01EY05216 and the Jules Stein Professor Endowment (W.L.H.), National Eye Institute Core Grant P30EY00331 and a Research to Prevent Blindness Unrestricted Grant (to the Jules Stein Eye Institute), and NIH grant R01NS028471 and the Mathers Foundation (B.K.K.). B.K.K. is a Chan Zuckerberg Biohub investigator.
Footnotes
Competing interest statement: B.K.K. is a cofounder of and consultant for ConfometRx, Inc.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013904117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Cherezov V., et al. , High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altenbach C., Kusnetzow A. K., Ernst O. P., Hofmann K. P., Hubbell W. L., High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl. Acad. Sci. U.S.A. 105, 7439–7444 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasmussen S. G. F., et al. , Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leurs R., Smit M. J., Alewijnse A. E., Timmerman H., Agonist-independent regulation of constitutively active G-protein-coupled receptors. Trends Biochem. Sci. 23, 418–422 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y.-Y., et al. , Spontaneous activation of β(2)- but not β(1)-adrenoceptors expressed in cardiac myocytes from β(1)β(2) double knockout mice. Mol. Pharmacol. 58, 887–894 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Bond R. A., et al. , Physiological effects of inverse agonists in transgenic mice with myocardial overexpression of the β 2-adrenoceptor. Nature 374, 272–276 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Engelhardt S., Grimmer Y., Fan G. H., Lohse M. J., Constitutive activity of the human β(1)-adrenergic receptor in β(1)-receptor transgenic mice. Mol. Pharmacol. 60, 712–717 (2001). [PubMed] [Google Scholar]
- 8.Seifert R., Wenzel-Seifert K., Constitutive activity of G-protein-coupled receptors: Cause of disease and common property of wild-type receptors. Naunyn Schmiedebergs Arch. Pharmacol. 366, 381–416 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Lefkowitz R. J., Cotecchia S., Samama P., Costa T., Constitutive activity of receptors coupled to guanine nucleotide regulatory proteins. Trends Pharmacol. Sci. 14, 303–307 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Leff P., The two-state model of receptor activation. Trends Pharmacol. Sci. 16, 89–97 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Ye L., Van Eps N., Zimmer M., Ernst O. P., Prosser R. S., Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature 533, 265–268 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Lamichhane R., et al. , Single-molecule view of basal activity and activation mechanisms of the G protein-coupled receptor β2AR. Proc. Natl. Acad. Sci. U.S.A. 112, 14254–14259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregorio G. G., et al. , Single-molecule analysis of ligand efficacy in β2AR-G-protein activation. Nature 547, 68–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baldwin A. J., Kay L. E., NMR spectroscopy brings invisible protein states into focus. Nat. Chem. Biol. 5, 808–814 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Akasaka K., Probing conformational fluctuation of proteins by pressure perturbation. Chem. Rev. 106, 1814–1835 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Fourme R., Girard E., Akasaka K., High-pressure macromolecular crystallography and NMR: Status, achievements and prospects. Curr. Opin. Struct. Biol. 22, 636–642 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Lerch M. T., et al. , Structure-relaxation mechanism for the response of T4 lysozyme cavity mutants to hydrostatic pressure. Proc. Natl. Acad. Sci. U.S.A. 112, E2437–E2446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roche J., et al. , Cavities determine the pressure unfolding of proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 6945–6950 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akasaka K., Kitahara R., Kamatari Y. O., Exploring the folding energy landscape with pressure. Arch. Biochem. Biophys. 531, 110–115 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Li H., Kamatari Y. O., Cavities and excited states in proteins. Subcell. Biochem. 72, 237–257 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Lerch M. T., Yang Z., Altenbach C., Hubbell W. L., “High-pressure EPR and site-directed spin labeling for mapping molecular flexibility in proteins” in Methods in Enzymology, Qin P. Z., Warncke K., Eds. (Academic Press, 2015), pp. 29–57. [DOI] [PubMed] [Google Scholar]
- 22.Jeschke G., DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 63, 419–446 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Lerch M. T., Yang Z., Brooks E. K., Hubbell W. L., Mapping protein conformational heterogeneity under pressure with site-directed spin labeling and double electron-electron resonance. Proc. Natl. Acad. Sci. U.S.A. 111, E1201–E1210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ballesteros J. A., Weinstein H., Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 (1995). [Google Scholar]
- 25.Rasmussen S. G. F., et al. , Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature 469, 175–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manglik A., et al. , Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell 161, 1101–1111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casey T. M., Fanucci G. E., “Spin labeling and double electron-electron resonance (DEER) to deconstruct conformational ensembles of HIV protease” in Methods in Enzymology, Qin P. Z., Warncke K., Eds. (Academic Press, 2015), pp. 153–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collauto A., et al. , Rates and equilibrium constants of the ligand-induced conformational transition of an HCN ion channel protein domain determined by DEER spectroscopy. Phys. Chem. Chem. Phys. 19, 15324–15334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dror R. O., et al. , Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc. Natl. Acad. Sci. U.S.A. 106, 4689–4694 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohse M. J., Maiellaro I., Calebiro D., Kinetics and mechanism of G protein-coupled receptor activation. Curr. Opin. Cell Biol. 27, 87–93 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Kendrick B. S., et al. , Preferential exclusion of sucrose from recombinant interleukin-1 receptor antagonist: Role in restricted conformational mobility and compaction of native state. Proc. Natl. Acad. Sci. U.S.A. 94, 11917–11922 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López C. J., Fleissner M. R., Guo Z., Kusnetzow A. K., Hubbell W. L., Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 18, 1637–1652 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grossfield A., Pitman M. C., Feller S. E., Soubias O., Gawrisch K., Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. J. Mol. Biol. 381, 478–486 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prehoda K. E., Mooberry E. S., Markley J. L., Pressure denaturation of proteins: Evaluation of compressibility effects. Biochemistry 37, 5785–5790 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Chalikian T. V., Breslauer K. J., Compressibility as a means to detect and characterize globular protein states. Proc. Natl. Acad. Sci. U.S.A. 93, 1012–1014 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taulier N., Chalikian T. V., Compressibility of protein transitions. Biochim. Biophys. Acta 1595, 48–70 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Abiko L. A., Grahl A., Grzesiek S., High pressure shifts the β1-adrenergic receptor to the active conformation in the absence of G protein. J. Am. Chem. Soc. 141, 16663–16670 (2019). [DOI] [PubMed] [Google Scholar]
- 38.DeVree B. T., et al. , Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature 535, 182–186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swaminath G., et al. , Sequential binding of agonists to the β2 adrenoceptor. Kinetic evidence for intermediate conformational states. J. Biol. Chem. 279, 686–691 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Reiner S., Ambrosio M., Hoffmann C., Lohse M. J., Differential signaling of the endogenous agonists at the β2-adrenergic receptor. J. Biol. Chem. 285, 36188–36198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., et al. , Norepinephrine- and epinephrine-induced distinct β2-adrenoceptor signaling is dictated by GRK2 phosphorylation in cardiomyocytes. J. Biol. Chem. 283, 1799–1807 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Yao X. J., et al. , The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc. Natl. Acad. Sci. U.S.A. 106, 9501–9506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobilka B. K., Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal. Biochem. 231, 269–271 (1995). [DOI] [PubMed] [Google Scholar]
- 44.McHaourab H. S., Steed P. R., Kazmier K., Toward the fourth dimension of membrane protein structure: Insight into dynamics from spin-labeling EPR spectroscopy. Structure 19, 1549–1561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawidowski D., Cafiso D. S., Allosteric control of syntaxin 1a by Munc18-1: Characterization of the open and closed conformations of syntaxin. Biophys. J. 104, 1585–1594 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Vera I. M. S., et al. , Elucidating a relationship between conformational sampling and drug resistance in HIV-1 protease. Biochemistry 52, 3278–3288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dockter C., et al. , Rigid core and flexible terminus: Structure of solubilized light-harvesting chlorophyll a/b complex (LHCII) measured by EPR. J. Biol. Chem. 287, 2915–2925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Georgieva E. R., Borbat P. P., Ginter C., Freed J. H., Boudker O., Conformational ensemble of the sodium-coupled aspartate transporter. Nat. Struct. Mol. Biol. 20, 215–221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krishnamani V., Hegde B. G., Langen R., Lanyi J. K., Secondary and tertiary structure of bacteriorhodopsin in the SDS denatured state. Biochemistry 51, 1051–1060 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Sattig T., Rickert C., Bamberg E., Steinhoff H. J., Bamann C., Light-induced movement of the transmembrane helix B in channelrhodopsin-2. Angew. Chem. Int. Ed. Engl. 52, 9705–9708 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Van Eps N., et al. , Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc. Natl. Acad. Sci. U.S.A. 108, 9420–9424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Eps N., et al. , Conformational equilibria of light-activated rhodopsin in nanodiscs. Proc. Natl. Acad. Sci. U.S.A. 114, E3268–E3275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Georgieva E. R., et al. , Effect of freezing conditions on distances and their distributions derived from double electron electron resonance (DEER): A study of doubly-spin-labeled T4 lysozyme. J. Magn. Reson. 216, 69–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pannier M., Veit S., Godt A., Jeschke G., Spiess H. W., Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 142, 331–340 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Polyhach Y., et al. , High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Phys. Chem. Chem. Phys. 14, 10762–10773 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Doll A., Pribitzer S., Tschaggelar R., Jeschke G., Adiabatic and fast passage ultra-wideband inversion in pulsed EPR. J. Magn. Reson. 230, 27–39 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Stein R. A., Beth A. H., Hustedt E. J., “A straightforward approach to the analysis of double electron-electron resonance data” in Methods in Enzymology, Qin P. Z., Warncke K., Eds. (Academic Press, 2015), pp. 531–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.