Significance
Introgressive hybridization is prevalent in recent and rapid animal radiations, and emerging evidence suggests that it leads to the sharing of genetic variation that can facilitate adaptation to new environments and generate novel phenotypes. Here we study a recent and rapid radiation of African mosquitoes in which only one species, An. funestus, is a primary human malaria vector with a continent-wide geographic distribution. We trace the evolutionary history of the group and demonstrate introgression events between multiple species, the most recent of which involved substantial gene flow into An. funestus that preceded its range expansion across tropical Africa. Our findings point to introgression as an underappreciated factor contributing to the acquisition of high malaria vectorial capacity.
Keywords: adaptive radiation, Anopheles funestus, anopheline mosquito species complex, introgression, reticulate evolution
Abstract
Advances in genomics have led to an appreciation that introgression is common, but its evolutionary consequences are poorly understood. In recent species radiations the sharing of genetic variation across porous species boundaries can facilitate adaptation to new environments and generate novel phenotypes, which may contribute to further diversification. Most Anopheles mosquito species that are of major importance as human malaria vectors have evolved within recent and rapid radiations of largely nonvector species. Here, we focus on one of the most medically important yet understudied anopheline radiations, the Afrotropical Anopheles funestus complex (AFC), to investigate the role of introgression in its diversification and the possible link between introgression and vector potential. The AFC comprises at least seven morphologically similar species, yet only An. funestus sensu stricto is a highly efficient malaria vector with a pan-African distribution. Based on de novo genome assemblies and additional whole-genome resequencing, we use phylogenomic and population genomic analyses to establish species relationships. We show that extensive interspecific gene flow involving multiple species pairs has shaped the evolutionary history of the AFC since its diversification. The most recent introgression event involved a massive and asymmetrical movement of genes from a distantly related AFC lineage into An. funestus, an event that predated and plausibly facilitated its subsequent dramatic geographic range expansion across most of tropical Africa. We propose that introgression may be a common mechanism facilitating adaptation to new environments and enhancing vectorial capacity in Anopheles mosquitoes.
Once considered a rare anthropogenic aberration in animals, interspecific hybridization is now recognized to be both taxonomically widespread and pervasive, particularly in rapidly diversifying groups (1–3). Moreover, mounting genome-scale evidence suggests that introgression, the genetic exchange between species through hybridization and backcrossing, is also prevalent and may be consequential for evolution. Examples from fish, birds, mammals, and insects—including Anopheles mosquitoes—have shown that introgressed variation favored by natural selection can facilitate adaptation, enhance fitness, and drive evolutionary innovation and diversification (4–7). It has been postulated that introgressive hybridization is most prevalent in species-rich and rapidly diversifying radiations (2, 3, 8). Introgression in these groups may solely be opportunistic, given the multiplicity of young species in geographic proximity, but the process may also favor adaptive radiation through the generation of completely novel phenotypes (6, 9, 10).
There are three to four dozen Anopheles mosquito species that are of major importance as human malaria vectors, and all have evolved within recent and rapid radiations of morphologically cryptic species (informally classified as species complexes) (11, 12). Most members of these species complexes play no or very minor roles in disease transmission. The repeated de novo origin of major malaria vectors across these independent species radiations therefore holds clues about the nature of key evolutionary innovations that confer the ability to transmit disease widely and efficiently. However, most Anopheles species complexes are understudied. This is especially true of the secondary or nonvector species for which genomic resources are lacking, and basic knowledge of distribution, ecology, and behavior is scant.
Until now, the single best-studied group has been the Anopheles gambiae complex, composed of at least eight morphologically indistinguishable species that diversified rapidly and recently, likely within the last half-million years (7, 13, 14). Phylogenomic analysis revealed widespread genealogical discordance (7). Some discordance was due to incomplete lineage sorting as a result of both rapid radiation and large effective population sizes (7), but the majority was caused by massive introgression between the main vector species, involving both the autosomes and the centromere-proximal region of the X chromosome. So extensive was its impact that the inferred species branching order was evident in only 2% of the genome—mostly on the distal portion of the X chromosome, which is protected from introgression by a succession of fixed chromosomal inversion differences.
One of the most medically important of the understudied Anopheles species complexes is the Afrotropical Anopheles funestus complex (AFC). The AFC comprises at least seven morphologically similar species (15–18), yet only An. funestus sensu stricto (hereafter, An. funestus) is a highly efficient malaria vector, rivaled in importance solely by An. gambiae and its sister species Anopheles coluzzii in the An. gambiae complex (19–22). Comparative genomics of these two complexes may therefore be instructive with regard to malaria vectorial capacity. Both groups diversified in sub-Saharan Africa and may have experienced common geographic, ecoclimatic, and anthropogenic forces that shaped their history. In addition, the primary vector An. funestus broadly shares several characteristics with primary vectors in the An. gambiae complex: a geographic range that encompasses most of tropical Africa (Fig. 1A), high levels of chromosomal inversion polymorphism (23–25), large effective population size, and little population genetic structure across the continent (26, 27). Furthermore, the discovery of two very distantly related mitochondrial DNA (mtDNA) haplotypes (clades 1 and 2) segregating in An. funestus (27) raises the prospect of historical introgression analogous to that documented for An. gambiae, prompting an intriguing question: Can introgression be a source of evolutionary novelty leading to augmented vectoral capacity?
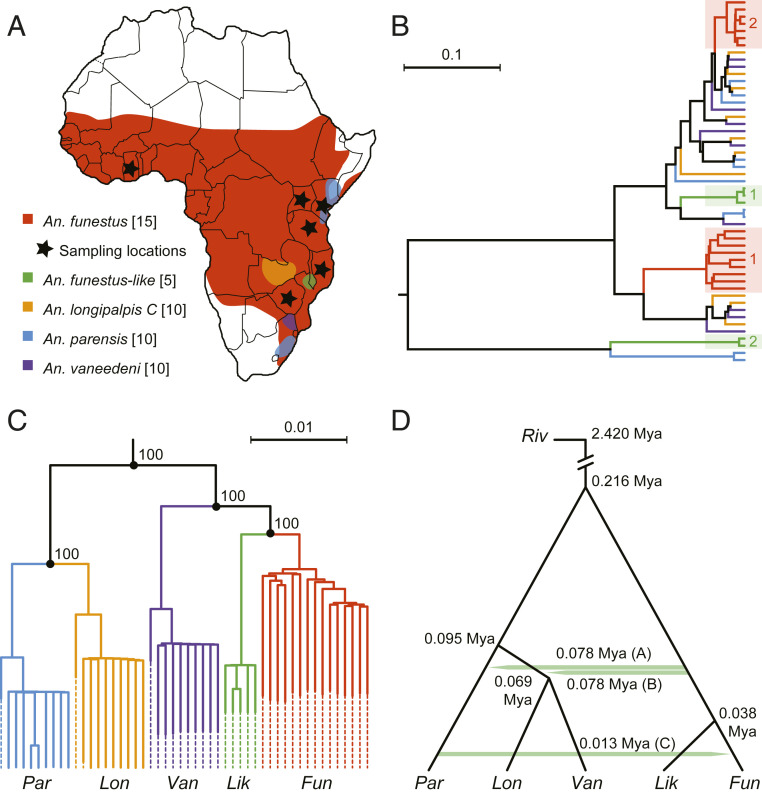
Fig. 1.
Distribution and genetic variation in the AFC. Color coding of species is consistent across panels. (A) Location and distribution of sampled species, adapted from ref. 21. Approximate sample locations for An. funestus are indicated by a black star. For full sample information, see SI Appendix, Table S1. (B) Phylogeny of complete mtDNA genomes constructed using BEAST2 indicating divergent clades of An. funestus (red shading) and An. funestus-like (green shading) (see SI Appendix, Fig. S12 for phylogeny with outgroup). (C) Neighbor-joining phylogeny averaged over the complete nuclear genome. (D) Summary evolutionary history displaying three introgression events as inferred by the methods described in the main text. Introgression events shown as green horizontal arrows between pairs of species indicate the majority direction of introgression. Median divergence and introgression times are displayed in millions of years ago (Mya). See SI Appendix, Table S11 for details. An. funestus (Fun), An. funestus-like (Lik), An. longipalpis C (Lon), An. parensis (Par), and An. vaneedeni (Van), An. rivulorum (Riv).
Here, we examine the role of introgression in the evolution of the AFC, using recent methods of phylogenetic network reconstruction that allow for divergence and reticulation to be inferred jointly. We use a combination of phylogenomic and population genomic analyses, based on de novo genome assemblies and additional whole genome resequencing, to: 1) establish species relationships, 2) determine the direction, extent, and genomic architecture of introgression across the complex, and 3) assess the role of introgression in the evolution of the primary vector An. funestus. We show that extensive interspecific gene flow involving multiple species pairs has shaped the evolutionary history of the AFC since its diversification ∼216 thousand years ago (Kya). The most recent introgression event ∼13 Kya involved a massive and asymmetrical movement of genes from a distantly related AFC lineage into An. funestus, an event that predated and plausibly facilitated its subsequent dramatic geographic range expansion across most of tropical Africa. We propose that introgression may be a common mechanism facilitating adaptation to new environments and enhancing vectorial capacity in Anopheles mosquitoes.
Results
Genome Sequencing and Assembly.
As a foundation for our analyses, we generated reference-assisted de novo genome assemblies (Table 1) from individual field-collected mosquitoes (Fig. 1A and SI Appendix, Texts S1–S3, Fig. S1, and Table S1). Augmenting the existing An. funestus AfunF3 reference from the FUMOZ colony (28), we assembled four new reference genomes from additional AFC species (An. funestus-like, Anopheles parensis, Anopheles vaneedeni, and Anopheles longipalpis type C—hereafter, An. longipalpis C). The only other AFC members, Anopheles confusus and Anopheles aruni, could not be obtained. Our de novo assemblies also included two outgroups: Anopheles rivulorum and Anopheles species A, the latter a previously recognized but formally undescribed species morphologically similar to An. funestus but distinctive in ITS2 sequence (>6% divergent from An. funestus; refs. 29, 30). Reference assemblies were contiguous on the X and chromosome arm 3R for all AFC species, while moderately fragmented scaffolds characterized chromosome arms 3L, 2R, and to a lesser extent, 2L (SI Appendix, Figs. S2 and S3 and Tables S2 and S3).
Table 1.
Genome assembly statistics
| Species | Country | Contigs | Size | N50 | Scafs* | Size | BUSCO† | Accession | |||
| Single | Duplicate | Fragmented | Missing | ||||||||
| An. funestus-like | Malawi | 15,489 | 209,420,710 | 67,275 | 9 | 201,162,353 | 95.5% | 2.6% | 1.1% | 0.8% | STHE00000000 |
| An. longipalpis C | Zambia | 33,338 | 323,670,219 | 30,221 | 7 | 220,976,958 | 75.1% | 18.2% | 3.3% | 3.4% | STHD00000000 |
| An. parensis | South Africa | 26,828 | 251,769,316 | 50,161 | 10 | 216,971,959 | 87.3% | 9.2% | 1.9% | 1.6% | STHC00000000 |
| An. vaneedeni | South Africa | 27,582 | 279,105,143 | 46,024 | 8 | 225,734,782 | 82.7% | 13.9% | 2.7% | 0.7% | STHA00000000 |
| An. species A | Kenya | 21,640 | 242,997,558 | 74,383 | 15 | 192,881,626 | 98.0% | 1.0% | 0.5% | 0.5% | STHF00000000 |
| An. rivulorum | South Africa | 37,847 | 273,938,921 | 26,085 | 14 | 199,718,359 | 92.3% | 4.6% | 1.7% | 1.4% | STHB00000000 |
Reference-assisted scaffolding with ragout and AfunF3.
Percent calculated out of 1,066 total BUSCOs.
In support of population genomic analyses and simulations, we also individually resequenced the genomes of 42 field-collected mosquitoes representing five AFC species (SI Appendix, Table S1). These included eight specimens each of An. longipalpis C, An. parensis, and An. vaneedeni; three of An. funestus-like; and 15 of An. funestus, six of which carried clade 2 mtDNA.
Resolving the Species Tree Despite a Complex History of Introgression.
Species relationships in the AFC have not been confidently resolved. Previous efforts to reconstruct phylogenies using fragments of two mtDNA genes uncovered widespread paraphyly (31), a pattern that we confirm here based on complete mtDNA genome sequences (Fig. 1B and SI Appendix, Fig. S12). A neighbor-joining tree averaged over the entire nuclear genome reveals reciprocal monophyly among species, while the mtDNA tree shows extensive paraphyly (Fig. 1 B and C and SI Appendix, Text S6). Moreover, An. funestus and An. funestus-like each contain two highly divergent mtDNA clades (Fig. 1B and SI Appendix, Fig. S12), consistent with the possibility of historical introgression events resulting in mitochondrial capture. Ribosomal DNA (rDNA) second internal transcribed spacer (ITS2) sequences, instrumental for taxonomic identification of morphologically cryptic species in the AFC (32, 33), also hint at possible historical introgression in this species group. Instead of the near-complete sequence identity expected among units of tandemly arrayed rDNA (34), we found that An. longipalpis C possesses two types of ITS2, one highly similar to An. parensis and the other highly similar to An. vaneedeni (SI Appendix, Fig. S1), in agreement with previous findings (15).
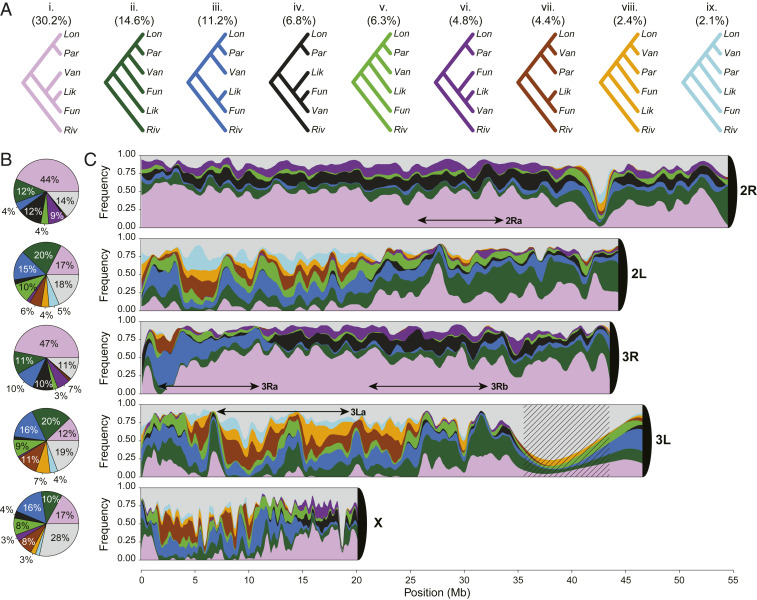
To explore phylogenetic relationships in the AFC, we used nonoverlapping windows 5 kb in length from the full five-species nuclear genome alignment (plus two outgroups; SI Appendix, Texts S4 and S6). Excluding masked heterochromatic, repetitive regions and windows not passing quality filters, this resulted in 24,556 windows spanning ∼123 Mb of aligned sequence. Reconstructing maximum likelihood phylogenies from each of these windows, we observed all possible topologies (n = 105) at least once. The most common topology (denoted tree i) was present at more than twice the frequency of the next most-common tree, having a genome-wide frequency of ∼30% (Fig. 2A and SI Appendix, Fig. S13). This topology is found in highest proportion across most of the length of chromosome arms 2R (44%) and 3R (47%) (Fig. 2 B and C). Its distribution is much more restricted on 2L, 3L, and especially the X chromosome where it is largely absent from positions ∼1.6 to ∼6.8 Mb (Fig. 2 B and C).
Fig. 2.
Frequency and distribution of gene trees. Phylogenetic trees were reconstructed in 5 kb nonoverlapping windows along the chromosomes using PhyML. Color coding of topologies is consistent across panels. (A) Nine major topologies (i–ix) found on any chromosome arm with a frequency of at least 5%. Normalized whole-genome frequencies are indicated in parentheses. (B) The frequency of each major topology on individual chromosome arms. Less frequent topologies are pooled together and displayed in gray in B and C. (C) Chromosome painting representing the frequency of topologies across chromosome arms. For display purposes the frequencies are averaged across adjacent windows. Approximate locations of common chromosomal inversions in An. funestus (3Ra, 3Rb, 3La, and 2Ra) are indicated by double-headed arrows. Centromeres are represented as black 1/4 circles. Hatching represents a masked region. An. funestus (Fun), An. funestus-like (Lik), An. longipalpis C (Lon), An. parensis (Par), An. vaneedeni (Van), and An. rivulorum (Riv).
Importantly, there are nine topologies observed frequently on at least one chromosome arm, reflecting substantial genealogical discordance (Fig. 2 and SI Appendix, Fig. S13). Their heterogeneous distribution along the genome appears idiosyncratic to individual chromosome arms rather than being driven by a common landscape of reduced recombination near centromeres or telomeres (Fig. 2C). Arm-specific topological patterns are not obviously related to the location of common chromosomal inversions known to segregate in An. funestus populations across tropical Africa (2Ra, 3Ra, 3Rb, and 3La; refs. 21, 24, 25) (Fig. 2C). Furthermore, in contrast to the pattern observed in the An. gambiae complex (7), we find no striking difference in the nature or frequency of autosomal versus X chromosome topologies; the most common trees on the autosomes (topologies i–iii) also are the most frequent on the X chromosome (SI Appendix, Fig. S14).
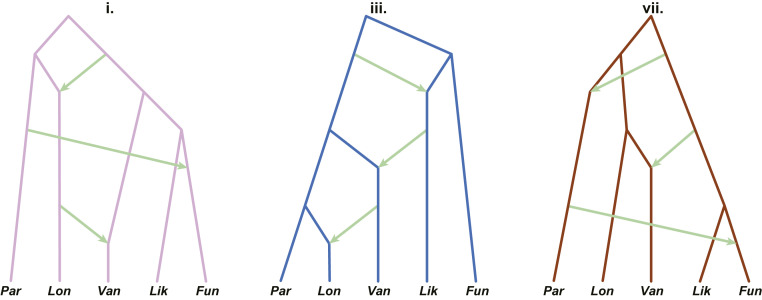
Facing phylogenetic uncertainty owing to incomplete lineage sorting (ILS) and/or introgression, we sought to resolve the true bifurcation history of the AFC by adopting an approach that allows for both divergence and reticulation using D-statistics (35, 36) and admixture graphs to evaluate the fit of each history to the data (SI Appendix, Text S7). To implement this approach, D-statistics (which are robust to the presence of natural selection, ref. 37) were calculated for all AFC species triplets using An. rivulorum as an outgroup (SI Appendix, Fig. S15 and Table S6). Starting with the nine most frequent window topologies from the whole-genome analysis (Fig. 2A), we built admixture graphs for each, adding up to three reticulations in all possible arrangements, and chose the graph with the highest likelihood (lowest cost function) (SI Appendix, Fig. S16). Remaining models based on competing topologies were compared in pairs using a likelihood ratio test. Any model that could not be rejected in favor of another was retained as an equally likely representation of AFC evolutionary history. This left three models (Fig. 3) whose backbones reflect branching patterns observed among the most common bifurcating trees inferred from the whole-genome analysis (trees i, iii, and vii; Fig. 2) but with three reticulations each.
Fig. 3.
Competing models representing the evolutionary history of the AFC inferred from admixture graphs. Lineage divergences correspond to major topologies i, iii, and vii in Fig. 2. Lineage reticulations and the inferred direction are indicated by green arrows. Models are not scaled to time. An. funestus (Fun), An. funestus-like (Lik), An. longipalpis C (Lon), An. parensis (Par), and An. vaneedeni (Van).
To identify the most likely evolutionary history among the three, we used approximate Bayesian computation (ABC) with a supervised machine learning model-selection procedure, a computationally tractable approach even for large datasets (38). For this analysis, we expanded our samples from a single reference genome for each species to multiple resequenced individuals from each AFC species (Fig. 1A and SI Appendix, Tables S1 and S4). Model selection was based on a random forest trained on data simulated under each competing model (SI Appendix, Text S8). Simulations drew from the observed empirical distribution of values of genome-wide recombination rates and nucleotide diversity and were initialized with demographic histories inferred from each species. Summary statistics were calculated using either the whole genome or the noncoding regions only, under the assumption that noncoding regions are less affected by selection; the inferred histories were robust to the choice of loci (SI Appendix, Texts S5 and S8). The best model, based on the backbone topology vii plus three reticulation events (Fig. 3), received an average of 600 out of 1,000 votes across the autosomes and a posterior probability of 0.68 (SI Appendix, Table S10). Model vii thus represents the species graph, our working hypothesis for the true species tree of the AFC and the reticulation events that punctuated its history. According to this hypothesis, An. funestus-like and An. funestus are sister taxa, as are An. vaneedeni and An. longipalpis C. An. parensis is sister to the latter clade. The backbone topology (vii in Fig. 2A) is represented by only ∼4% of the genome because the treelike history has been almost entirely overwritten by multiple introgression events involving multiple pairs of nonsister taxa.
Recent Introgression into An. funestus Preceded Its Continent-Wide Range Expansion.
To derive estimates of the timing of lineage splitting and introgression events, we used ABC with simulations under model vii (SI Appendix, Text S9). Estimates are summarized in generation times (SI Appendix, Table S11) and years (Fig. 1D), the latter assuming a mutation rate of 2.8 × 10−9 (39) and 11 generations per year. Under these assumptions, the initial radiation of the AFC—the divergence of the ([Van + Lon]Par) clade from the (Fun + Lik) clade—occurred ∼216 Kya (95% CI, 213–222 Kya). Next was the split between An. parensis and the (Van + Lon) clade at ∼95 Kya (91–100 Kya). There followed two temporally indistinguishable introgression events (A and B in Fig. 1D) dating to ∼78 Kya (68–87 Kya), both involving the ancestor of the (Fun + Lik) clade. Events A and B (Fig. 1D) featured gene flow from that ancestor into An. parensis and the ancestor of the (Van + Lon) clade, respectively. These introgression events closely preceded or overlapped the splitting of this lineage into An. vaneedeni and An. longipalpis C at ∼69 Kya (62–79 Kya). The most recent species split, leading to the sister taxa An. funestus-like and An. funestus, occurred only ∼38 Kya (31–45 Kya).
We estimate that the third introgression event from An. parensis into An. funestus (labeled “C” in Fig. 1D), occurred considerably more recently, only ∼13 Kya (12–15 Kya). Two lines of evidence suggest that this introgression preceded the range expansion of An. funestus into its current continent-wide distribution across tropical Africa. First, we sequenced An. funestus genomes sampled from six geographic localities spanning West, East, and Southern Africa (SI Appendix, Tables S1 and S4). The median divergence time among An. funestus populations inferred from two different approaches was ∼1.18 Kya (0.29–7.00 Kya) and ∼0.945 Kya (0.019–2.2 Kya) (SI Appendix, Text S5 and Table S5), estimates that are not significantly different (Wilcoxon rank-sum test, P value = 0.47). Both dates are substantially younger than the estimated introgression from An. parensis into An. funestus. Second, we verified that each An. funestus population was equally distant from An. parensis using pairwise genetic distance (dXY; SI Appendix, Text S9). Pairwise distances were highly similar between each An. funestus population and An. parensis (dXY average 0.026, standard deviation 0.0004), whether or not population samples came from localities where the species potentially co-occur, consistent with historical rather than contemporary localized gene flow.
Introgression Involved Substantial Fractions of Autosomes and the X Chromosome.
We next analyzed the distribution and directionality of introgression along the genome between pairs of AFC species using a supervised machine learning framework in the software package FILET, developed for this purpose (40). An extra trees classifier was trained on data simulated under our model of AFC evolutionary history to identify 10 kb windows along the genome with a high probability of introgression (SI Appendix, Text S10). We tested all 10 pairwise combinations of species, not only the pairs implicated by D-statistics, as D-statistics cannot detect gene flow between sister lineages and may also lack power to detect minor gene flow events. Moreover, because gene flow was inferred between the lineages leading to An. funestus and An. parensis at two separate time periods (Fig. 1D, events A and C), we attempted to distinguish these events by training the classifier on simulated data under two exclusive scenarios, one that allowed migration at event A but barred it at event C and a second under the converse. In all cases, we retained only those windows classified as introgressed with ≥90% probability.
To corroborate our results from FILET, an independent test of introgression based on alternative evidence was also applied (SI Appendix, Text S11). This statistical test (QuIBL) employs the distribution of internal branch lengths of triplet topologies discordant with the species tree to distinguish between ILS and introgression (4).
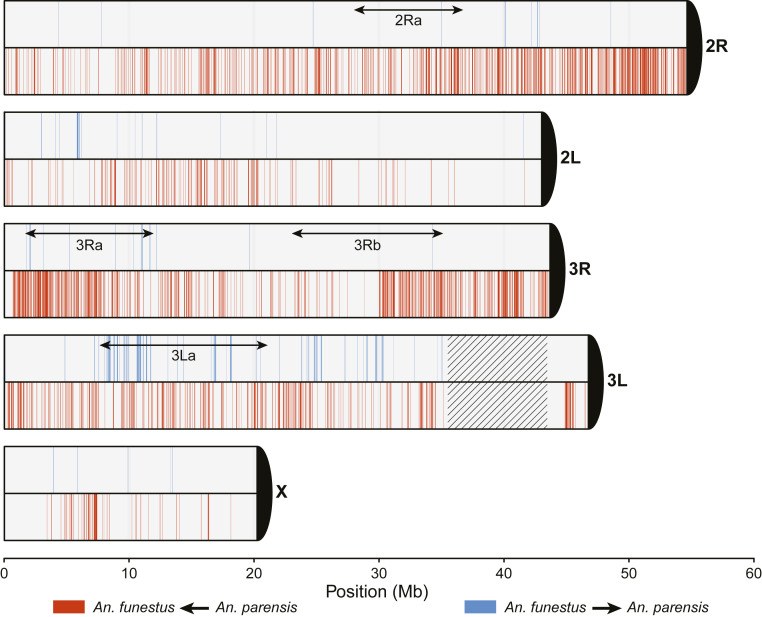
The results from both methods validate our inference of the three introgression events depicted in Fig. 1D (SI Appendix, Tables S13 and S14). Genomic regions predicted to be introgressed were heterogeneously distributed along the genome (Fig. 4 and SI Appendix, Figs. S18–S22). Furthermore, the directionality of gene flow was highly asymmetric (SI Appendix, Table S13). For example, we detected no introgression into An. funestus resulting from events A or B; the majority of introgression from these events was detected in the genomes of An. parensis and An. vaneedeni (at least 11 and 20 Mb, respectively; Fig. 1D and SI Appendix, Table S13). Notably, the most recent introgression event C was strongly biased in the direction of An. funestus, accounting for 31.6 Mb (22.5% of the accessible genome; SI Appendix, Table S13). FILET detected substantial introgression between some species pairs on the X chromosome as well as the autosomes (Fig. 4 and SI Appendix, Table S13). This was partly corroborated by QuIBL, but the power of this test to statistically distinguish ILS from introgression was limited on the X chromosome due to short branches and low counts (SI Appendix, Table S14).
Fig. 4.
Genomic regions of introgression between An. funestus and An. parensis. Windows classified as introgressed between An. funestus and An. parensis with >90% probability are represented on each chromosome arm. Blue indicates introgression from An. funestus into An. parensis; red indicates introgression from An. parensis into An. funestus. Empty areas were classified either with lower confidence or as not introgressed. Approximate locations of common chromosomal inversions in An. funestus (3Ra, 3Rb, 3La, and 2Ra) are indicated by double-headed arrows. Centromeres are represented as black 1/4 circles. Hatching represents a masked region.
We detected a fourth introgression event not uncovered in our earlier inferences of divergence and reticulation based on admixture graphs, as we had limited those analyses to only three reticulations. Both methods of detecting introgression applied here supported this event, which involved substantial X chromosome and autosomal gene flow mainly from An. parensis into An. longipalpis C (SI Appendix, Fig. S22; event D in SI Appendix, Tables S13 and S14). We also confirmed the absence of substantial introgression between sister taxa (events G and H, SI Appendix, Table S13), including An. funestus and An. funestus-like, which is particularly noteworthy in contrast to the prevalence of introgression between nonsister groups.
Discussion
Africa bears >90% of the world’s burden of morbidity and mortality attributable to malaria principally because it is home to the most important Anopheles mosquito vectors. The most obvious attributes shared by these major vectors, which set them apart from their closely related minor or nonvector sibling species, are a high degree of anthropophily, a nearly pan-African species range, and abundant levels of both chromosomal inversion polymorphism and nucleotide diversity. In the historical literature there has been a supposition that these highly anthropophilic malaria vectors should be the most recently radiated members of species complexes, given their dependence on a resource—the human species—that was neither abundant nor widespread until relatively recently. For example, Coluzzi et al. (41) noted that “A. gambiae seems to be the least likely candidate for the ancestral line, as this highly anthropophilic species appears to be the product of a speciation process driven by human impact on the environment subsequent to the Neolithic revolution.” Yet when the elusive species tree for the An. gambiae complex was finally confidently resolved (7), this expectation did not necessarily fit the data: The lineage leading to the two most efficient vectors in the complex was one of the earliest to split (509 Kya according to the most recent estimate; ref. 13) not long after the initial radiation of the complex. Even in the AFC, where our data suggest that the divergence of An. funestus from An. funestus-like was indeed the most recent split, the divergence time of 38 Kya is not consistent with a human-influenced speciation process subsequent to the ∼5 Kya expansion of the Bantu-speaking agriculturalists from Central Africa across sub-Saharan Africa (42). More plausibly, the Bantu expansion could have helped to promote both the demographic and the geographic range expansions of both An. funestus and the primary vectors in the An. gambiae complex (43). What is striking in both species groups is the strongly asymmetric gene flow from nonsister taxa into the lineages that lead to major vectors, species whose invasive and synanthropic phenotypes seem to have emerged following hybridization upon secondary contact. In An. gambiae, because migration included a 21 Mb inversion on chromosome arm 2L (7, 13) that is a known target of spatially varying selection (44), it is likely that at least some of the introgression was adaptive. In the An. gambiae complex introgression of inversions and other variation is thought to have facilitated expansion of the species range, allowed more efficient exploitation of different niches, and led to increased population density and longevity—characteristics of vectorial capacity (41). Evidence for adaptive introgression is lacking in An. funestus thus far, but our data suggest that the expansion of this species into its current geographic range across most of tropical Africa was subsequent to receiving 31.6 Mb of introgressed variation from An. parensis ∼13 Kya. For An. funestus, and maybe malaria vectors or disease vectors more broadly (45), this injection of genetic diversity—which plausibly has greater phenotypic consequences when the donor is a nonsister species—may have facilitated adaptation to new and anthropogenically modified environments, leading to geographic range expansion and enhanced vectorial capacity.
In animals with heteromorphic sex chromosomes, the X chromosome tends to be more resistant to introgression than the autosomes, owing in part to higher densities of incompatibility and local adaptation loci on the X (or Z) chromosome (46–50). Historical introgression in the An. gambiae complex conforms to this pattern as massive gene exchange between one species pair (the ancestor of the An. gambiae clade and An. arabiensis) was mainly autosomal (7). The distal X chromosome—a region distinguished by five overlapping fixed inversion differences between the An. gambiae clade and An. arabiensis—was protected from gene exchange, presumably due to both suppressed recombination conferred by the inversions and selection against introgression of incompatibility loci. These results are consistent with laboratory crossing experiments showing that certain autosomal chromosomal inversions can be introgressed between species and subsequently maintained as stable heterotic polymorphisms, while heterospecific X chromosome inversions are rapidly eliminated (51). In light of this general trend and our previous findings in the An. gambiae complex, we were surprised to find no strong topological discordance nor striking quantitative differences in introgression between the X chromosome and the autosomes in the AFC. One factor that may help account for this difference is that in all AFC species studied here whose karyotypes have been characterized, the X chromosomes are homosequential (16, 23, 24, 52), which should allow for greater recombination on the X chromosome relative to chromosomes that differ by fixed inversions. The absence of fixed inversion differences on the X chromosome between AFC species is suggestive. In the An. gambiae complex, extensively sympatric species differ by fixed inversions on the X chromosome, while species with nonoverlapping distributions and possible vicariant origins, harbor homosequential X chromosomes (53).
Speculation about the role of geography in species divergence is difficult even in the An. gambiae complex, but much more so in the AFC where almost all foundational knowledge about historical biogeography, current species distributions, and bionomics is absent or scant and outdated. With this important caveat, we present a working hypothesis consistent with historical climatic patterns in Africa at the time of the AFC radiation and the known biology of AFC species. During the Middle and Late Pleistocene, the climate of Africa featured repeated oscillations in temperature and rainfall linked to glacial–interglacial cycles (54). The climatic shifts between humid-warm phases (pluvials) and arid-cool phases (interpluvials) were especially intense between 115 and 90 Kya, resulting in megadroughts (55, 56) that repeatedly expanded and contracted Africa’s biomes and initiated population vicariance events (57). Taking into account the shared preference of AFC species for breeding among vegetation at the edges of lakes or slow-moving streams (21, 22, 58)—habitats already quite patchily distributed even in a mesic climate—we suppose that speciation in the AFC was allopatric and driven by arid interpluvials, but that alternating episodes of mesic pluvials could have facilitated secondary contact and hybridization and contributed to the long-term persistence of variation due to admixture (59).
In contrast to sex chromosomes, mtDNA commonly crosses species boundaries even in the absence of detectable nuclear introgression (60–62). Given this tendency, it is not surprising that we found evidence consistent with mtDNA introgression in the AFC. Although other explanations are possible and not mutually exclusive, introgression probably contributes to mtDNA paraphyly (Fig. 1B), and mtDNA capture most likely explains the coexistence of distant mtDNA lineages within the same species, observed both in An. funestus and An. funestus-like (shaded boxes 1 and 2, Fig. 1B). In the absence of alternative genomic resources, it has been common among vector biologists to employ mtDNA to make inferences about anopheline population structure, phylogeography, and even interspecific species relationships. Recently, the complete mtDNA genomes of 43 mosquitoes morphologically identified as An. funestus were sequenced and assembled from three localities in Southern and Central Africa (63). Bayesian phylogenetic reconstruction of these sequences revealed two deeply diverged lineages, coexisting in two of the sampling locations. The authors interpreted their findings in terms of intraspecies genetic relationships and population differentiation (63), but a reanalysis of these data together with our own reveals that the lineages described in the former study are representative of interspecific mtDNA divergences in the AFC (SI Appendix, Text S12 and Fig. S23). The knowledge that there has been extensive introgression between species in the An. gambiae complex (7) and now the AFC cautions against exclusive use of mtDNA to infer intraspecies or even interspecific relationships in closely related anopheline mosquitoes.
As African countries progress along the road toward malaria elimination, there is a growing recognition that control of the major vector species did not interrupt local transmission but instead uncovered persistent “residual” malaria transmitted by lesser-known outdoor biting species (64, 65). By itself, this situation emphasizes the importance of expanding the research emphasis to lesser vectors. Our study provides further impetus for broadening the focus, showing that the evolutionary history of a major vector species in an understudied species complex has been strongly impacted by introgression from minor and nonvector species with major consequences for malaria transmission. The time is now ripe to pivot from asking “What makes the world’s primary malaria vectors so good?” (sensu, ref. 66) to asking “What makes the difference between a good and a bad vector?” from a genomic and evolutionary perspective (67). High-quality reference genomes for all members of malaria vector species complexes, not only the primary vectors, is a tractable first step in that direction. Here we leveraged the recently upgraded An. funestus genome assembly AfunF3 (28) to generate de novo reference assemblies from species inside and outside the group that lacked these genomic resources and performed additional genome sequencing. Beyond our immediate results, these resources will support much needed future studies of the AFC. New advances in sequencing technologies (e.g., ref. 68) will lead to improved assemblies that may allow further insights into the distribution of introgression blocks along the genome and the identification of adaptive introgression. Whether introgression has played a wider role in the origin of other dominant malaria vectors beyond An. gambiae and An. funestus remains to be investigated. Our current hypothesis concerning the species branching order and reticulations in the AFC satisfactorily explains conflicts in previous mitochondrial phylogenies and provides a testable framework to underpin a deeper understanding of the origin of vectorial capacity in the AFC.
Materials and Methods
Please see the SI Appendix for detailed information about: 1) sample information; 2) de novo genome assembly; 3) mitochondrial genome assembly; 4) whole genome alignments; 5) population genomics and variant calling; 6) phylogenetic reconstruction; 7) species networks using D-statistics and admixture graphs; 8) model selection of introgression hypotheses using random forests; 9) estimating introgression and divergence timing using ABC; 10) identifying genomic regions of introgression by machine learning; 11) detecting introgression using branch lengths; 12) introgression and inference from mtDNA.
Supplementary Material
Acknowledgments
We thank D. Schrider for help with the FILET analyses of introgression and M. Kern for assistance with DNA extraction. This work was supported by NIH Grant R21 AI123491 (to N.J.B. and D.E.N.), the NSF Grant DEB-1936187 (to M.W.H.), and the Adaptive Life Program (GELIFES) of the University of Groningen (to M.C.F.). N.J.B., S.T.S., and F.L. also were supported by the Bill & Melinda Gates Foundation OPP1141988 Target Malaria.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018142117/-/DCSupplemental.
Data Availability.
DNA sequence data have been deposited in GenBank, https://www.ncbi.nlm.nih.gov/genbank. Genome assemblies and sequence reads have been submitted to the National Center for Biotechnology Information, NIH, https://www.ncbi.nlm.nih.gov/ (accession nos. PRJNA646526 and PRJNA531511). The mitochondrial genome alignments have been submitted as a pop set for each species (An. funestus [MT917167–MT917182], An. funestus-like [MT917158–MT917162], An. vaneedeni [MT917128–MT917137], An. longipalpis C [MT917148–MT917157], An. parensis [MT917138–MT917147], An. rivulorum [MT917163–MT917164], An. species A [MT917165–MT917166]). VCF files of field-collected individual samples (DOI: 10.6084/m9.figshare.13017518), phylogenetic trees (DOI: 10.6084/m9.figshare.13017488), and feature vectors (DOI: 10.6084/m9.figshare.13017506) are available on https://figshare.com/. The complete models (with all of the demographic data), distributions of both rho and pi (in text and graphic formats), the observed statistics and all custom python scripts, including those used to run the ABC and ABCrf analyses conducted in this study are available at GitHub, https://github.com/stsmall/abc_scripts/.
References
- 1.Mallet J., Besansky N., Hahn M. W., How reticulated are species? BioEssays 38, 140–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarzer J., et al. , Repeated trans-watershed hybridization among haplochromine cichlids (Cichlidae) was triggered by Neogene landscape evolution. Proc. Biol. Sci. 279, 4389–4398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seehausen O., Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Edelman N. B., et al. , Genomic architecture and introgression shape a butterfly radiation. Science 366, 594–599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meier J. I., et al. , Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8, 14363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamichhaney S., et al. , Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518, 371–375 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Fontaine M. C., et al. , Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347, 1258524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seehausen O., Conditions when hybridization might predispose populations for adaptive radiation. J. Evol. Biol. 26, 279–281 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Enciso-Romero J., et al. , Evolution of novel mimicry rings facilitated by adaptive introgression in tropical butterflies. Mol. Ecol. 26, 5160–5172 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Grant P. R., Grant B. R., 40 Years of Evolution: Darwin’s Finches on Daphne Major Island (Princeton University Press, Princeton, NJ, 2014). [Google Scholar]
- 11.Harbach R. E., “The phylogeny and classification of Anopheles” in Anopheles Mosquitoes–New Insights into Malaria Vectors, Manguin S., Ed. (InTech, 2013), pp. 55. [Google Scholar]
- 12.Manguin S., et al. , Biodiversity of Malaria in the World (John Libbey Eurotext, Paris, 2008), pp. 427. [Google Scholar]
- 13.Thawornwattana Y., Dalquen D., Yang Z., Coalescent analysis of phylogenomic data confidently resolves the species relationships in the Anopheles gambiae species complex. Mol. Biol. Evol. 35, 2512–2527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White B. J., Collins F. H., Besansky N. J., Evolution of Anopheles gambiae in relation to humans and malaria. Annu. Rev. Ecol. Evol. Syst. 42, 111–132 (2011). [Google Scholar]
- 15.Koekemoer L. L., et al. , Cryptic species within Anopheles longipalpis from southern Africa and phylogenetic comparison with members of the An. funestus group. Bull. Entomol. Res. 99, 41–49 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spillings B. L., et al. , A new species concealed by Anopheles funestus Giles, a major malaria vector in Africa. Am. J. Trop. Med. Hyg. 81, 510–515 (2009). [PubMed] [Google Scholar]
- 17.Harbach R. E., Subgenus cellia classification. Mosquito Taxonomic Inventory, mosquito-taxonomic-inventory.info/. Accessed 28 February 2020.
- 18.Garros C., Harbach R. E., Manguin S., Morphological assessment and molecular phylogenetics of the Funestus and Minimus groups of Anopheles (Cellia). J. Med. Entomol. 42, 522–536 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Coetzee M., Fontenille D., Advances in the study of Anopheles funestus, a major vector of malaria in Africa. Insect Biochem. Mol. Biol. 34, 599–605 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Coetzee M., Koekemoer L. L., Molecular systematics and insecticide resistance in the major African malaria vector Anopheles funestus. Annu. Rev. Entomol. 58, 393–412 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Dia I., Guelbeogo M. W., Ayala D., “Advances and perspectives in the study of the malaria mosquito Anopheles funestus” in Anopheles Mosquitoes–New Insights into Malaria Vectors, Manguin S., Ed. (InTech, 2013), chap. 7. [Google Scholar]
- 22.Gillies M. T., De Meillon B., The Anophelinae of Africa South of the Sahara (South African Institute for Medical Research, Johannesburg, ed. 2, 1968). [Google Scholar]
- 23.Green C. A., Cladistic analysis of mosquito chromosome data (Anopheles (Cellia) Myzomyia. J. Hered. 73, 2–11 (1982). [PubMed] [Google Scholar]
- 24.Green C. A., Hunt R. H., Interpretation of variation in ovarian polytene chromosomes of Anopheles funestus Giles, A. parensis Gillies, and A. aruni? Genetica 51, 187–195 (1980). [Google Scholar]
- 25.Sharakhov I. V., Sharakhova M. V., Mbogo C. M., Koekemoer L. L., Yan G., Linear and spatial organization of polytene chromosomes of the African malaria mosquito Anopheles funestus. Genetics 159, 211–218 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michel A. P., et al. , Effective population size of Anopheles funestus chromosomal forms in Burkina Faso. Malar. J. 5, 115 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michel A. P., et al. , Rangewide population genetic structure of the African malaria vector Anopheles funestus. Mol. Ecol. 14, 4235–4248 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Ghurye J., et al. , A chromosome-scale assembly of the major African malaria vector Anopheles funestus. Gigascience 8, giz063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevenson J., et al. , Novel vectors of malaria parasites in the western highlands of Kenya. Emerg. Infect. Dis. 18, 1547–1549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobo N. F., et al. , Unexpected diversity of Anopheles species in Eastern Zambia: Implications for evaluating vector behavior and interventions using molecular tools. Sci. Rep. 5, 17952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi K. S., Koekemoer L. L., Coetzee M., Population genetic structure of the major malaria vector Anopheles funestus s.s. and allied species in southern Africa. Parasit. Vectors 5, 283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koekemoer L. L., Kamau L., Hunt R. H., Coetzee M., A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am. J. Trop. Med. Hyg. 66, 804–811 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Choi K. S., Coetzee M., Koekemoer L. L., Simultaneous identification of the Anopheles funestus group and Anopheles longipalpis type C by PCR-RFLP. Malar. J. 9, 316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stage D. E., Eickbush T. H., Sequence variation within the rRNA gene loci of 12 Drosophila species. Genome Res. 17, 1888–1897 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green R. E., et al. , A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson N., et al. , Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hahn M. W., Molecular Population Genetics (Sinauer Associates/Oxford University Press, 2018). [Google Scholar]
- 38.Pudlo P., et al. , Reliable ABC model choice via random forests. Bioinformatics 32, 859–866 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Keightley P. D., Ness R. W., Halligan D. L., Haddrill P. R., Estimation of the spontaneous mutation rate per nucleotide site in a Drosophila melanogaster full-sib family. Genetics 196, 313–320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrider D. R., Ayroles J., Matute D. R., Kern A. D., Supervised machine learning reveals introgressed loci in the genomes of Drosophila simulans and D. sechellia. PLoS Genet. 14, e1007341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coluzzi M., Sabatini A., della Torre A., Di Deco M. A., Petrarca V., A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298, 1415–1418 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Li S., Schlebusch C., Jakobsson M., Genetic variation reveals large-scale population expansion and migration during the expansion of Bantu-speaking peoples. Proc. Biol. Sci. 281, 20141448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miles A. et al.; Anopheles gambiae 1000 Genomes Consortium; Data analysis group; Partner working group; Sample collections—Angola; Burkina Faso; Cameroon; Gabon; Guinea; Guinea-Bissau; Kenya; Uganda; Crosses; Sequencing and data production; Web application development; Project coordination , Genetic diversity of the African malaria vector Anopheles gambiae. Nature 552, 96–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng C., et al. , Ecological genomics of Anopheles gambiae along a latitudinal cline: A population-resequencing approach. Genetics 190, 1417–1432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett K. L., et al. , Historical environmental change in Africa drives divergence and admixture of Aedes aegypti mosquitoes: A precursor to successful worldwide colonization? Mol. Ecol. 25, 4337–4354 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Ellegren H., et al. , The genomic landscape of species divergence in Ficedula flycatchers. Nature 491, 756–760 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Garrigan D., et al. , Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22, 1499–1511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janoušek V., et al. , Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol. Ecol. 21, 3032–3047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin S. H., Davey J. W., Salazar C., Jiggins C. D., Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 17, e2006288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muirhead C. A., Presgraves D. C., Hybrid incompatibilities, local adaptation, and the genomic distribution of natural Introgression between species. Am. Nat. 187, 249–261 (2016). [DOI] [PubMed] [Google Scholar]
- 51.della Torre A., Merzagora L., Powell J. R., Coluzzi M., Selective introgression of paracentric inversions between two sibling species of the Anopheles gambiae complex. Genetics 146, 239–244 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamau L., Koekemoer L. L., Hunt R. H., Coetzee M., Anopheles parensis: The main member of the Anopheles funestus species group found resting inside human dwellings in Mwea area of central Kenya toward the end of the rainy season. J. Am. Mosq. Control Assoc. 19, 130–133 (2003). [PubMed] [Google Scholar]
- 53.Ayala F. J., Coluzzi M., Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 102 (suppl. 1), 6535–6542 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoag C., Svenning J.-C., African environmental change from the Pleistocene to the Anthropocene. Annu. Rev. Environ. Resour. 42, 27–54 (2017). [Google Scholar]
- 55.Cohen A. S., et al. , Ecological consequences of early Late Pleistocene megadroughts in tropical Africa. Proc. Natl. Acad. Sci. U.S.A. 104, 16422–16427 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scholz C. A., et al. , East African megadroughts between 135 and 75 thousand years ago and bearing on early-modern human origins. Proc. Natl. Acad. Sci. U.S.A. 104, 16416–16421 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lorenzen E. D., Heller R., Siegismund H. R., Comparative phylogeography of African savannah ungulates. Mol. Ecol. 21, 3656–3670 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Evans A. M., Leeson H. S., The funestus series of Anopheles in southern Rhodesia, with description of a new variety. Ann. Trop. Med. Parasitol. 29, 33–47 (1935). [Google Scholar]
- 59.Kagawa K., Seehausen O., The propagation of admixture-derived adaptive radiation potential. Proc. Biol. Sci. 287, 20200941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Good J. M., Vanderpool D., Keeble S., Bi K., Negligible nuclear introgression despite complete mitochondrial capture between two species of chipmunks. Evolution 69, 1961–1972 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Toews D. P., Brelsford A., The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 21, 3907–3930 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Bachtrog D., Thornton K., Clark A., Andolfatto P., Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution 60, 292–302 (2006). [PubMed] [Google Scholar]
- 63.Jones C. M., et al. , Complete Anopheles funestus mitogenomes reveal an ancient history of mitochondrial lineages and their distribution in southern and central Africa. Sci. Rep. 8, 9054 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burke A., et al. , Anopheles parensis contributes to residual malaria transmission in South Africa. Malar. J. 18, 257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burke A., et al. , A new malaria vector mosquito in South Africa. Sci. Rep. 7, 43779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohuet A., Harris C., Robert V., Fontenille D., Evolutionary forces on Anopheles: What makes a malaria vector? Trends Parasitol. 26, 130–136 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Ruzzante L., Reijnders M. J. M. F., Waterhouse R. M., Of genes and genomes: Mosquito evolution and diversity. Trends Parasitol. 35, 32–51 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Kingan S. B., et al. , A high-quality de novo genome assembly from a single mosquito using PacBio sequencing. Genes (Basel) 10, 62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequence data have been deposited in GenBank, https://www.ncbi.nlm.nih.gov/genbank. Genome assemblies and sequence reads have been submitted to the National Center for Biotechnology Information, NIH, https://www.ncbi.nlm.nih.gov/ (accession nos. PRJNA646526 and PRJNA531511). The mitochondrial genome alignments have been submitted as a pop set for each species (An. funestus [MT917167–MT917182], An. funestus-like [MT917158–MT917162], An. vaneedeni [MT917128–MT917137], An. longipalpis C [MT917148–MT917157], An. parensis [MT917138–MT917147], An. rivulorum [MT917163–MT917164], An. species A [MT917165–MT917166]). VCF files of field-collected individual samples (DOI: 10.6084/m9.figshare.13017518), phylogenetic trees (DOI: 10.6084/m9.figshare.13017488), and feature vectors (DOI: 10.6084/m9.figshare.13017506) are available on https://figshare.com/. The complete models (with all of the demographic data), distributions of both rho and pi (in text and graphic formats), the observed statistics and all custom python scripts, including those used to run the ABC and ABCrf analyses conducted in this study are available at GitHub, https://github.com/stsmall/abc_scripts/.