Significance
Guinea yam is an important staple tuber crop in West Africa, where it contributes to the sustenance and sociocultural lives of millions of people. Understanding the genetic diversity of Guinea yam and its relationships with wild relatives is important for improving this important crop using genomic information. A recent genomics study proposed that Guinea yam originated from a wild relative, the rainforest species Dioscorea praehensilis. Our results based on sequencing of 336 Guinea yam accessions do not support this notion; rather, our results indicate a hybrid origin of Dioscorea rotundata from crosses between the savannah species Dioscorea abyssinica and D. praehensilis.
Keywords: domestication, Guinea yam, hybrid, population genomics, wild progenitors
Abstract
White Guinea yam (Dioscorea rotundata) is an important staple tuber crop in West Africa. However, its origin remains unclear. In this study, we resequenced 336 accessions of white Guinea yam and compared them with the sequences of wild Dioscorea species using an improved reference genome sequence of D. rotundata. In contrast to a previous study suggesting that D. rotundata originated from a subgroup of Dioscorea praehensilis, our results suggest a hybrid origin of white Guinea yam from crosses between the wild rainforest species D. praehensilis and the savannah-adapted species Dioscorea abyssinica. We identified a greater genomic contribution from D. abyssinica in the sex chromosome of Guinea yam and extensive introgression around the SWEETIE gene. Our findings point to a complex domestication scenario for Guinea yam and highlight the importance of wild species as gene donors for improving this crop through molecular breeding.
Yams (Dioscorea spp.) are major starchy tuber crops that are widely consumed in the tropics. Ten yam species are cultivated worldwide, including Dioscorea alata in Southeast Asia, Dioscorea trifida in South America, and Dioscorea rotundata in West and Central Africa (1). D. rotundata, also known as white Guinea yam, is the most important species in West and Central Africa, an area accounting for 92.5% of global yam production in 2018 (http://www.fao.org/statistics). Beyond its nutritional and food value, Guinea yam is also important for the culture of West African people (2).
Despite the considerable importance of Guinea yam, its origin has been elusive. There are two types of Guinea yam: white Guinea yam (D. rotundata) and yellow Guinea yam (Dioscorea cayenensis). D. cayenensis is thought to be a triploid species of hybrid origin, with D. rotundata as the maternal parent and Dioscorea burkilliana as the paternal parent (3, 4). In turn, the triploid D. rotundata is thought to be a hybrid between D. rotundata and Dioscorea togoensis (4). However, the origin of diploid D. rotundata, which accounts for the majority of Guinea yam production (4), has been ambiguous. Two wild species are candidate progenitors of diploid D. rotundata: the savannah-adapted wild species Dioscorea abyssinica and the rainforest-adapted wild species Dioscorea praehensilis (3–10). The geographical distributions of D. abyssinica and D. praehensilis overlap slightly (SI Appendix, Fig. S1). Based on morphological evaluation, Coursey proposed that D. rotundata might be a hybrid between the two species (8). However, other reports have indicated that the origin of Guinea yam is ambiguous due to the small number of markers (3–7), introgression (6, 7), or incomplete lineage sorting (7).
The whole-genome sequence of Guinea yam has been reported (11). A recent genome study involving 86 D. rotundata, 47 D. praehensilis, and 34 D. abyssinica accessions suggested that diploid D. rotundata was domesticated from D. praehensilis (10). Here we addressed this hypothesis using an expanded set of genomes from cultivated and wild Dioscorea species.
In this study, we generated an improved version of the Guinea yam reference genome and used it to analyze the genomes of 336 accessions of D. rotundata and its wild relatives. Based on these analyses, we attempted to reveal the history of Guinea yam domestication. Our results suggest that diploid D. rotundata was most likely derived from homoploid hybridization between D. abyssinica and D. praehensilis. By evaluating the genomic contributions of each parental species to D. rotundata, we revealed greater representation of the D. abyssinica genome in the sex chromosome of D. rotundata and a signature of extensive introgression in the SWEETIE gene on chromosome 17.
Genetic Diversity of Guinea Yam
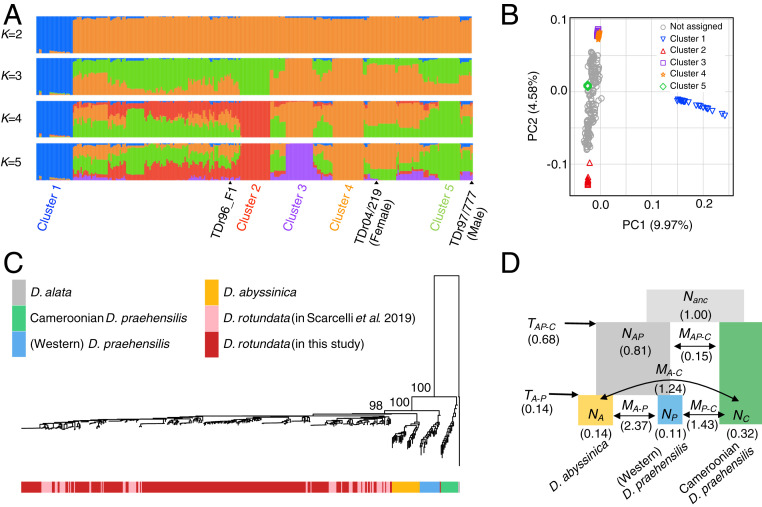
We obtained DNA samples from 336 accessions of D. rotundata maintained at the International Institute of Tropical Agriculture (IITA) in Nigeria, representing the genetic diversity of Guinea yam landraces and improved lines from West Africa. We subjected these samples to whole-genome resequencing on the Illumina sequencing platform. We aligned the resulting short reads to the newly assembled reference genome (SI Appendix, sections S1 and S2) and extracted single nucleotide polymorphism (SNP) information for use in genetic diversity studies (SI Appendix, Table S1 and section S3). Based on admixture analysis with the sNMF program (12), we defined five major clusters (Fig. 1A). When K = 2, cluster 1 was clearly separated from the other accessions. Principal component analysis (PCA) also separated cluster 1 from the rest of the clusters (Fig. 1B). Accessions in cluster 1 had significantly higher heterozygosity and ∼10-fold more unique alleles than those in the four remaining clusters (SI Appendix, Figs. S2 and S3 and Table S2). Because flow cytometry analysis confirmed that all 10 accessions analyzed in cluster 1 were triploids (Dataset S1), we hypothesized that cluster 1 represents triploid D. rotundata, a hybrid of D. rotundata and D. togoensis (4). After removing the cluster 1 accessions, the nucleotide diversity of D. rotundata was estimated as 14.83 × 10−4 (SI Appendix, Table S3), which is ∼1.5-fold larger than that reported previously (10), presumably because we used a larger number of samples with diverse genetic backgrounds in our study. Linkage disequilibrium of diploid D. rotundata showed a decay of r2 = 0.13 in a 200-kb genomic region (SI Appendix, Fig. S4), which is slower than that of cassava, another clonally propagated crop (13).
Fig. 1.
Genetic diversity and phylogenomics of Guinea yam and its wild relatives. (A) Ancestry proportions of each Guinea yam accession with 6,124,093 SNPs. TDr96_F1 is the sample used as the reference genome. (B) PCA result of the 336 Guinea yam accessions. (C) NJ tree of four African yam lineages reconstructed using D. alata as an outgroup based on 463,293 SNPs. The numbers indicate bootstrap values after 100 replications. The sequences of D. rotundata in the previous study (10) were included in the tree. (D) Evolutionary relationship of three African wild yam lineages (D. abyssinica, Western D. praehensilis, and Cameroonian D. praehensilis) as inferred by ∂a∂i (15) using 17,532 SNPs. N, M, and T represent the relative population size from Nanc, migration rate, and divergence time, respectively.
Phylogenomic Analysis of African Yam
Using the SNP information, we constructed a rooted neighbor-joining (NJ) tree (14) based on 308 Guinea yam accessions sequenced in the present study (excluding cluster 1 triploid accessions), as well as 80 D. rotundata, 29 D. abyssinica, 21 Western D. praehensilis, and 18 Cameroonian D. praehensilis accessions that were sequenced in a previous study (10) using two accessions of Asian species D. alata as an outgroup (Fig. 1C). Throughout the analyses described below, we used 388 D. rotundata accessions by combining our samples and those used previously (10). According to this NJ tree, the D. rotundata accessions sequenced in this study are genetically close to the D. rotundata accessions reported previously (10) (Fig. 1C). However, the NJ tree showed that D. rotundata is more closely related to D. abyssinica than to Western D. praehensilis (Fig. 1C), which is inconsistent with a previous finding (10) that D. rotundata is most closely related to Western D. praehensilis.
To elucidate the evolutionary relationships of the three wild Dioscorea species that are closely related to D. rotundata—D. abyssinica (designated as A), Western D. praehensilis (P), and Cameroonian D. praehensilis (C)—we performed diffusion approximations for demographic inference (∂a∂i) analysis (15), which allows for estimation of demographic parameters based on an unfolded site frequency spectrum. First, we tested three phylogenetic models—{{A, P}, C}, {{P, C}, A}, and {{C, A}, P}—using 17,532 SNPs that were polarized using D. alata as an outgroup without considering migration among the species. Of the three models, {{A, P}, C} had the highest likelihood (SI Appendix, Table S4).
This result is not consistent with the previous finding that {{P, C}, A} had the highest likelihood (10), as determined using a different method with fastsimcoal2 software (16). To exactly repeat the previous analysis, we tested these three models with fastsimcoal2 (16) using the previous reference genome (11), which indicated that {{A, P}, C} had the highest likelihood (SI Appendix, Table S5). Taken together, our results are inconsistent with the previous report (10) but are consistent with the PCA result from the same report, which separated Cameroonian D. praehensilis from the other African yams in PC1 (figure 2A of ref. 10).
Based on the assumption that {{A, P}, C} describes the true evolutionary relationship among the three wild Dioscorea species, we reestimated the evolutionary parameters with ∂a∂i, allowing symmetric migration (gene flow) among the species (Fig. 1D). Since the results indicated that Cameroonian D. praehensilis is distantly related to D. rotundata and was not likely involved in genetic exchange with D. rotundata (Fig. 1C), we focused on Western D. praehensilis, which we refer to as D. praehensilis hereinafter for brevity.
Hybrid Origin of Guinea Yam
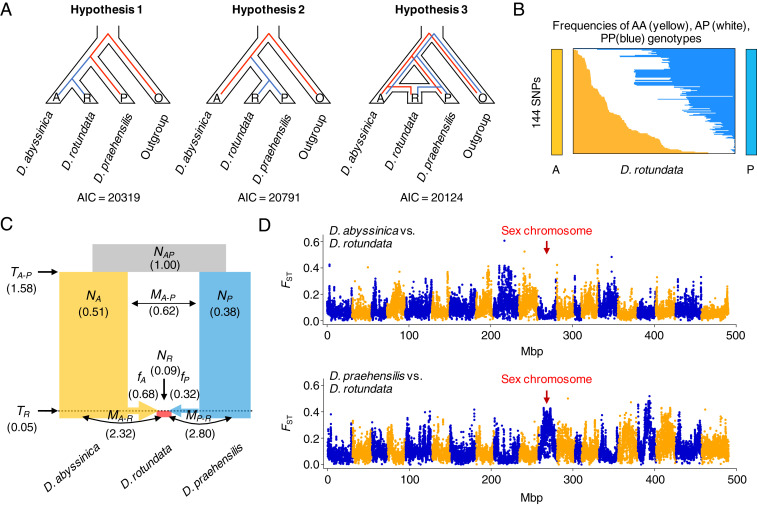
We propose three hypotheses for the origin of Guinea yam (D. rotundata) based on the NJ tree (Fig. 1C) and ∂a∂i (15) (Fig. 1D). The first hypothesis is that D. rotundata was derived from D. abyssinica (hypothesis 1 in Fig. 2A); the second is that D. rotundata was derived from D. praehensilis (hypothesis 2 in Fig. 2A). However, in hypotheses 1 and 2, the divergence time of D. rotundata from the wild species might not be sufficient to separate the three lineages, and there may be incomplete lineage sorting among the species. The third hypothesis is that D. rotundata originated as an admixture between D. abyssinica and D. praehensilis (hypothesis 3 in Fig. 2A).
Fig. 2.
Evidence for the hybrid origin of Guinea yam. (A) Hypotheses for the domestication of Guinea yam (D. rotundata). Hypothesis 1 assumes that D. rotundata diverged from D. abyssinica. Hypothesis 2 assumes that D. rotundata diverged from D. praehensilis. Hypothesis 3 assumes that D. rotundata was derived from a hybrid between D. abyssinica and D. praehensilis. D. alata served as an outgroup. (B) Frequencies of individuals homozygous for D. abyssinica allele (A, in yellow), homozygous for D. praehensilis allele (P, in blue), and heterozygous for A and P (in white) among the 388 D. rotundata sequences as studied for 144 SNPs. (C) Evolutionary parameters related to the hybrid origin of Guinea yam as inferred by ∂a∂i (15) using 15,461 SNPs. N, M, and T represent the relative population size from NAP, migration rate, and divergence time, respectively. fA and fp indicate the genomic contributions from D. abyssinica and D. praehensilis when the hybridization occurred, respectively. (D) FST between the wild and cultivated yams. This was conducted with a 100-kb window and a 20-kb step. Chromosome 11 of D. rotundata containing the sex-determining locus shows a shorter distance to that of D. abyssinica and a longer distance to that of D. praehensilis.
Before estimating the evolutionary parameters for the three hypotheses, we studied the allele frequencies of the 388 D. rotundata sequences, focusing on 144 SNPs that are positioned over the entire genome and are oppositely fixed in the two candidate progenitors (Fig. 2B and SI Appendix, Fig. S5). If hypothesis 1 or 2 is correct, then the allele frequencies in these 144 SNPs should be highly skewed to either of the progenitors. The patterns of allele contributions from the two candidate species to D. rotundata were nearly identical, however. This result suggests that hypothesis 3, the admixture origin of Guinea yam, is most likely correct.
We tested the three hypotheses by ∂a∂i (15) with symmetric migration (gene flow) rates using 15,461 SNPs polarized by D. alata (Fig. 2A), which showed that hypothesis 3 had the highest likelihood and the lowest Akaike information criterion value (Fig. 2C and SI Appendix, Table S4). This result supports the admixture hypothesis, that D. rotundata was derived from crosses between D. abyssinica and D. praehensilis. The parameters estimated by ∂a∂i indicate that the hybridization between D. abyssinica and D. praehensilis was relatively recent in relation to the divergence between the two wild species. This analysis also indicated that the genomic contributions from D. abyssinica and D. praehensilis during the hybridization period were ∼68% and 32%, respectively. Introgression generally results in highly asymmetric genomic contributions from the parental species, whereas hybridization shows symmetric genomic contributions (17). The intermediate genomic contributions revealed by this analysis support the hybridization hypothesis rather than the introgression hypothesis. Our findings are in line with the hybrid origin of the Guinea yam proposed by Coursey in 1976 based on morphology (8) and supports his speculation that spontaneous hybridization between wild yams could have occurred at the artifactual “dump heaps” created by people living in the savannah between the forest and the Sahara (9).
To evaluate the genetic distances of D. rotundata from the two parental species for each chromosome, we calculated fixation index (FST) values (18) (Fig. 2D and SI Appendix, Table S6). The genetic distances from the two parents varied across the different chromosomes, and the overall genetic distance of D. rotundata from D. abyssinica was shorter than that from D. praehensilis (SI Appendix, Table S6). Intriguingly, chromosome 11, to which we previously mapped the candidate locus for sex determination (11), had the shortest genetic distance from D. abyssinica and the longest genetic distance from D. praehensilis among all chromosomes, indicating that chromosome 11 of D. rotundata is highly skewed to D. abyssinica (Fig. 2D and SI Appendix, Table S6). Similarly, interspecies divergence is different between the autosomes and sex chromosome of the dioecious plant species Silene (19).
Evolutionary History of Guinea Yam
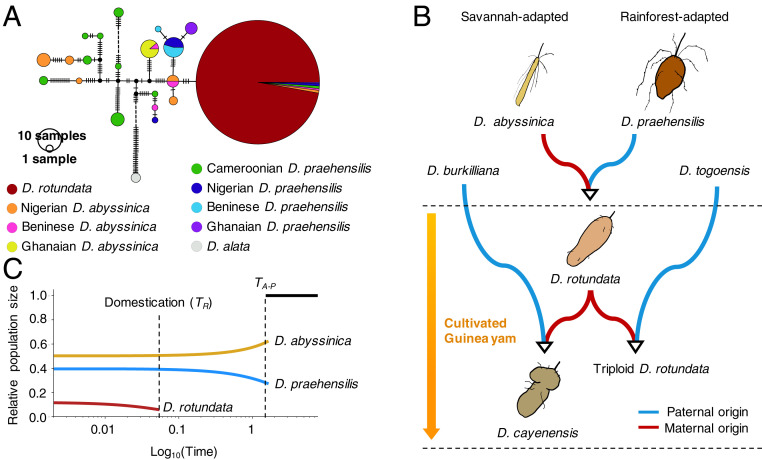
In angiosperms, plastid genomes are predominantly inherited maternally (20), making them useful for studying maternal lineages. To infer the maternal history of Guinea yam, we constructed a haplotype network of the whole plastid genome with all samples used in the NJ tree (Fig. 1C), as well as the triploid accessions in cluster 1 (Fig. 3A and SI Appendix, section S6). According to this haplotype network, Cameroonian D. praehensilis has the longest genetic distance from D. rotundata. This result is in line with the phylogenomic trees of African yam (Fig. 1 C and D). Strikingly, the plastid genomes of diploid and triploid D. rotundata are uniform and very similar to those of Nigerian or Beninese D. abyssinica, although the latter has another plastid genome lineage distant from that of D. rotundata. The plastid genomes of D. praehensilis from Nigeria, Benin, and Ghana appear to be derived from Nigerian or Beninese D. abyssinica. These results indicate that D. abyssinica is an older lineage than D. praehensilis, and that the places of origin of D. rotundata and D. praehensilis are probably around Nigeria or Benin. Based on the whole-genome diversity of D. rotundata, a recent study (10) hypothesized that the origin of D. rotundata was around north Benin, as supported by the current results. The plastid genomes of some wild species are identical to those of cultivated Guinea yams. Hybridization between cultivated yams and wild yams may account for this observation (7).
Fig. 3.
Evolutionary scenario of African yam origins. (A) Haplotype network of the whole plastid genomes of 416 D. rotundata (including the triploid accessions), 68 wild relatives, and two D. alata accessions as the outgroup. The number of vertical dashes represents the number of mutations. Western (Nigerian, Beninese, and Ghanaian) D. praehensilis and D. rotundata seem to have diverged from Nigerian and Beninese D. abyssinica. (B) Possible scenario of domestication of Guinea yam. The blue line represents paternal origin, and the red line represents maternal origin. (C) Changes in population sizes of D. rotundata and its wild relatives as inferred by ∂a∂i (15). The parameters except population size were identical to those used in Fig. 2C. After the domestication of D. rotundata, the population size of D. rotundata has increased with migration from the wild progenitors.
The results of nuclear genome admixture (Fig. 2) and plastid haplotype network (Fig. 3A) analyses indicate that the maternal origin of diploid D. rotundata is D. abyssinica and its paternal origin is D. praehensilis (Fig. 3B). Hybridization between D. abyssinica and D. praehensilis is rare (10), but such rare hybrids appear to have been domesticated by humans. The triploid D. rotundata shares its plastid haplotype with diploid D. rotundata, indicating that diploid D. rotundata served as the maternal parent and D. togoensis was the paternal parent. D. cayenensis is reported to have D. rotundata as the maternal parent and D. burkilliana as the paternal parent (3, 4). All cultivated Guinea yams are hybrids containing D. abyssinica plastid genomes.
To explore the changes in population size, we reinferred the demographic history of African yam by ∂a∂i (15), allowing migration (Fig. 3C and SI Appendix, section S7). We used the same dataset as in Fig. 2C. By fixing the parameters predicted in Fig. 2C except population size, we reestimated each population size at the start and end points after the emergence of these species, assuming an exponential increase/decrease in population size. According to this analysis, since the emergence of the wild progenitors of Guinea yam, the population size of D. abyssinica has been decreasing, while that of D. praehensilis has been increasing (Fig. 3C). This finding suggests that the D. praehensilis population was derived from D. abyssinica, which is consistent with the results of haplotype network analysis (Fig. 3A).
Extensive Introgression at the SWEETIE Locus
To explore multiple introgression to D. rotundata from the two wild species, we analyzed the f4 statistic (21) using four groups: D. rotundata clusters 2 and 5, D. rotundata cluster 4, D. abyssinica, and D. praehensilis (SI Appendix, section S8). The f4 statistic reveals the representation of two alternative discordant genealogies (Fig. 4A). The f4 value is close to zero if the first two groups of D. rotundata show a concordant genealogy in relation to D. abyssinica and D. praehensilis. In contrast, the f4 value diverges from zero if the two groups of D. rotundata exhibit discordant genealogy and a large genetic distance to each other. We obtained the f4 statistic f4 (P25, P4, PP, PA) for each SNP and performed sliding window analysis (Fig. 4B). The f4 value was close to zero across the genome, indicating that overall, we cannot decide between topology 1 and topology 2. However, the genomic regions around the SWEETIE gene showed the lowest f4 (P25, P4, PP, PA) [Z(f4) = −5.66], with overrepresentation of topology 2 in the SWEETIE gene (DRNTG_01731) (SI Appendix, Table S7).
Fig. 4.
Signature of extensive introgression around the SWEETIE gene. (A) Topology of f4 (P25, P4, PP, PA) in clusters 2, 4, and 5 and wild yams. Positive f4 values represent the long internal branch of the upper tree (topology 1), and negative f4 values represent the long internal branch of the bottom tree (topology 2). (B) f4 values across the genome. This was conducted with a 250-kb window and a 25-kb step. Red dots indicate outliers of the sliding window, which have |Z(f4)| > 5. The locus around the SWEETIE gene shows extraordinarily negative f4 values. (C) Neighbor-Net around the SWEETIE gene (4∼4.15 Mb on chromosome 17). This was constructed by SplitsTree (22) using a total of 458 SNPs.
To explore the genealogical relationships around the SWEETIE gene, we constructed a Neighbor-Net (22) around this locus (4.00 to 4.15 Mb on chromosome 17) (Fig. 4C). The Neighbor-Net showed that the locus of cluster 4 was close to that of D. praehensilis, while the loci of clusters 2 and 5 and some other accessions were close to those of D. abyssinica. These results indicate that the SWEETIE gene was introgressed from the wild species more than once. The SWEETIE gene encodes a membrane protein involved in the general control of sugar flux (23). The Arabidopsis thaliana sweetie mutant shows pronounced changes in the accumulation of sugar, starch, and ethylene along with significant changes in growth and development (24). We still do not know the effect of this introgression on the phenotype of Guinea yam, but this locus appears to be a target of selection.
Homoploid Hybrid Formation as the Trigger of Domestication
The importance of hybridization and polyploidization for crop domestication is well documented (25, 26), including in bread wheat (27) and banana (28). Compared with allopolyploidy, only a limited number of homoploid hybridizations have been reported in plants (29), and homoploid hybridizations have rarely contributed to the origin of crops (30). Homoploid hybridization can increase genetic variation via recombination between distantly related species, and it often allows the hybrid to adapt to unexploited niches (31). In the case of Guinea yam, the savannah-adapted wild species D. abyssinica and the rainforest-adapted wild species D. praehensilis are not suitable for agriculture; however, their hybrid, D. rotundata, could have been adopted for cultivation by humans. Gene combinations from different wild yams might have contributed to the domestication of Guinea yam. The present study provides an example of the origin of a crop through homoploid hybridization.
Use of Wild Species to Improve Guinea Yam
A project for the improvement of Guinea yam by crossbreeding has been initiated (AfricaYam: https://africayam.org). However, the current breeding projects depend solely on D. rotundata genetic resources. Systematic efforts are needed to introgress beneficial alleles from wild species into crops; these alleles will increase disease resistance and abiotic stress tolerance to improve crop resiliency and productivity (32). Our study revealed that the two wild progenitor species (D. abyssinica and D. praehensilis) of Guinea yam contain much greater genetic diversity than D. rotundata (Fig. 2C), suggesting that these wild species could be useful sources for alleles of agricultural importance. However, the D. abyssinica and D. praehensilis accessions in IITA GenBank account for only 1.6% of the total Dioscorea accessions maintained as of 2018 (33). Therefore, it will be important to collect and preserve wild Dioscorea species as genetic resources for improving Guinea yam. Our findings suggest that new alleles of loci, such as the SWEETIE gene, were introgressed from wild yams into cultivated Guinea yams multiple times, which likely conferred the plants with phenotypes preferred by humans. Many more alleles from wild species remain to be exploited for systematic breeding. Our findings highlight the need to consider how to effectively leverage the gene pools of wild species from different habitats for the rapid breeding of Guinea yam using genomic information.
Supplementary Material
Acknowledgments
We thank Sophien Kamoun for valuable comments on the manuscript. This study was carried out under the auspices of the AfricaYam Project funded by the Bill and Melinda Gates Foundation and the EDITS-Yam project funded by the Japan International Research Center for Agricultural Sciences.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015830117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and supporting information.
References
- 1.Hancock J. F., “Starch staples and sugar” in Plant Evolution and the Origin of Crop Species (CABI Publishing, ed. 3, 2012), pp. 168–169. [Google Scholar]
- 2.Obidiegwu J. E., Akpabio E. M., The geography of yam cultivation in southern Nigeria: Exploring its social meanings and cultural functions. J. Ethnic. Foods 4, 28–35 (2017). [Google Scholar]
- 3.Terauchi R., Chikaleke V. A., Thottappilly G., Hahn S. K., Origin and phylogeny of Guinea yams as revealed by RFLP analysis of chloroplast DNA and nuclear ribosomal DNA. Theor. Appl. Genet. 83, 743–751 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Girma G., et al. , Next-generation sequencing-based genotyping, cytometry and phenotyping for understanding diversity and evolution of Guinea yams. Theor. Appl. Genet. 127, 1783–1794 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Magwé‐Tindo J., et al. , Guinea yam (Dioscorea spp., Dioscoreaceae) wild relatives identified using whole plastome phylogenetic analyses. Taxon 67, 905–915 (2018). [Google Scholar]
- 6.Scarcelli N., et al. , Farmers’ use of wild relative and sexual reproduction in a vegetatively propagated crop. The case of yam in Benin. Mol. Ecol. 15, 2421–2431 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Scarcelli N., et al. , Crop wild relative conservation: Wild yams are not that wild. Biol. Conserv. 210, 325–333 (2017). [Google Scholar]
- 8.Coursey D. G., “Yams, Dioscorea spp. (Dioscoreaceae)” in Evolution of Crop Plants, Simmonds N. W., Ed. (Longman Group, 1976), pp. 70–74. [Google Scholar]
- 9.Coursey D. G., “The origins and domestication of yams in Africa” in Origins of African Plant Domestication, Harlan J. R., Wet J. M. J. D., Stemler A. B. L., Eds. (De Gruyter Mouton, 1976), pp. 383–408. [Google Scholar]
- 10.Scarcelli N., et al. , Yam genomics supports West Africa as a major cradle of crop domestication. Sci. Adv. 5, eaaw1947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamiru M., et al. , Genome sequencing of the staple food crop white Guinea yam enables the development of a molecular marker for sex determination. BMC Biol. 15, 86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frichot E., Mathieu F., Trouillon T., Bouchard G., François O., Fast and efficient estimation of individual ancestry coefficients. Genetics 196, 973–983 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramu P., et al. , Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat. Genet. 49, 959–963 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Saitou N., Nei M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 15.Gutenkunst R. N., Hernandez R. D., Williamson S. H., Bustamante C. D., Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Excoffier L., Dupanloup I., Huerta-Sánchez E., Sousa V. C., Foll M., Robust demographic inference from genomic and SNP data. PLoS Genet. 9, e1003905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folk R. A., Soltis P. S., Soltis D. E., Guralnick R., New prospects in the detection and comparative analysis of hybridization in the tree of life. Am. J. Bot. 105, 364–375 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Wright S., The genetical structure of populations. Ann. Eugen. 15, 323–354 (1951). [DOI] [PubMed] [Google Scholar]
- 19.Hu X.-S., Filatov D. A., The large-X effect in plants: Increased species divergence and reduced gene flow on the Silene X-chromosome. Mol. Ecol. 25, 2609–2619 (2016). [DOI] [PubMed] [Google Scholar]
- 20.McCauley D. E., The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends Ecol. Evol. 10, 198–202 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Reich D., Thangaraj K., Patterson N., Price A. L., Singh L., Reconstructing Indian population history. Nature 461, 489–494 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huson D. H., Bryant D., Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Veyres N., Aono M., Sangwan-Norreel B. S., Sangwan R. S., Has Arabidopsis SWEETIE protein a role in sugar flux and utilization? Plant Signal. Behav. 3, 722–725 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veyres N., et al. , The Arabidopsis sweetie mutant is affected in carbohydrate metabolism and defective in the control of growth, development and senescence. Plant J. 55, 665–686 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Hughes C. E., et al. , Serendipitous backyard hybridization and the origin of crops. Proc. Natl. Acad. Sci. U.S.A. 104, 14389–14394 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salman-Minkov A., Sabath N., Mayrose I., Whole-genome duplication as a key factor in crop domestication. Nat. Plants 2, 16115 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Peng J. H., Sun D., Nevo E., Domestication evolution, genetics and genomics in wheat. Mol. Breed. 28, 281–301 (2011). [Google Scholar]
- 28.Heslop-Harrison J. S., Schwarzacher T., Domestication, genomics and the future for banana. Ann. Bot. 100, 1073–1084 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rieseberg L. H., Homoploid reticulate evolution in Helianthus (Asteraceae): Evidence from ribosomal genes. Am. J. Bot. 78, 1218–1237 (1991). [Google Scholar]
- 30.Zhang B.-W., et al. , Phylogenomics reveals an ancient hybrid origin of the Persian walnut. Mol. Biol. Evol. 36, 2451–2461 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Mallet J., Hybrid speciation. Nature 446, 279–283 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Warschefsky E., Penmetsa R. V., Cook D. R., von Wettberg E. J. B., Back to the wilds: Tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am. J. Bot. 101, 1791–1800 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Darkwa K., Olasanmi B., Asiedu R., Asfaw A., Review of empirical and emerging breeding methods and tools for yam (Dioscorea spp.) improvement: Status and prospects. Plant Breed. 139, 474–497 (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and supporting information.