Significance
Regulation of quiescence is critical for the maintenance of adult HSCs, and the underlying mechanisms are poorly understood. Using a novel mouse conditional knockout allele of transcription factor gene Prdm16, we show that its deletion in the adult hematopoietic system led to a gradual loss of adult HSCs over time. This loss of adult HSCs was associated with their significantly increased cycling. We further found that Prdm16 promotes the quiescence of adult HSCs by activating the transcription of HSC cell cycle inhibitors including Cdkn1a and Egr1. Our study identifies Prdm16 as a critical regulator of adult HSC quiescence.
Keywords: hematopoietic stem cells, Prdm16, quiescence
Abstract
Regulation of quiescence is critical for the maintenance of adult hematopoietic stem cells (HSCs). Disruption of transcription factor gene Prdm16 during mouse embryonic development has been shown to cause a severe loss of fetal liver HSCs; however, the underlying mechanisms and the function of Prdm16 in adult HSCs remain unclear. To investigate the role of Prdm16 in adult HSCs, we generated a novel conditional knockout mouse model and deleted Prdm16 in adult mouse hematopoietic system using the IFN-inducible Mx1-Cre. Our results show that Prdm16 deletion in the adult mouse hematopoietic system has a less severe effect on HSCs, causing a gradual decline of adult HSC numbers and a concomitant increase in the multipotent progenitor (MPP) compartment. Prdm16 deletion in the hematopoietic system following transplantation produced the same phenotype, indicating that the defect is intrinsic to adult HSCs. This HSC loss was also exacerbated by stress induced by 5-fluorouracil injections. Annexin V staining showed no difference in apoptosis between wild-type and knockout adult HSCs. In contrast, Bromodeoxyuridine analysis revealed that loss of Prdm16 significantly increased cycling of long-term HSCs (LT-HSCs) with the majority of the cells found in the S to G2/M phase. Consistently, RNA sequencing analysis of mouse LT-HSCs with and without Prdm16 deletion showed that Prdm16 loss induced a significant decrease in the expression of several known cell cycle regulators of HSCs, among which Cdkn1a and Egr1 were identified as direct targets of Prdm16. Our results suggest that Prdm16 preserves the function of adult LT-HSCs by promoting their quiescence.
Cell cycle control plays an important role in maintaining adult hematopoietic stem cells (HSCs). Adult HSCs are largely quiescent, and increased cycling has been thought to cause their loss by inducing premature differentiation. In support of this notion, gene ablation studies in mice have shown that cell cycle inhibitors, particularly cyclin-dependent kinase (CDK) inhibitors including CDKN1A (p21) and CDKN1C (p57), are critical for the maintenance of adult HSCs (1, 2). A number of transcription factors known to be essential for HSC maintenance, such as Gfi1, Pbx1, and Mecom, were also found to negatively regulate their proliferation (3–5). Elucidating the molecular mechanisms governing adult HSC quiescence will greatly enhance our capability to harness the power of these cells in regenerative medicine. However, the current understanding of the regulation of HSC quiescence in response to various intrinsic and extrinsic signals remains very limited and will be significantly advanced by identifying additional pathways and regulators.
PRDM16 encodes a zinc-finger transcription factor homologous to MECOM and was first cloned from chromosome translocations in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) patients (6). Two major isoforms of PRDM16 protein have been reported due to usage of different transcriptional start sites: a longer variant containing the N-terminal PR domain, which is homologous to the SET domain found in lysine methyltransferases and has been shown to methylate H3K4 and H3K9 (7, 8), and a shorter variant that lacks the PR domain (6). The same DNA-binding specificity is expected for the two isoforms, as both contain the entire 10 C2H2-type zinc fingers that are divided into two domains: the N-terminal DNA binding domain 1 (DBD1), containing seven zinc fingers, and the C-terminal DBD2, containing three zinc fingers. Increasing evidence in the leukemia field suggests that significant functional differences exist between the two isoforms. The chromosome rearrangements involving PRDM16 in MDS and AML patients cause preferential activation over the full-length protein of the shorter isoform of PRDM16, which is also the only form capable of inducing myeloid leukemia development in mice (9). This shorter isoform of Prdm16 is also preferentially activated by retroviral insertions causing immortalization of myeloid progenitors (10). Mechanistically, direct activation of Spi1 by the shorter isoform has been recently shown to be critical for its transformation of megakaryocyte-erythroid progenitors into leukemia stem cells for AML development (11). In contrast, the full-length form has been shown to antagonize leukemia development through activating Gfi1b (8, 12).
Previous studies have also identified Prdm16 as an important regulator for the generation/maintenance of fetal liver HSCs; however, the underlying mechanisms and the role of Prdm16 in adult HSC function remain unclear. Homozygous mice for the Prdm16 gene trap allele die shortly after birth, precluding analysis of the role of Prdm16 in adult HSCs (13, 14). Although a Prdm16 conditional allele was recently reported, Vav-Cre was used to induce hematopoietic-specific deletion (12). Due to the expression of Vav-Cre during fetal hematopoiesis, it is unclear whether the observed severe HSC defects in adult mice in this study are partly due to possible generation of abnormal adult HSCs in the absence of Prdm16.
To address the function of Prdm16 in adult HSCs, we generated a different Prdm16 conditional allele and deleted the gene in the hematopoietic system in adult mice using the inducible Mx1-Cre (15). In contrast to the acute loss of HSCs observed in previous studies, our results show that Prdm16 deletion caused a gradual reduction in adult HSC numbers that was exacerbated under stress. Consistent with this phenotype, we found that Prdm16 plays a critical role in maintaining the quiescence of adult HSCs. Downstream targets of Prdm16 involved in this regulation also were identified.
Results
Generation of Prdm16 Knockout and Conditional Alleles.
To determine the role of Prdm16 in adult HSCs, we generated a conditional allele of Prdm16 through gene targeting in mouse embryonic stem (ES) cells. In order to completely knock out Prdm16 function, two loxP sites were inserted sequentially before exon 6 and after exon 12 by two consecutive gene-targeting experiments in CJ7 mouse ES cells. This design allows removal of the entire first and 50% of the second zinc finger domain of Prdm16, the only two domains responsible for its DNA binding capability (SI Appendix, Fig. S1 A and B). Two ES cell clones harboring the doubly targeted Prdm16 allele were confirmed by Southern blotting analysis and subsequently used for blastocyst injection. A Prdm16 conditional allele was subsequently generated by breeding mice carrying the targeted allele with ACTB-Flpe mice to remove the neomycin and blasticidin resistance cassettes used for selection in ES cells. A Prdm16 conventional knockout also was generated by crossing the targeted mice to ACTB-Cre mice. As expected, mice homozygous for the Prdm16 conditional allele are healthy and fertile. In contrast, the homozygous Prdm16 knockout mice die within 24 h of birth, likely due to cleft palate development. Prdm16 messenger RNA (mRNA) was not detected by RT-PCR in fetal livers from e18.5 Prdm16−/− embryos (SI Appendix, Fig. S1C), nor was PRDM16 protein detected in fetal brain nuclear extracts isolated from e15.5 Prdm16−/− embryos (SI Appendix, Fig. S1D), confirming loss of Prdm16 expression in the knockout animals.
Prdm16−/− Fetal Livers Have Severe Reduction of Long-Term HSCs.
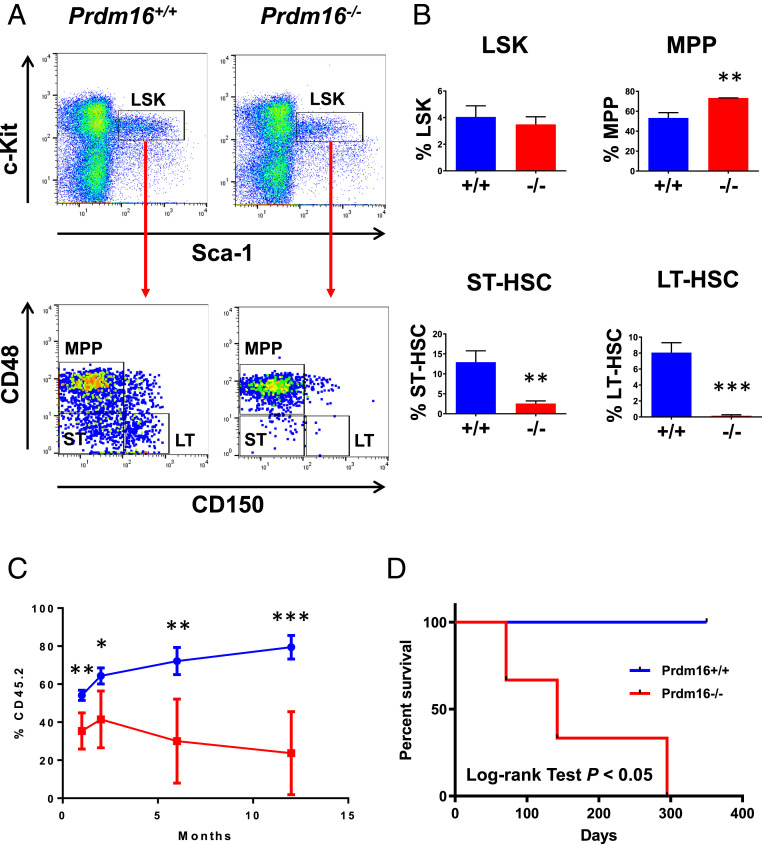
Previous studies, using a Prdm16 gene trap allele (Prdm16LacZ), have shown that Prdm16LacZ/LacZ animals have severe reduction of fetal liver HSCs but grossly normal hematopoiesis with no significant changes in the frequency of myeloid, B, or T cells (13, 14). In agreement with the gene trap model, analysis of e18.5 fetal thymuses and livers from our Prdm16−/− embryos demonstrated that hematopoiesis was essentially normal, with similar frequencies of myeloid and B cells detected in fetal livers (SI Appendix, Fig. S2A) and of T cells detected in fetal thymuses (SI Appendix, Fig. S2B). Flow cytometry analysis showed that the frequency of the Lin-Sca-1+c-Kit+ (LSK) population enriched for HSCs was similar in the e18.5 Prdm16−/− livers in comparison to Prdm16+/+ fetal livers (Fig. 1 A and B). More detailed analysis of the LSK compartment showed a significant increase in multipotent progenitors (MPPs), approximately fivefold reduction in short-term HSCs (ST-HSCs, Lin-Sca-1+c-Kit+CD48−CD150−) and over 50-fold reduction in long-term HSCs (LT-HSCs) (Lin-Sca-1+c-Kit+CD48−CD150+) in the Prdm16−/− fetal livers, indicating that the HSC compartment is severely compromised upon loss of Prdm16 (Fig. 1 A and B). Similar to the e18.5 Prdm16−/− fetal livers, MPPs also were significantly increased and ST-HSCs and LT-HSCs were significantly decreased in e14.5 Prdm16−/− fetal livers (SI Appendix, Fig. S3 A and B). However, the reduction in LT-HSCs at e14.5 was not to the same extent as at e18.5 (SI Appendix, Fig. S3C), suggesting that LT-HSCs seed the liver normally and that the effects of Prdm16 loss on LT-HSCs become apparent subsequently, most likely due to defects in HSC expansion which is essentially a continuous self-renewal cycle (16). Consistent with this hypothesis, e18.5 Prdm16−/− fetal liver cells expanded poorly in culture under a condition shown to promote self-renewal of LT-HSCs (SI Appendix, Fig. S4A) (17). Immature clonogenic cells also decreased rapidly in the Prdm16−/− culture over time (SI Appendix, Fig. S4B). To test the self-renewal capability of Prdm16−/− fetal liver HSCs in vivo, 1 × 106 fetal liver cells from e18.5 Prdm16+/+ or Prdm16−/− embryos were competed against the same number of wild-type CD45.1+ bone marrow cells after transplantation into lethally irradiated congenic C57BL/6-Ly5.2 recipient mice. At 1-mo posttransplantation, the percentage of peripheral blood CD45.2+ cells was significantly lower in the recipients of Prdm16−/− fetal liver cells than in mice receiving Prdm16+/+ fetal liver cells (Fig. 1C). At 2, 6, and 12 mo, the percentage of CD45.2 cells was reduced even further in the recipients of Prdm16−/− fetal liver cells (Fig. 1C). Analysis of the HSC compartment further showed a complete loss of Prdm16−/− LT-HSCs in the recipients of Prdm16−/− fetal liver cells at 12 mo (SI Appendix, Fig. S4C). We also tested the self-renewal capability of Prdm16−/− fetal liver HSCs by serial transplantation. Interestingly, all primary recipients of 2 × 106 Prdm16−/− fetal liver cells were viable at 4 mo posttransplantation. However, upon secondary transplantation, the recipients of Prdm16−/− fetal liver cells started dying at 3 mo after transplantation and did not survive beyond 10 mo, while recipients of wild-type fetal liver cells remained healthy (Fig. 1D). Taken together, these results suggest that the loss of Prdm16 negatively affects the self-renewal of fetal liver HSCs.
Fig. 1.
Significant reduction in fetal liver HSCs after Prdm16 deletion. (A) Representative FACS analysis of LSK compartments including LT-HSC (LT), ST-HSC (ST), and MPP in fetal livers of e18.5 Prdm16−/− versus Prdm16+/+ embryos. (B) Quantification of results in A (n = 3 for each group). The frequencies (mean ± SD) of LSK cells in total bone marrow nucleated cells and of LT-HSC/ST-HSC/MPP populations in LSK cells are shown. (C) Competition of 1 × 106 e18.5 Prdm16−/− or Prdm16+/+ fetal liver cells against equal number of wild-type C57BL/6-Ly5.2 bone marrow cells in lethally irradiated C57BL/6-Ly5.2 recipient mice. The frequencies (mean ± SD) of fetal liver cells (CD45.2+) in peripheral blood of recipient mice at 1, 2, 6, and 12 mo after transplantation are shown (n = 5 for each fetal liver genotype). (D) Survival curves of irradiated C57BL/6-Ly5.2 mice receiving 1 × 106 bone marrow cells from primary recipient mice of e18.5 Prdm16−/− or Prdm16+/+ fetal liver cells (n = 3 for each genotype). *P < 0.05; **P < 0.01; ***P < 0.001.
Deletion of Prdm16 in Adult Mice Leads to Gradual Reduction of LT-HSCs due to Self-Renewal Defects.
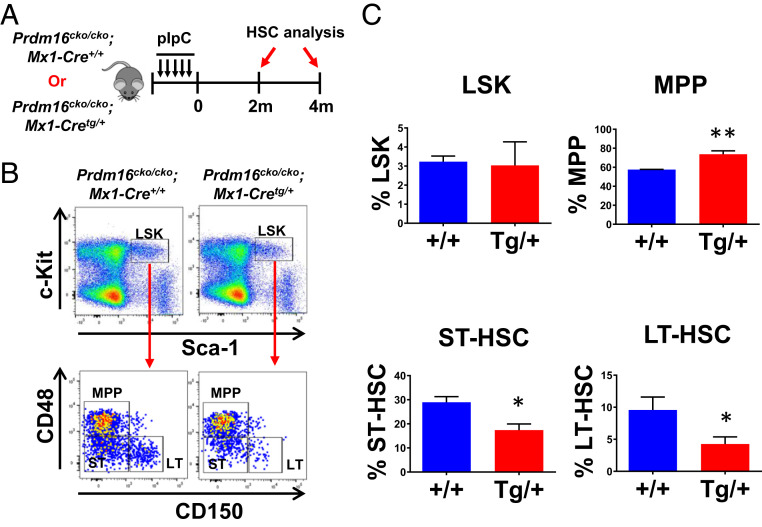
In contrast to the highly proliferative fetal liver HSCs, adult LT-HSCs in the bone marrow are mostly quiescent (18). Upon hematopoietic injury or stress, for example, massive bleeding, LT-HSCs exit quiescence and contribute significantly to the production of more-mature hematopoietic cells (19). The role of Prdm16 in adult HSCs remains unclear, as previous studies disrupted the gene before the establishment of these cells in the bone marrow (12–14). To answer this question, we bred Prdm16cko/cko mice with the IFN-inducible Mx1-Cretg/+ mice to generate adult Prdm16cko/cko;Mx1-Cretg/+ and control Prdm16cko/cko;Mx1-Cre+/+ mice and analyzed their HSC compartments after inducing Prdm16 deletion by polyinosinic–polycytidylic acid (pIpC) injections (SI Appendix, Fig. S5). At 2 mo post pIpC injections, the percentages of LT-HSCs and ST-HSCs in 8- to 12-wk-old Prdm16cko/cko;Mx1-Cretg/+ mice were significantly reduced in comparison to control mice, whereas the percentage of MPPs was increased (Fig. 2A). Similar changes also were observed at 4 mo without detection of any significant changes in total bone marrow cellularity (SI Appendix, Fig. S6). These data suggest that deletion of Prdm16 in adult bone marrow leads to a gradual but not acute loss of HSCs in our model, potentially due to their failure to self-renew. In order to determine that the reduction was intrinsic to the HSCs and not due to confounding effects from the microenvironment, we transplanted bone marrow cells from control mice and Prdm16cko/cko;Mx1-Cretg/+ into lethally irradiated C57BL/6-Ly5.2 mice. We subsequently induced Prdm16 deletion at 2 mo posttransplantation and analyzed donor HSCs at 4 mo afterward (Fig. 3A). Prdm16 deletion in the transplanted cells led to a reduction in the percentages of donor LT-HSCs and ST-HSCs and an increase in the percentage of donor MPPs, confirming an intrinsic role for Prdm16 in adult HSCs (Fig. 3B). In addition, Prdm16 deletion did not have any obvious effects on hematopoietic development in the periphery, since no difference was found in the percentages of donor-derived myeloid or lymphoid cells in the recipient mice at 2 mo after pIpC injections (SI Appendix, Fig. S7). Since Prdm16 deletion in primary or transplanted animals did not lead to acute bone marrow failure, we performed serial transplantations to test the self-renewal capability of Prdm16-deleted HSCs at 4 mo after pIpC injections. Interestingly, recipients of Prdm16cko/cko;Mx1-Cretg/+ bone marrow survived secondary transplantation despite significant reduction in LT-HSCs but succumbed to bone marrow failure in a tertiary transplantation (Fig. 3 C and D). Overall, these data strongly suggest that Prdm16 is an important intrinsic regulator for the self-renewal of adult LT-HSCs but not their differentiation into mature hematopoietic lineages.
Fig. 2.
Conditional Prdm16 deletion induces gradual and significant reduction in adult HSCs. (A) Schematic overview of Prdm16 deletion induction in adult Prdm16cko/cko;Mx1-Cretg/+ mice. (B) Representative FACS analysis of LSK compartments including LT-HSC (LT), ST-HSC (ST), and MPP in bone marrow of Prdm16cko/cko;Mx1-Cretg/+ versus Prdm16cko/cko;Mx1-Cre+/+ mice at 2 mo after pIpC injections. (C) Quantification of results in B (n = 4 for each group). The frequencies (mean ± SD) of LSK cells in total bone marrow nucleated cells and LT-HSC/ST-HSC/MPP populations in LSK cells are shown. *P < 0.05; **P < 0.01.
Fig. 3.
Reduction in adult HSCs induced by Prdm16 deletion is cell-intrinsic. (A) Schematic overview of Prdm16 deletion in a transplantation assay to analyze the intrinsic role of Prdm16 in LT-HSCs. Lethally irradiated C57BL/6-Ly5.2 recipient mice were transplanted with 1 × 106 bone marrow cells from Prdm16cko/cko;Mx1-Cretg/+ or Prdm16cko/cko;Mx1-Cre+/+ mice. After a 2-mo recovery period, recipient mice were injected with pIpC every other day for a total of five injections and analyzed 4 mo post pIpC injections. (B) The frequencies (mean ± SD) of CD45.2+LSK cells in CD45.2+ bone marrow nucleated cells and LT-HSC/ST-HSC/MPP populations in CD45.1+LSK cells in primary recipient mice at 4 mo after pIpC injections are shown (n = 5 for each genotype). (C) Representative FACS analysis of CD45.2+LSK and CD45.2+LT-HSC populations in secondary recipient mice at 4 mo posttransplantation. (D) Survival curves of irradiated tertiary recipient mice receiving 1 × 106 bone marrow cells from secondary recipients (n = 5 for each genotype). *P < 0.05.
Further Reduced Self-Renewal Capability of Prdm16-Deficient Adult HSCs under Stress.
We also examined the self-renewal capability of Prdm16-deficient adult HSCs by performing competitive repopulation assay. No significant difference in reconstitution was detected between bone marrow cells from Prdm16cko/cko;Mx1-Cre+/+ and Prdm16cko/cko;Mx1-Cretg/+ mice (SI Appendix, Fig. S8), potentially due to the slow and gradual reduction in adult LT-HSCs upon Prdm16 deletion. We therefore tested whether stress could accelerate their loss by injecting the recipient mice with three separate doses of 5-fluorouracil (5-FU) 2 wk apart after inducing Prdm16 deletion (Fig. 4A). Since 5-FU is toxic for actively proliferating hematopoietic cells, quiescent LT-HSCs are forced to divide in order to respond to their loss (20). Analysis of peripheral blood from the recipients of Prdm16cko/cko;Mx1-Cre+/+ or control Prdm16cko/cko;Mx1-Cretg/+ bone marrow before pIpC injections showed that the reconstitution by donor (CD45.2) and competitor (CD45.1) cells was essentially equal between the groups (Fig. 4B). In contrast, at 1 mo post pIpC injections, the percentage of Prdm16cko/cko;Mx1-Cretg/+ donor cells in recipient mice was notably lower than that of Prdm16cko/cko;Mx1-Cre+/+ cells, suggesting that repeated stress following loss of Prdm16 significantly reduces their ability to compete with wild-type HSCs (Fig. 4B). At 2 and 6 mo, the difference increased further (Fig. 4B), with significant reductions in granulocytes (CD45.2+Gr-1+) and macrophages (CD45.2+Mac-1+) (Fig. 4C). In addition, analysis of the bone marrow of these mice at 6 mo showed a drastic reduction in the number of Prdm16-deleted LT-HSCs, demonstrating that the cells were efficiently exhausted and outcompeted after 5-FU−induced stress (SI Appendix, Fig. S9).
Fig. 4.
Competitive reconstitution capability of Prdm16-deficient HSCs was significantly reduced after 5-FU treatments. (A) Schematic overview of a competitive reconstitution assay to analyze the intrinsic role of Prdm16 in LT-HSCs. Lethally irradiated C57BL/6-Ly5.2 mice were transplanted with 1 × 106 bone marrow cells from Prdm16cko/cko;Mx1-Cretg/+ or Prdm16cko/cko;Mx1-Cre+/+ mice plus 1 × 106 CD45.1+ competitor bone marrow cells from healthy C57BL/6-Ly5.2 mice. After a 2-mo recovery period, mice were treated with five pIpC injections followed by three 5-FU injections 2 wk apart and analyzed at 1, 2, and 6 mo post 5-FU injections for donor (CD45.2+) reconstitution. (B) Frequencies (mean ± SD) of CD45.2+ cells in peripheral blood of recipient mice at indicated time points after 5-FU treatments. (C) Frequencies (mean ± SD) of CD45.2+Gr-1+, CD45.2+Mac-1+, CD45.2+CD19+, and CD45.2+CD3+ in peripheral blood of recipient mice at indicated time points after 5-FU treatments (n = 5 for each genotype). *P < 0.05; **P < 0.01.
Increased Cycling in Adult LT-HSCs Lacking Prdm16.
Loss of Prdm16 resulted in a significant decrease in the self-renewal capability of adult LT-HSCs, suggesting that Prdm16 deletion could be decreasing their survival and/or causing the cells to differentiate, possibly by reducing their quiescence. To determine the cell cycle status and survival of LT-HSCs upon Prdm16 loss, we performed cell cycle analysis by Bromodeoxyuridine (BrdU) incorporation (21) and apoptosis analysis by Annexin V staining (22) on LT-HSCs from Prdm16cko/cko;Mx1-Cre+/+ and Prdm16cko/cko;Mx1-Cretg/+ mice at 2 mo after pIpC injections. The BrdU analysis demonstrated significantly increased cycling of LT-HSCs in the pIpC-treated Prdm16cko/cko;Mx1-Cretg/+ mice compared to control mice (Fig. 5 A and B). In contrast, no significant increase in Annexin V staining was detected in Prdm16-deleted LT-HSCs upon induction of Cre expression (Fig. 5C). Taken together, our data suggest that the reduced self-renewal capability and premature differentiation of Prdm16-deleted adult LT-HSCs is likely due to their decreased ability to maintain quiescence.
Fig. 5.
Prdm16 deletion promoted cycling of LT-HSCs without affecting apoptosis. (A) Representative cell cycle analysis of LT-HSCs in Prdm16cko/cko;Mx1-Cretg/+ or Prdm16cko/cko;Mx1-Cre+/+ mice after pIpC injections. (B) Quantification of results in A (n = 4 for each group). (C) Frequencies of Annexin V positive apoptotic LT-HSCs in Prdm16cko/cko;Mx1-Cretg/+ or Prdm16cko/cko;Mx1-Cre+/+ mice after pIpC injections (n = 4 for each group). *P < 0.05.
Deletion of Prdm16 Decreases the Expression of Genes Critical for LT-HSC Quiescence and Self-Renewal.
To investigate the mechanism by which Prdm16 regulates LT-HSC function, we performed RNA sequencing (RNA-seq) analysis of sorted LT-HSCs from Prdm16cko/cko;Mx1-Cretg/+ and Prdm16cko/cko;Mx1-Cre+/+ mice treated with pIpC (Fig. 6A). To minimize short-term effects of pIpC on gene expression, these cells were purified at 5 wk after the last pIpC injection for the analysis. Deletion of Prdm16 was confirmed by genotyping and sequencing reads mapped to the locus (SI Appendix, Fig. S10). We found that 1,578 genes were differentially expressed between the two populations, with 564 genes down-regulated in the Prdm16-deleted LT-HSCs and 1,014 genes up-regulated using 1.5-fold change in gene expression as the cutoff (Fig. 6B and Dataset S1). As expected from the defective maintenance of these cells after Prdm16 deletion, gene set enrichment analysis (GSEA) demonstrated up-regulation of gene sets down-regulated in normal HSCs and leukemia stem cells and gene signatures associated with more differentiated cell types (Fig. 6C and SI Appendix, Fig. S11). Consistent with the increased proliferation of these cells upon Prdm16 loss, genes associated with cell cycle arrest were found to be mostly down-regulated upon Prdm16 deletion (Fig. 6D). Importantly, several genes previously known to be critical for maintaining the quiescence and self-renewal function of HSCs were significantly down-regulated in Prdm16-deleted cells, including Fos, Egr1, Cdkn1a, and Fosb (Fig. 6E and SI Appendix, Fig. S12) (1, 23–25). However, we did not detect any significant correlation with gene signatures associated with senescence and oxidative stress previously linked to Prdm16 disruption.
Fig. 6.
RNA-seq analysis of LT-HSCs before and after Prdm16 deletion. (A) Schematic overview of collection of LT-HSCs for RNA-seq analysis. (B) Pie chart showing the numbers of differentially expressed genes induced by Prdm16 deletion. (C) GSEA analysis of differentially expressed genes in LT-HSCs after Prdm16 deletion using gene sets down-regulated in stem cells. (D) GSEA analysis of differentially expressed genes in LT-HSCs after Prdm16 deletion using a gene set associated with cell cycle arrest. (E) Heat map showing reduced expression of regulator genes of HSC quiescence and maintenance after Prdm16 deletion.
Cell Cycle Regulators Cdkn1a and Egr1 Are Direct Targets of Prdm16.
Cell cycle control plays an important role in maintaining the self-renewal function of adult LT-HSCs. Our cell cycle analysis of Prdm16-deleted adult LT-HSCs suggests that Prdm16 is a critical regulator of quiescence in these cells. Among genes whose expression was significantly changed by Prdm16 deletion, both Cdkn1a and Egr1 have been shown to be important for cell cycle regulation and maintenance of HSCs in vivo, by gene targeting studies (1, 23). Analysis of the Cdkn1a and Egr1 locus using the Genomatix software revealed several potential PRDM16 binding sites, suggesting that both could be direct transcriptional targets of PRDM16. To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) analysis in BM2 myeloid progenitor cells which were immortalized due to retroviral insertional activation of Prdm16 (10). Significant PRDM16 binding was readily detected at regions containing potential PRDM16 binding sites (Fig. 7), suggesting that both Cdkn1a and Egr1 represent direct targets of PRDM16. Taken together, our data suggest that Prdm16 plays a critical role in maintaining quiescence of adult LT-HSCs by directly regulating Cdkn1a and Egr1 expression.
Fig. 7.
Cdkn1a and Egr1 represent direct targets of Prdm16. ChIP analysis of regions of (A) Cdkn1a or (B) Egr1 locus containing predicted PRDM16 binding sites (1, 2) and negative control regions (N) in BM2 myeloid progenitors using a PRDM16-specific antibody and control IgG (Upper, mean ± SD). Diagrams of tested region of Cdkn1a and Egr1 locus are also shown (Lower). Locations of PCR amplicons are indicated as black bars. Transcriptional start sites are indicated as arrows. Exons are indicated as white boxes with coding regions highlighted in gray.
Discussion
In this study, we found that Prdm16 is a critical regulator of adult HSC quiescence but is dispensable for the generation of downstream progenitors and mature cells. We showed that loss of Prdm16 in adult HSCs leads to increased cycling and gradual loss of the cells over time. We also showed that HSC loss was accelerated upon 5-FU−induced stress, underscoring an increased importance of Prdm16 for HSCs under stress conditions. Our data further uncovered several key downstream targets of Prdm16 that likely contribute to its regulation of HSC dormancy. We additionally identified the CDK inhibitor gene Cdkn1a and the transcription factor gene Egr1 as direct transcriptional targets of Prdm16.
Prdm16 is likely a specific regulator of HSCs, which could explain why the hematopoietic phenotype of our adult mouse model is only acute under severe stress such as serial transplantation or serial 5-FU treatments. A recent study has suggested that hematopoiesis in the adult is mostly maintained by short-term HSCs and MPPs but not LT-HSCs (26). LT-HSCs rarely contribute to the hematopoietic pool except under stress, for example, with 5-FU injections. Therefore, it is challenging to detect a phenotype in adult HSCs that have lost Prdm16 expression, since the gene appears to be critical only for that compartment. Many knockout models claiming an HSC phenotype also have significant effects on downstream progenitors, which makes the phenotype more severe (27, 28). It could be argued that, in those cases, it is difficult to distinguish between true HSC defects in vivo and a failure of more committed progenitors.
According to our data, apoptosis was not affected in adult LT-HSCs after Prdm16 deletion, whereas cycling was significantly increased compared to controls. Stringent regulation of the cell cycle is critical for maintaining adult LT-HSCs in a quiescent state and in allowing self-renewal to occur to preserve the HSC pool (29). It is therefore likely that increased cycling of adult LT-HSCs upon Prdm16 deletion is responsible for their gradual loss through differentiation. Fetal liver HSCs, unlike adult HSCs, proliferate extensively. It is also possible that Prdm16 loss in fetal liver HSCs could further increase their proliferation, leading to an accelerated exhaustion of these cells. In line with these phenotypic findings, one of the significantly down-regulated genes upon Prdm16 loss is the CDK inhibitor Cdkn1a, which maintains LT-HSCs as quiescent by preventing the cell from exiting the G0 phase of the cell cycle (1). Another significantly down-regulated gene is Egr1, loss of which has been shown to increase cycling of LT-HSCs, in part, by decreasing Cdkn1a expression (23). Our ChIP results in BM2 cells further suggest that both are direct targets of PRDM16. It will be important to validate the role of Cdkn1a in Prdm16-mediated regulation of LT-HSC quiescence by restoring its expression in Prdm16-deleted LT-HSCs; however, such experiments will be difficult to perform due to the need to precisely control the level of Cdkn1a expression. Overexpression of Cdkn1a will likely cause a complete block in the proliferation of LT-HSCs, rendering the detection of their engraftment difficult.
Both our Prdm16 germline and conditional knockout models displayed less severe hematopoietic phenotypes than previously reported models. Our germline knockout model recapitulates the previously described gene trap model in regard to the perinatal lethality, cleft palate development, loss of phenotypic fetal liver HSCs, and lack of significant changes in mature blood cell numbers (13, 14). However, our transplantation studies, where the intrinsic function of HSCs is measured, did not show the same acute phenotype as the previous gene trap and knockout models where Prdm16-deficient fetal liver cells competed very poorly with wild-type BM cells. Similarly shown by competition assay, the adult HSC defect in our conditional knockout model is also significantly less severe than a recently reported conditional model (12). One main reason for these differences could be our Prdm16 targeting strategy, which likely results in a more complete ablation of Prdm16 expression. Based on mouse expressed sequence tags mapped to the Prdm16 locus (genome.ucsc.edu), shorter Prdm16 transcripts potentially initiating from the region between exons 11 and 12 likely exist. Indeed, by performing 5′ rapid amplification of complementary DNA (cDNA) ends (5′RACE) in mouse LSK cells, we have identified two such novel transcripts of Prdm16, both predicted to produce a significantly shorter PRDM16 isoform (SI Appendix, Fig. S13). These mRNAs should be inactivated by Cre recombination in our model, while likely remaining intact in the previously reported gene trap and conditional models. Although the function of this significantly shorter isoform is unclear, it is possible that it may normally antagonize the activity of the two longer isoforms, due to its truncated nature. Therefore, the loss of just the longer isoforms in previous models may disrupt a critical balance, which may translate into a more severe phenotype. Future studies will be needed to clarify the function of this shorter isoform. Another possibility may also contribute to the phenotypic difference between our conditional model using Mx1-Cre and the previously reported conditional model using Vav-Cre. As Vav-Cre is expressed in fetal liver HSCs, it is possible that loss of Prdm16 in these cells may affect their transition into fully functional adult HSCs.
Materials and Methods
Mice.
All mice were maintained in the animal facility of Laboratory of Animal Medicine at Uniformed Services University of the Health Sciences (USUHS). Female C57BL/6-Ly5.2 mice (Charles River, 8 wk to 12 wk old) were used as recipients for all transplantation experiments. All mouse experiments were carried out according to protocols approved by USUHS Institutional Animal Care and Use Committee.
Generation of Prdm16 Conditional and Knockout Alleles in Mice.
A 129 bacterial artificial chromosome (BAC) library (CT7, Invitrogen) was screened with a Prdm16 genomic probe to identify a BAC clone containing the Prdm16 genomic locus (clone no. 264O8). Two targeting constructs were generated to insert one loxP site into intron 5 and another in intron 12 of Prdm16. Two fragments (7.9 and 8.4 kb) containing intron 5 and intron 12 of Prdm16, respectively, were first retrieved from the BAC clone in EL350 cells through recombineering into an ES cell targeting vector PL253 as previously described (30). A loxP plus Frt-flanked neocassette was targeted to the resulting intron 5 retrieval construct in EL350 cells through recombineering. First, a targeting cassette containing Pgk-em7-neo flanked by homology arms to intron 5 of Prdm16 was constructed in PL451. The homology arms are PCR amplified using the following primers: 5′ arm sense, 5′-CGG CGG CCG CGT ACC CAC ACT GAC AAG TAG-3′; 5′ arm antisense, 5′-CGC GGG ATC CAG CTA GCC TGC CTG GAG AGA GAA CAC C-3′; 3′ arm sense, 5′-CGC GGA ATT CAA GCT TGG GTG AAA GCT GAA GGG CCT-3′; 3′ arm antisense, 5′-CGC GGT CGA CTG AGC AAA GTC AGG CTC CTC-3′. The homology arms were sequence verified, restriction digested, and cloned into PL451 via four-way ligation. The targeting cassette was released by NotI/SalI double digest and targeted through coelectroporation into heat shock-induced EL350 cells. To insert the second loxP site in intron 12 of Prdm16, a targeting cassette containing frt5-Pgk-Em7-blast-frt5-loxP flanked by homology arms to targeting site was constructed in a modified version of PL451. Homology arms were amplified using the following primers: 5′ arm sense, 5′-CGG CGG CCG CGG AGC AGC CAT ACA GGT AG-3′; 5′ arm antisense, 5′-CGC GGG ATC CCA GGC TCT CTC ACA TCC TCT-3′; 3′ arm sense, 5′-CGC GGA ATT CTC TCC TGT ATC CAA AGT GTC-3′; 3′ arm antisense, 5′-CGC GGT CGA CTA GCC CCT CGT TTT TGA CAC-3′. The targeting cassette was released by NotI/SalI double digest and targeted similarly as described above. The intron 5 targeting vector after linearization by NotI digestion was first electroporated into 129-derived CJ7 ES cells, using standard procedures. G418 (180 μg/mL) and Ganciclovir (2 μM) double-resistant clones were analyzed by Southern blotting hybridization, using both 5′ and 3′ external probes. One of the correctly targeted clones was expanded and subsequently electroporated with the intron 12 targeting vector linearized also by NotI digestion. Clones resistant to Blasticidin (12.5 μg/mL) and Ganciclovir (2 μM) were also analyzed by Southern blotting hybridization, using both 5′ and 3′ external probes. External probes were PCR amplified using the following primers: loxP1 5′ probe, sense, 5′-CCT AAG GTG GTC CTT GAG AG-3′; loxP1 5′ probe, antisense, 5′-CAA CTT TCC CGG CTG AAC AG-3′; loxP1 3′ probe (loxP2 5′ probe), sense, 5′-TCT AAG TCT GCT CCC TGA TG-3′; loxP1 3′ probe (loxP2 5′ probe), antisense, 5′-TTT CCC TCT CTG GAG CCT TG-3′; loxP2 3′ probe, sense, 5′-TCT CTG CTG TTA GGG CCA AC-3′; loxP2 3′ probe, antisense, 5′-ATC AAG TGC CGT GGG AGC TG-3′. The loxP1 3′ probe (loxP2 5′ probe) was also used after the second targeting and HpaI/MfeI double digestion of DNA to identify ES clones with the two loxP sites targeted to the same Prdm16 locus. Correctly targeted clones were then injected into C57BL/6 blastocysts using standard procedures, and resulting chimeras were mated with C57BL/6 females to obtain germline transmission of the targeted allele. A Prdm16 conventional knockout and a conditional allele were subsequently generated by breeding mice carrying the targeted allele with ACTB-Cre and ACTB-Flpe mice, respectively. Both heterozygous knockout and conditional mice were backcrossed to C57BL/6J mice for 7 generations.
Bone Marrow Transplantation.
For transplantation, fetal liver (e18.5) or BM cells were injected into the tail vein of each lethally irradiated (1,100 rads from 137Cs source) C57BL/6-Ly5.2 female mouse without supporting bone marrow cells from unirradiated C57BL/6-Ly5.2mice. Retroorbital bleeding was performed at 1, 2, and 6 mo to analyze the short-term and long-term engraftment of the donor cells by fluorescence-activated cell sorting (FACS). For secondary or tertiary transplantation, 1 × 106 cells from bone marrow of primary or secondary recipients were injected into lethally irradiated secondary or tertiary recipients without supporting bone marrow cells. To induce Prdm16 deletion in conditional cells at 2 mo after transplantation, recipient mice were injected with five injections of 400 µg of pIpC (Calbiochem) dissolved in phosphate-buffered saline at 2-d intervals.
Flow Cytometry.
Flow cytometry analyses of mouse fetal tissues (liver and thymus) and adult tissues (peripheral blood and bone marrow) were performed using BD LSRII flow cytometer. After sample collection and ACK lysis of red blood cells, cells were blocked by incubation with anti-FcγR-II/III and subsequently stained with antibodies against markers for myeloid (Gr-1, Mac-1), erythroid (Ter-119), B (CD19), and T (CD3) lineages (Biolegend). Dead cells were excluded by staining with Sytox Blue (Invitrogen). For analysis of LT-HSCs (Lin−Sca-1+c-Kit+CD48−CD150+), ST-HSCs (Lin−Sca-1+c-Kit+CD48−CD150−), and MPPs (Lin−Sca-1+c-Kit+CD48+CD150−), after ACK lysis of red blood cells, lineage-positive cells were first labeled by incubation with a mixture of purified rat anti-mouse antibodies specific for Gr-1, Mac-1 (not included for fetal liver cells), CD4, CD8, B220, CD127, and Ter-119, and subsequently labeled by incubation with sheep anti-rat PE-Cy5 (Invitrogen). The Lin− cells were then stained with allophycocyanin (APC)-conjugated anti-mouse Sca-1 (D7, Biolegend), APC-eFluor 780-conjugated anti-mouse c-Kit (ACK2, eBioscience), PE-Cy7-conjugated anti-mouse CD150 (TC15-12F12.2, Biolegend), biotinylated anti-mouse CD48 (HM48-1, Biolegend) antibodies, and Streptavidin-Pacific Orange (Invitrogen). All samples were analyzed using a BD LSRII flow cytometer, and data were analyzed using FlowJo software.
Cell Cycle and Apoptosis Analysis.
Cell cycle status of LT-HSC was analyzed in Prdm16cko/cko;Mx1-Cre+/+ and Prdm16cko/cko;Mx1-Cretg/+ mice by BrdU incorporation as previously described (31). Briefly, 2 mo post pIpC injections (5×), mice were injected intraperitoneally with 5 mg of BrdU and maintained on 1 mg/mL BrdU in the drinking water for 72 h. Bone marrow cells were subsequently harvested and stained with antibodies against lineage markers, c-kit, Sca-1, CD150, and CD48 for LT-HSC detection and analyzed using the fluorescein isothiocyanate (FITC) BrdU Flow Kit (BD Pharmingen). Apoptosis in LT-HSC was analyzed 2 mo post pIpC injections using the FITC Annexin-V staining kit (BD Pharmingen) according to the manufacturer’s instructions.
RNA-seq and Bioinformatics Analysis.
Bone marrow was harvested from adult (8 wk to 12 wk old) Prdm16cko/cko;Mx1-Cre+/+ and Prdm16cko/cko;Mx1-Cretg/+ (five each genotype) 5 wk post pIpC injections for LT-HSCs (Lin−Sca-1+c-Kit+CD48−CD150+) sorting. LT-HSCs were sorted from pooled bone marrow directly into 100 μL of RNA lysis buffer (from PicoPure RNA Isolation Kit, Thermo Cat. No. KIT0204) in three technical replicates for each genotype and stored at −80 °C. Low-input RNA purification, library generation, and Illumina sequencing were performed at Cofactor Genomics. The RNA integrity number values for purified RNA were ≥9. Sequencing produced ∼30,000,000 single-end reads per sample with minimum read length of 1 × 75 bp. Processed and trimmed FASTQ sequencing files were uploaded and analyzed using the Galaxy platform (https://usegalaxy.org). Briefly, to ensure sequence quality, FASTQ files were analyzed by FastQC and then aligned to the mouse genome (mm10) using HISAT2. Read counts were determined from aligned binary alignment map files with FeatureCounts, and differential gene expression was performed using the Limma Bioconductor package with a cutoff of P < 0.05 and 1.5-fold change. Heatmaps were generated from normalized count files using Heatmap2. GSEA was performed using the GSEA v4.0.3 application (https://www.gsea-msigdb.org/gsea/index.jsp).
ChIP.
ChIP analyses were performed using ChIP-IT Express kit (Active Motif). Immunoprecipitations were performed using sheep anti-PRDM16 antibody (10E2, #5356, Cell Signaling Technologies) and sheep IgG (#P120-101, Bethyl Laboratories). Chromatin DNA was purified using MinElute PCR Purification Kit (QIAGEN) and quantified by real-time PCR. The following locus-specific primers were used:
| Cdkn1a-1 | 5′ GCC GCG GTG TCA GAG TCT AG 3′ |
| 5′ CGA AGC TCT CAC CTC TGA ATG TC 3′ | |
| Cdkn1a-2 | 5′ TTT AGG AAG CCT GGG CTC ATC 3′ |
| 5′ GGT GGA GAG AGG AGA GGA AAG G 3′ | |
| Cdkn1a-N | 5′ GGG AAG GTA GGA AAG GAT ATT TGG 3′ |
| 5′ CCG CCC GTC GAC CAT 3′ | |
| Egr1-1 | 5′ CTC TCA GCC ATC GGA AAG TGT 3′ |
| 5′ TAG GGC TCA GGA AGT GAC TCT CTT 3′ | |
| Egr1-N | 5′ AGC CTG GGC TGA ATC AAA AG 3′ |
| 5′ TCA GTG GTA GGA AAT CAG AGA AGA GA 3′ |
Real-Time RT-PCR.
Total RNA was extracted using RNAeasy Plus mini kit (Qiagen). For real-time RT-PCR, oligo-dT-primed cDNA samples were prepared from total RNA using SuperScript III (Invitrogen), and real-time PCR analysis was performed in triplicate using SYBR green detection reagents (Invitrogen) according to the manufacturer's instructions in 20-µL final volume in 96-well plates on a QuantStudio real-time PCR system (Applied Biosystems). The cycling conditions are 50 °C for 2 min followed by 95 °C for 2 min, and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Primer specificity was verified by sequencing PCR products and performing melting curve analysis after PCR. Relative changes in expression were calculated according to the ∆∆Ct method. Gene-specific primer sequences are as follows:
| Prdm16 | 5′ TCC GCG GTC AGC AAT AGC 3′ |
| 5′ CCG ACA TGT CAG GGC TCC TA 3′ | |
| Cdkn1a | 5′ CGA GAA CGG TGG AAC TTT GAC 3′ |
| 5′ CCA GGG CTC AGG TAG ACC TT 3′ | |
| Egr1 | 5′ AGA CGA GTT ATC CCA GCC AAA 3′ |
| 5′ GGT CGG AGG ATT GGT CAT GC 3′ | |
| Rpl4 | 5′ ATG ATG AAC ACC GAC CTT AGC A 3′ |
| 5′ CGG AGG GCT CTT TGG ATT TC 3′ |
5′RACE.
The 5′RACE was performed on total RNA extracted from mouse LSK cells using the SMARTer RACE 5′/3′ Kit (Takara) following user instructions. Prdm16-specific primer used for PCR amplification was: 5′ CTG GTG CCC AGG TGG TTC GTG AGC AC 3′.
Statistical Analysis.
Sample sizes and animal numbers were determined by previous experiences. No samples were excluded from analyses. All data were analyzed by two-tailed Student’s t test, except that survival curves were compared by log-rank test. The researchers were not blinded during sample collection and analysis.
Supplementary Material
Acknowledgments
This work was supported by USUHS Research Accelerating Military Pediatrics Grant 310534 (Y.D.). The authors would like to thank USUHS flow cytometry core facility for assistance in data analysis. The views presented in this manuscript are those of the authors; no endorsement by USUHS or the Department of Defense has been given or should be inferred.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017626117/-/DCSupplemental.
Data Availability.
RNA-seq data that support the findings of this study are available in the Gene Expression Omnibus (GEO) under accession number GSE154011.
References
- 1.Cheng T., et al. , Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287, 1804–1808 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Zou P., et al. , p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell 9, 247–261 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Hock H., et al. , Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431, 1002–1007 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Ficara F., Murphy M. J., Lin M., Cleary M. L., Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell 2, 484–496 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., et al. , PR-domain-containing Mds1-Evi1 is critical for long-term hematopoietic stem cell function. Blood 118, 3853–3861 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishikata I., et al. , A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood 102, 3323–3332 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Pinheiro I., et al. , Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150, 948–960 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Zhou B., et al. , PRDM16 suppresses MLL1r leukemia via intrinsic histone methyltransferase activity. Mol. Cell 62, 222–236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shing D. C., et al. , Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J. Clin. Invest. 117, 3696–3707 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Y., Jenkins N. A., Copeland N. G., Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood 106, 3932–3939 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu T., et al. , PRDM16s transforms megakaryocyte-erythroid progenitors into myeloid leukemia-initiating cells. Blood 134, 614–625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corrigan D. J., et al. , PRDM16 isoforms differentially regulate normal and leukemic hematopoiesis and inflammatory gene signature. J. Clin. Invest. 128, 3250–3264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuikov S., Levi B. P., Smith M. L., Morrison S. J., Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat. Cell Biol. 12, 999–1006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguilo F., et al. , Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood 117, 5057–5066 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kühn R., Schwenk F., Aguet M., Rajewsky K., Inducible gene targeting in mice. Science 269, 1427–1429 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Ema H., Nakauchi H., Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 95, 2284–2288 (2000). [PubMed] [Google Scholar]
- 17.Zhang C. C., Lodish H. F., Murine hematopoietic stem cells change their surface phenotype during ex vivo expansion. Blood 105, 4314–4320 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikkola H. K., Orkin S. H., The journey of developing hematopoietic stem cells. Development 133, 3733–3744 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Zhao J. L., Baltimore D., Regulation of stress-induced hematopoiesis. Curr. Opin. Hematol. 22, 286–292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison D. E., Lerner C. P., Most primitive hematopoietic stem cells are stimulated to cycle rapidly after treatment with 5-fluorouracil. Blood 78, 1237–1240 (1991). [PubMed] [Google Scholar]
- 21.Jankovic V., et al. , Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 1260–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermes I., Haanen C., Steffens-Nakken H., Reutelingsperger C., A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods 184, 39–51 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Min I. M., et al. , The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell 2, 380–391 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Okada S., Fukuda T., Inada K., Tokuhisa T., Prolonged expression of c-fos suppresses cell cycle entry of dormant hematopoietic stem cells. Blood 93, 816–825 (1999). [PubMed] [Google Scholar]
- 25.McKinney-Freeman S., et al. , The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell 11, 701–714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Busch K., et al. , Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Jude C. D., et al. , Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell 1, 324–337 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMahon K. A., et al. , Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 1, 338–345 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Rossi L., et al. , Less is more: Unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell 11, 302–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P., Jenkins N. A., Copeland N. G., A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13, 476–484 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semerad C. L., Mercer E. M., Inlay M. A., Weissman I. L., Murre C., E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. U.S.A. 106, 1930–1935 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data that support the findings of this study are available in the Gene Expression Omnibus (GEO) under accession number GSE154011.