Significance
FGF23 is a circulating hormone produced in bone tissue that acts in the kidney to increase phosphate excretion and vitamin D metabolism essential for calcium absorption. Mutations that elevate FGF23 abundance cause autosomal dominant hypophosphatemic rickets or tumor-induced osteomalacia. Loss of FGF23, on the other hand, causes increased phosphate levels, such as in familiar tumor calcinosis. Here we show that the region in FGF23 responsible for KLA binding contains two distinct binding sites for KLA, demonstrating that FGF23 is able to simultaneously bind and assemble two KLA molecules. An engineered protein containing the identified KLA ligand acts as a potent FGF23 inhibitor offering new pharmacological intervention for treating diseases caused by excessive FGF23 abundance or activity.
Keywords: endocrine FGF, phosphorylation, cell signaling, surface receptors, biological inhibitor
Abstract
The three members of the endocrine-fibroblast growth factor (FGF) family, FGF19, 21, and 23 are circulating hormones that regulate critical metabolic processes. FGF23 stimulates the assembly of a signaling complex composed of α-Klotho (KLA) and FGF receptor (FGFR) resulting in kinase activation, regulation of phosphate homeostasis, and vitamin D levels. Here we report that the C-terminal tail of FGF23, a region responsible for KLA binding, contains two tandem repeats, repeat 1 (R1) and repeat 2 (R2) that function as two distinct ligands for KLA. FGF23 variants with a single KLA binding site, FGF23-R1, FGF23-R2, or FGF23-wild type (WT) with both R1 and R2, bind to KLA with similar binding affinity and stimulate FGFR1 activation and MAPK response. R2 is flanked by two cysteines that form a disulfide bridge in FGF23-WT; disulfide bridge formation in FGF23-WT is dispensable for KLA binding and for cell signaling via FGFRs. We show that FGF23-WT stimulates dimerization and activation of a chimeric receptor molecule composed of the extracellular domain of KLA fused to the cytoplasmic domain of FGFR and employ total internal reflection fluorescence microscopy to visualize individual KLA molecules on the cell surface. These experiments demonstrate that FGF23-WT can act as a bivalent ligand of KLA in the cell membrane. Finally, an engineered Fc-R2 protein acts as an FGF23 antagonist offering new pharmacological intervention for treating diseases caused by excessive FGF23 abundance or activity.
The large family of fibroblast growth factors (FGFs) has been known for its important roles in regulating critical cellular processes during embryonic development and homeostasis of normal tissues (1–3). While most FGFs act as cytokines or hormonelike proteins that mediate their pleiotropic cellular processes by binding to cell surface receptors endowed with intrinsic tyrosine kinase activity (FGFRs), a subfamily of FGFs (FGF 11–14) was shown to be uniquely expressed intracellularly. The mechanism of action and physiological roles of intracellular FGFs are poorly understood (4–6).
In contrast to most receptor tyrosine kinases (RTKs) that are activated by a single ligand molecule that binds with high affinity to the extracellular domain of its cognate RTK with a dissociation constant in the subnanomolar range, the binding affinities of FGFs to FGFRs are, at least, 1,000–10,000 fold weaker with dissociation constants in the submicromolar range (7–9). The weak binding affinities toward FGFRs of the largest subfamily of FGF molecules designated canonical FGFs are offset by interactions with cell surface heparan sulfate proteoglycans (HSPGs). Both biochemical and structural studies revealed how multiple interactions between heparin or HSPG with both FGF and FGFR mediate tight association enabling robust receptor dimerization and tyrosine kinase activation (10, 11).
The three endocrine FGFs, FGF19, 21, and 23 are part of an additional subfamily of FGF molecules. Endocrine FGFs function as circulating hormones that play essential roles in the control of various metabolic processes (12). In addition to the conserved FGF-domain found in all FGF ligands, endocrine FGFs contain unique C-terminal tails (CTs) composed of 46 (FGF19), 34 (FGF21), or 89 (FGF23) amino acids that serve as specific and high-affinity ligands for the two members of the Klotho family of surface receptors. It was shown that KLA serves as a high-affinity receptor for FGF23 while β-Klotho (KLB) functions as a high-affinity surface receptor for both FGF19 and FGF21 (13–16). Structural analyses of free and ligand-occupied Klotho proteins revealed the molecular basis underlying the specificity and high affinity of KLA and KLB toward endocrine FGFs. It also showed that Klotho proteins function as the primary receptors for endocrine FGFs whereas FGFR functions as a catalytic subunit that mediates cell signaling via its tyrosine kinase domain (8, 17, 18). Accordingly, endocrine FGFs stimulate their cellular responses by forming a ternary complex with Klotho proteins and FGFRs to induce receptor dimerization, tyrosine kinase activation, and cell signaling. Unlike FGFRs that are ubiquitously expressed, the expressions of KLA and KLB are restricted to specific tissues and organs to enable precise targeting of endocrine FGFs to stimulate their physiological responses in specific cells and tissues (19–22). The ability of endocrine FGFs to circulate is attributed to the loss of conserved heparin binding sites that are essential for the function of canonical FGFs (23).
FGF23 is a 32-kDa glycoprotein, mainly produced in the bone by osteoblasts and osteocytes, that serve as a key hormone in regulating phosphate homeostasis, vitamin D, and calcium metabolism (24, 25). Circulating levels of physiologically active FGF23 are regulated by proteolytic cleavage to produce a FGF23 molecule lacking its unique CT (26, 27). The cleavage resulting in FGF23 inactivation prevents assembly and stimulation of the FGF23/FGFR/KLA complex. Additionally, the processing of FGF23 includes several posttranslational modifications which affect its stability and susceptibility toward proteolysis. Secreted FGF23 was shown to be O-glycosylated in its C-terminal cleavage site (28, 29) to protect the protein from C-terminal cleavage. In order for the cleavage site to be exposed, FGF23 has to be first phosphorylated in this region (30). Phosphorylation prevents glycosylation and exposes the cleavage site to proteolysis.
In this paper, we demonstrate that the CT of FGF23 contains two tandem repeats and that each repeat binds with high affinity to KLA. This contrasts with FGF19 and FGF21, whose CTs contain a single binding site to KLB. Engineered FGF23 variants containing each of the two repeats individually or both repeats bind specifically to KLA and stimulate cell signaling to a similar extent. We also demonstrate that two cysteine residues flanking the second repeat form a disulfide bridge in FGF23 secreted by mammalian cells. However, both oxidized or unbridged forms of FGF23 exhibit similar KLA binding characteristics and similar cellular stimulatory activities. We also show that FGF23-WT induces mitogen-activated protein kinase (MAPK) activation in cells expressing chimeric KLA-FGFR proteins and use TIRFM imaging of individual KLA molecules on the cell surface to demonstrate that FGF23 has the capacity for simultaneous binding to two KLA molecules. These insights reveal the complexity of FGF23 regulation and its role in assembling the FGF23/FGFR/KLA signaling complex.
Results
The C-Terminal Tail of Mammalian FGF23 Contains Two Separate KLA Binding Regions.
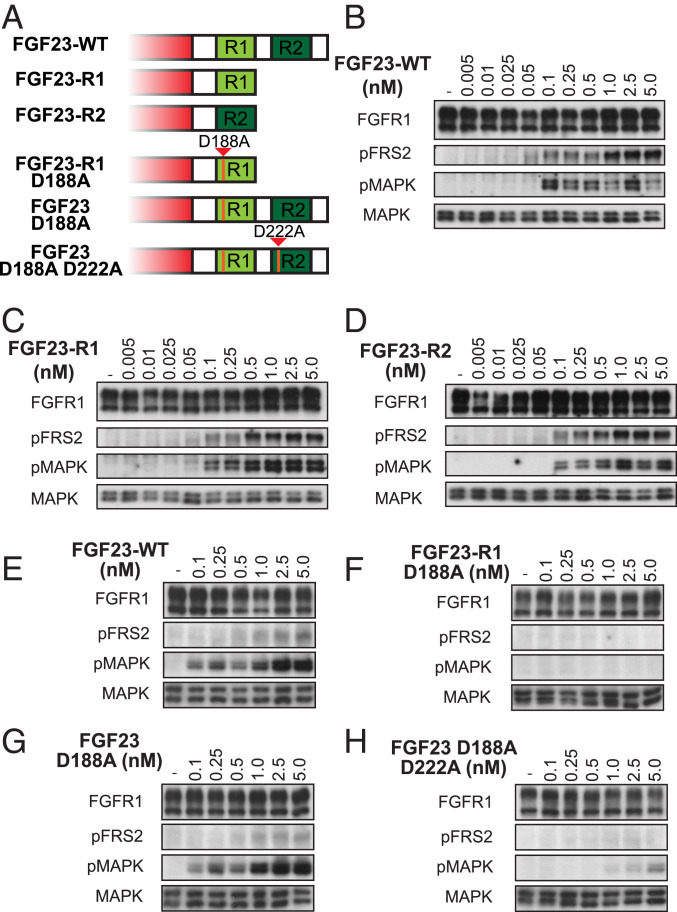
The crystal structures of the CTs of FGF19 or FGF21 bound to the extracellular region of KLB revealed conserved interactions along elongated interfaces that span both glycoside hydrolase like domains D1 and D2 (also designated KL1 and KL2 domains) of KLB (8, 18). A FGF23 deletion mutant lacking 46 C-terminal amino acids was previously shown to be biologically active (31). This deletion mutant was applied in the structural analysis of a complex containing the extracellular region of KLA (sKLA) and FGFR1c extracellular domain that revealed conserved interactions primarily with D1 of sKLA (17). Comparison of the primary structures of FGF19, FGF21, and FGF23 (Fig. 1A) shows that the C-terminal tails of FGF19 and 21 contain 46 and 34 amino acids, respectively, while FGF23 contains a longer CT of 89 amino acids. Inspection of the primary structures shows that unlike FGF19 and FGF21, the CT of FGF23 contains two homologous tandem repeats (Fig. 1B). Each repeat contains a DPL/F motif which is crucial for maintaining the compact and rigid structure necessary for binding to the D1 (KL1) site as well as a cluster of basic residues that bind to the D2 (KL2) site, suggesting that a single FGF23 molecule may possess two separate binding regions for KLA. It is noteworthy that, while all vertebrate FGF23 proteins have long CTs, only mammals have a second repeat homologous to the Klotho binding regions of FGF19, 21, and 23 (Fig. 1B and SI Appendix, Fig. S1).
Fig. 1.
The CT of FGF23 contains two distinct regions that specifically bind to KLA. (A) Schematic of FGF19, FGF21, FGF23, and FGF1. Signal peptide (SP) is colored in orange, the FGFR binding domain in red, the KLB binding regions in the CTs of FGF19 and 21 are shown in cyan, and the tandem repeats in the CT of FGF23 are shown in light green and dark green, respectively. (B) Sequence alignments and comparison of the CTs of human FGF19 and FGF21 to the first (R1) and second repeats (R2) of human FGF23 CT. The DPL motifs of FGF19, 21, and 23, critical for binding to D1 of KLA and KLB are highlighted in red and the sucrose phosphate synthase sugar mimicking SepProSer (SPS) motifs in FGF19 and FGF21 critical for binding to the pseudosubstrate binding pocket in D2 of KLB are highlighted in green. (C) Representative BLI sensorgrams illustrating the binding of GST fusion of the CT of FGF23 (GST-FL), R1 (GST-R1), and R2 (GST-R2) to sKLA. Biosensors coated with anti-GST antibodies were used to capture GST-fused FGF23 peptide fragments and dipped into solutions containing a series of concentrations of sKLA (6.25, 12.5, 25, 50, 100, and 200 nM). Sensorgrams were fitted with a 1:1 ligand:receptor binding model (colored lines) to calculate dissociation constants and kinetic parameters. (D) Summary of kinetic parameters and dissociation constants of BLI measurements. Data are presented as mean values ± SD from three independent experiments.
To examine the significance of each of the two FGF23 repeats in KLA binding and receptor activation, we applied biolayer interferometry (BLI) analyses to measure the kinetic parameters and dissociation constants of each repeat alone or the entire CT of FGF23 toward sKLA. To that end, glutathione S-transferase (GST)-fusion proteins expressing the full-length (FL) tail of FGF23 (amino acids S180–I251), the first repeat R1 (amino acids S180–S205), or the second repeat R2 (amino acids S212–T239) were produced in Escherichia coli (SI Appendix, Fig. S2) and immobilized on BLI sensors. sKLA was produced in human embryonic kidney 293 (HEK293) Epstein–Barr nuclear antigen (EBNA) cells and used as an analyte in the BLI measurements (see Materials and Methods).
The results from the BLI measurements (Fig. 1C) show that GST fusion proteins with each single repeat or both repeats bind to sKLA with similar kinetic parameters and dissociation constants (Kd) of 15–20 nM (Fig. 1D), indicating that both R1 and R2 function as distinct bona fide ligands of KLA and that FGF23-WT possesses two distinct binding sites for KLA. Since the BLI measurements clearly show that the FL tail of FGF23 as well as R1 and R2 form stable complexes with sKLA, we next expressed and purified FGF23 with the FL tail (FGF23-WT), FGF23 variants containing only one of the two repeats, FGF23-R1, and FGF23-R2, as well as FGF23 variants with one or both repeats inactivated by a point mutation in the DPL motif (D188A in R1 and D222A in R2) (Fig. 2A) and examined their ability to stimulate cell signaling. HEK293 cells coexpressing FGFR1c and KLA were stimulated with increasing concentrations of the different FGF23 variants (as indicated in Fig. 2 B–H) for 10 min at 37 °C and lysates of unstimulated or ligand-stimulated cells were subjected to immunoblotting with anti-pFRS2α antibodies to monitor its phosphorylation as well as with anti-pMAPK antibodies and MAPK antibodies to monitor MAPK stimulation and MAPK expression, respectively. The results presented in Fig. 2 B–D show that FGF23-WT, FGF23-R1, and FGF23-R2 activate cell signaling to a similar extent as revealed by tyrosine phosphorylation of FRS2α and the activation of MAPK response (saturation is reached at 0.5–1.0-nM and at 0.1–0.5-nM ligand concentration, respectively). By contrast, tyrosine phosphorylation of FRS2 and MAPK response were not detected in cells stimulated with a FGF23-R1 D188A mutant (Fig. 2F). Interestingly, FGF23 D188A, a mutant with inactive R1 and a functional R2, stimulated tyrosine phosphorylation of FRS2α and activation of MAPK response to the same extent as FGF23-WT (Fig. 2G) indicating that FGF23 is capable of utilizing R2 alone, in the context of FL CT (separated by 50 amino acids from the FGF moiety) for KLA binding and FGFR activation. This finding also raises questions whether the crystal structure of the ternary FGF23-R1/sKLA/FGFR1c complex (17) may represent an oversimplified picture which does not depict the heterogeneity in the interactions between FGF23 and KLA. Finally, in cells treated with FGF23 in which both R1 and R2 are inactivated by D188A/D222A double mutations, tyrosine phosphorylation of FRS2α is completely abolished, and MAPK activation is barely detectable (Fig. 2H).
Fig. 2.
FGF23-WT and FGF23 variants with either R1 or R2 induce similar cellular responses. (A) Schematic of the CTs of FGF23 variants used for cell stimulation. R1 and R2 are shown in light green and dark green, respectively. Mutations in R1 and/or R2 of the FGF23 CT that abolish binding to KLA, are marked in orange. (B–H) Comparison of FGF23-WT or FGF23 variants induced tyrosine phosphorylation of FRS2α and mitogen-activated protein kinase (MAPK) response. HEK293 cells stably expressing FGFR1c together with KLA were left unstimulated or stimulated with increasing concentrations of FGF23-WT or FGF23 variants as indicated for 10 min at 37 °C. Cell lysates were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) and analyzed for tyrosine phosphorylation of FRS2α and MAPK activation by immunoblotting with antibodies for pFRS2 and pMAPK, respectively, and with anti-MAPK as a control.
R2 of FGF23 Functions as an Antagonist of FGF23-Induced Cell Signaling.
Since the C-terminal region of FGF23 binds tightly to KLA and does not interact with FGFR, it may function as a competitor of FGF23 binding to KLA and, consequently, as an inhibitor of FGF23-induced cell signaling. Previous studies have shown that a FL C-terminal peptide (31, 32) and a R1 peptide (31) can antagonize FGF23 activation both in vitro and in vivo.
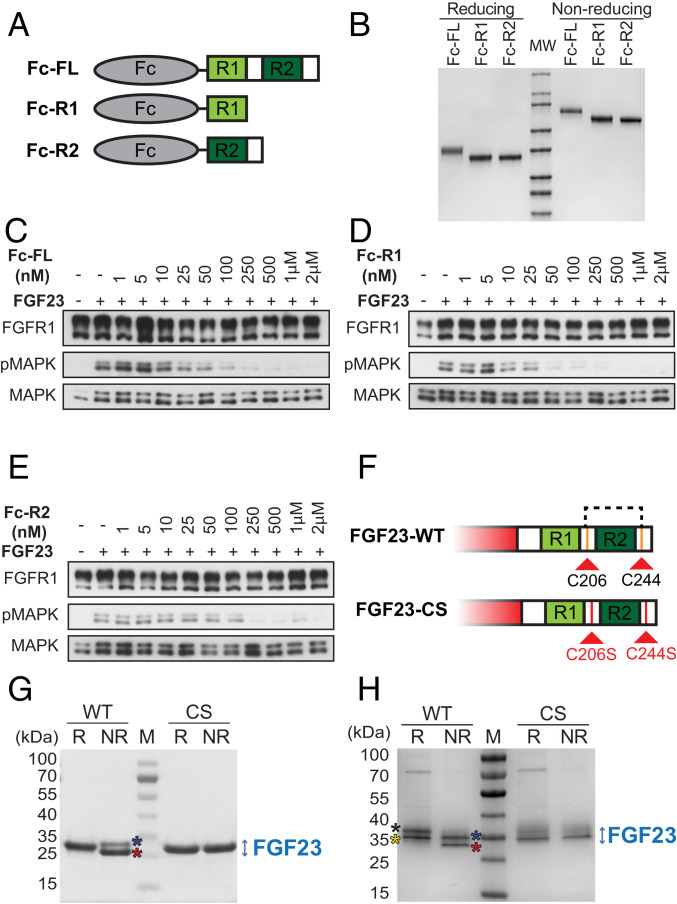
To test whether R2 (S212–T239) exerts an antagonistic activity on FGF23 signaling and to compare its efficiency to those of the FL FGF23 C-tail (S180–I251) or R1 peptide (S180–S205), we expressed and purified these peptides in a form of Fc fusion proteins, designated Fc-FL, Fc-R1, and Fc-R2 (Fig. 3 A and B) and explored their effect upon FGF23-induced stimulation of HEK293 cells coexpressing KLA and FGFR1c. Cells were incubated with increasing concentrations (as indicated, Fig. 3 C–E) of individual Fc-fusion protein followed by stimulation with FGF23-WT for 10 min. Lysates from unstimulated or FGF23-stimulated cells were subjected to immunoblotting with anti-pMAPK antibodies to determine MAPK response or antibodies to FGFR1 and MAPK as controls for protein loading. The experiment presented in Fig. 3 C–E shows that Fc-FL, Fc-R1, and Fc-R2 were able to inhibit FGF23-induced MAPK stimulation with a similar concentration that inhibits response by 50% within the 25–50-nM range. These results demonstrate that Fc-R2 antagonizes FGF23-WT-induced MAPK response similar to the antagonistic activities of Fc-R1 or Fc-FL. The ability of Fc-R2 to inhibit the formation of the KLA-FGFR signaling complex establishes it as a potential therapeutic for diseases resulting from increased FGF23 signaling.
Fig. 3.
Similar inhibition of FGF23-induced stimulation of cells treated with Fc-R2 to cells treated with Fc-FL or Fc-R1 and cysteine residues flanking R2 in FGF23 CT form an intramolecular disulfide bridge. (A and B) Schematic (A) and SDS/PAGE analyzes (B) under reducing (R) or nonreducing (NR) conditions of Fc-FL, Fc-R1, or Fc-R2. The Fc moiety is colored in tan. R1 and R2 are colored in light green and dark green, respectively. (C–E) HEK 293 cells stably expressing FGFR1c and KLA were incubated with increasing concentrations (as indicated) of Fc-FGF23 FL tail (Fc-FL), Fc-R1, or Fc-R2 for 45 min at 37 °C. Cells were then stimulated with FGF23-WT for additional 10 min, and cell lysates were subjected to SDS/PAGE and analyzed for MAPK stimulation by immunoblotting with anti-pMAPK antibodies. Anti-FGFR1 and anti-MAPK antibodies were used as control for protein loading. (F) Schematic of CTs of FGF23-WT and FGF23-CS. R1 and R2 are shown in light green and dark green, respectively. Cysteine residues (C) are highlighted in orange. Serine residues (S) are highlighted in red. (G) SDS/PAGE analyses of FGF23-WT and FGF23-CS mutant expressed in E. coli, under R and NR conditions. FGF23-WT and FGF23-CS were expressed and purified as described in Materials and Methods. While FGF23-CS migrates on SDS/PAGE as a single band under both R and NR conditions, FGF23-WT migrates as two distinct bands under NR conditions. Both proteins were excised from the gel and subjected to mass-spectrometric (MS) analysis. (H) SDS/PAGE analyses of FGF23-WT or FGF23-CS mutant expressed in Expi293F cells under R and NR conditions. Unlike E. coli produced FGF23, mammalian produced FGF23 is O-linked glycosylated. Under R conditions, the upper band (black asterisk) and lower band (yellow asterisk) present an O-linked glycosylated form and a non- or poorly glycosylated form of FGF23, respectively (SI Appendix, Fig. S3B). Under NR conditions, both glycosylated (blue asterisk) and nonglycosylated FGF23 (red asterisk) migrate faster than the reduced proteins on SDS/PAGE due to the formation of intramolecular disulfide bridge. Both FGF23-WT and FGF23-CS (expressed in Expi293F cells) migrate on SDS/PAGE under R and NR conditions as two distinct bands. The upper and lower bands represent O-linked glycosylated and nonglycosylated forms of the ligand, respectively.
Cysteine Residues Flanking R2 of FGF23 Form a Disulfide Bridge.
Amino acid sequence alignments of FGF23 from different species, see SI Appendix, Fig. S1) show that, in mammals, R2 is flanked by two cysteine residues, e.g., Cys206 and Cys244 in human FGF23 (Fig. 3F). To determine whether these cysteine residues form a disulfide bridge, we expressed FGF23-WT in E. coli and analyzed the refolded and purified protein by SDS/PAGE under both R and NR conditions in comparison to those of a mutant FGF23 in which both cysteines are substituted by serine residues (FGF23-CS).
The experiment presented in Fig. 3G shows that FGF23-WT migrates on SDS/PAGE as a distinct single band under the R condition and as two bands (marked with blue and red asterisks) under the NR condition. The FGF23-CS mutant, on the other hand, migrates on SDS/PAGE as a single distinct band under both reducing (R) and non-reducing (NR) conditions. To determine if either of the two bands of FGF23-WT contains intramolecular disulfide bonds under the NR condition, each of the two bands were excised from the gel, subjected to trypsin and endoproteinase GluC digestion, and analyzed by MS to detect disulfide-linked peptides (33, 34). The MS analysis revealed that the lower band (Fig. 3G, red asterisk, see SI Appendix, Fig. S3 A and B) contains peptides with an intramolecular disulfide bond between Cys206 and Cys244. Only trace amounts of these peptides were detected in the proteolytic digest of the upper band (Fig. 3G, blue asterisk, see SI Appendix, Fig. S3B). Based on these results we conclude that while the majority of FGF23-WT expressed in bacteria becomes oxidized during refolding to form a disulfide bond between Cys206 and Cys244, the two cysteines are not bridged in a subpopulation of refolded FGF23 molecules expressed in E. coli.
We next expressed FLAG-tagged FGF23-WT and its CS mutant in Expi293F cells (Fig. 3H) and purified both ligands using affinity chromatography followed by size exclusion chromatography (Materials and Methods). The SDS/PAGE analysis presented in Fig. 3H shows that FGF23-WT produced in mammalian cells (FGF23-WT) migrates as two distinct bands under both R and NR conditions. Unlike bacterially expressed protein, FGF23 expressed in Expi293F cells is O-glycosylated and the two distinct bands visualized by SDS/PAGE under R and NR conditions reveal different glycosylation forms of FGF23-WT which was confirmed by in vitro treatment with O-glycosidase and α-(2→3,6,8,9)-neuraminidase (SI Appendix, Fig. S4 and Materials and Methods). We also showed that the FGF23-CS mutant expressed in Expi293F cells (FGF23-CS) migrated on SDS/PAGE as two distinct bands under both R and NR conditions due to differential glycosylation (SI Appendix, Fig. S4B). The upper band of FGF23-CS is more smeared (Fig. 3H) than the corresponding band of FGF23-WT, suggesting potential heterogeneity in glycosylation patterns. MS analyses showed that FGF23 produced in Expi293F cells contains peptides with a disulfide bridge connecting Cys206 and Cys244. As was proposed that O-linked glycosylation protects FGF23 from proteolysis, we asked whether Cys206–Cys244 disulfide bridging affects FGF23 accessibility to proteolytic digestion. The experiment presented in SI Appendix, Fig. S5 shows the results of limited proteolysis experiment with FGF23-WT and the CS mutant. Limited proteolysis of both proteins with various enzymes (as indicated) was performed using the Proti-Ace Kit (Hampton Research) under the manufacturer’s protocol. Digested samples were subjected to SDS/PAGE followed by Coomassie blue staining to visualize the digested products. Based on the pattern of the bands visualized by SDS/PAGE, we concluded that the protease-digested products of FGF23-WT and FGF23-CS are similar to each other and, therefore, Cys206–Cys244 disulfide bridging does not have a major impact on FGF23 accessibility to proteolytic digestion.
To explore the role of Cys206 to Cys244 disulfide formation on FGF23 binding to soluble KLA, we used BLI measurements to compare the kinetic parameters and dissociation constants of FGF23-WT to those of FGF23-CS, FGF23 D188A, and FGF23-CS D188A expressed in Expi293F cells. The experiment presented in SI Appendix, Fig. S6A shows that all four FGF23 variants exhibit similar binding kinetics and dissociation constants toward sKLA in the range of 13–18 nM (Table 1). Furthermore, stimulation of HEK293 cells expressing KLA and FGFR1c with increasing concentrations of FGF23-WT or FGF23-CS revealed similar profiles of tyrosine phosphorylation of FRS2α, MAPK response, as well as similar serine phosphorylation of FGFR1c by activated MAPK, a feedback mechanism that leads to the attenuation of ligand stimulation (SI Appendix, Fig. S6 B and C). These results emphasize the ability of R2 in its oxidized form to create a ternary active complex with KLA and FGFR1c.
Table 1.
Binding of FGF23 variants to sKLA
| KD (nM) | kon (×105 M−1 s−1) | koff (×10−3 s−1) | |
| FGF23-WT | 14 ± 3 | 2.6 ± 0.5 | 3.4 ± 0.1 |
| FGF23-CS | 18 ± 4 | 2.3 ± 0.4 | 4.2 ± 0.4 |
| FGF23 D188A | 13 ± 3 | 2.8 ± 0.5 | 3.6 ± 0.2 |
| FGF23-CS D188A | 18 ± 3 | 2.5 ± 0.3 | 4.4 ± 0.3 |
| FGF23 D188A D222A* | n/a | n/a | n/a |
No binding detected (no change in BLI signal).
FGF23 Can Act as a Bivalent Ligand of KLA Molecules Expressed on the Cell Membrane.
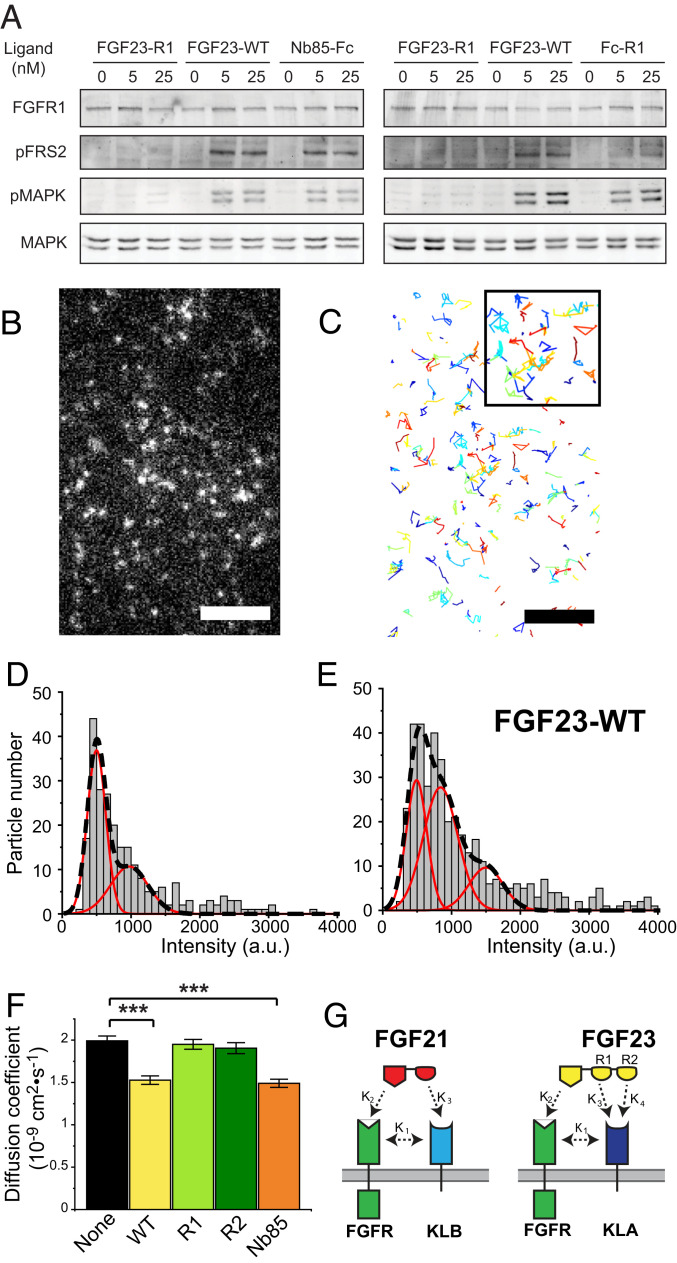
We next examined the possibility of whether a single FGF23-WT molecule is capable of binding via its R1 and R2 regions of the CT to two KLA molecules. In other words, the aim of this experiment is to test the ability of FGF23-WT to function as a bivalent ligand of KLA molecules located on the cell membrane. To address this question, we constructed and expressed a chimeric receptor molecule composed of the extracellular domain of KLA fused to the transmembrane and cytoplasmic domain of FGFR1 in L6 cells. We hypothesized that FGF23-WT may function as a bivalent ligand capable of inducing dimerization of the chimeric receptor molecules, stimulating their tyrosine kinase activity and subsequent activation of downstream signaling. As positive controls we analyzed the activity of a dimeric Fc-nanobody that binds specifically to the extracellular domain of KLA and a dimeric Fc-R1 fusion protein for their ability to stimulate tyrosine phosphorylation of FRS2 and MAPK responses in these cells. Cells expressing the chimeric KLA-FGFR1c receptor were stimulated with 5 or 25 nM of FGF23-WT, FGF23-R1, the bivalent anti-KLA nanobody (Nb85-Fc), and Fc-R1 for 10 min at 37 °C. Lysates from unstimulated or ligand-stimulated cells were subjected to SDS/PAGE analysis followed by immunoblotting with anti-pFRS2α antibodies to monitor its phosphorylation, anti-pMAPK antibodies to monitor MAPK activation, or anti-MAPK antibodies and anti-FGFR1 antibodies as controls for protein loading. The experiment presented in Fig. 4A shows that both mammalian (Left) and E. coli (Right) produced FGF23-WT as well as bivalent KLA nanobody and Fc-R1 protein induce robust activation of MAPK response. By contrast the monovalent FGF23-R1 variant (produced in E. coli or mammalian cells) failed to simulate MAPK response. These experiments demonstrate that FGF23-WT is capable of stimulating the dimerization of KLA molecules located on the cell membrane via its CT (Fig. 4D).
Fig. 4.
FGF23-WT binds simultaneously to two KLA molecules, expressed on cell membranes. (A) L6 cells stably expressing KLA-FGFR1c chimeric receptors were left unstimulated or stimulated with FGF23-WT or FGF23-R1 expressed in Expi293F cells (Left), or FGF23-WT and FGF23-R1 expressed in E. coli (Right), Nb85-Fc fused (Left) or Fc-R1 (Right) for 10 min at 37 °C. Cell lysates of unstimulated or ligands stimulated cells were subjected to SDS/PAGE and analyzed for FRS2α phosphorylation and the activation of MAPK by immunoblotting with anti-pFRS2 and anti-pMAPK antibodies, respectively, and with anti-MAPK as a control. (B) Expanded view of single HaloTag-KLA particles on the surface of living L6 cells imaged by TIRFM. The HaloTag on the extracellular side of KLA was labeled with a cell-impermeant Alexa488 HaloTag ligand. Particle density is 0.21 particles/μm2. A single frame (100-ms exposure) at the start of a 10-Hz recording is shown. (Scale bar, 5 μm.) (C) Automated detection and tracking of moving HaloTagKLA particles during a 10-s recording period. Single-particle tracking was performed as described in Materials and Methods. Inset, higher magnification. (D and E) Representative intensity distributions of HaloTag-KLA in a cell left unstimulated (Left) or stimulated (Right) with FGF23-WT (25 nM) for ∼10 min, and particle densities were 0.26 and 0.05 particles/μm2 for unstimulated and stimulated conditions, respectively. Intensities represent the volume under two-dimensional (2D) Gaussian fits of the fluorescence of particles. Intensities were taken from the beginning (three frames) of each recording, and their distribution was fitted with a mixed Gaussian model. Black dashed lines, mixed fit. Red lines, individual components. (F) Diffusion coefficient of HaloTag-KLA particles calculated from their mean square displacement (MSD) in unstimulated cells (18 cells, five transfections) and cells stimulated with FGF23-WT (25 nM; 16 cells, four transfections), FGF23-R1 (25 nM; 14 cells, three transfections), FGF23-R2 (25 nM; 16 cells, three transfections), and Nb85-Fc (2.5 nM; 16 cells, three transfections). Error bars indicate mean ± SE, ***P < 0.0001 by Student’s t test. (G) A schematic depicting the bivalency of the CT of FGF23 binding to KLA.
We next investigated FGF23 stimulation of KLA dimerization using a single-molecule imaging approach. We sought to visualize KLA molecules on the cell membrane by labeling KLA fused to an N-terminal (extracellular) HaloTag with a cell-impermeant fluorescent HaloTag ligand Alexa488. L6 cells expressing low levels of HaloTag-KLA were briefly labeled (15 min at 37 °C) with Alexa488, and individual fluorescent particles were imaged using TIRF microscopy to visualize individual KLA molecules on the cell surface (35–37). Fig. 4B shows a representative TIRF microscopy image of a low expressing cell with a particle density of 0.21 particles/μm2, which is similar to the densities reported in single-molecule imaging studies of receptor dimerization (<0.45 particles/μm2; refs. 38, 39). Particles were automatically detected and tracked (40, 41) to delineate their movements on the cell surface (Fig. 4C). Consistent with the particles representing single molecules, they often photobleached in a single step. The intensity distribution of particles in unstimulated cells could be fitted with a mixed Gaussian model, comprising a major peak with an intensity (498 ± 16 a.u.) similar to that of free dye absorbed to glass (554 ± 16 a.u.; see SI Appendix, Fig. S7)—thus, likely corresponding to monomeric HaloTag-KLA–and a minor peak with roughly twice the intensity (973 ± 129 a.u). This second smaller peak may (Fig. 4D) reflect the dynamic equilibrium between monomers and dimers based on the intensity of individual tracks over time, which occasionally showed transient doubling (SI Appendix, Fig. S8). Visual inspection of recordings also showed transient merging of particles (SI Appendix, Fig. S9), although it is possible that these apparent merging events merely reflected the colocalization of particles rather than their association due to the diffraction limit of light. In contrast, when cells were stimulated with FGF23-WT, the intensity distribution became shifted to the right with the second (i.e., dimer) peak (840 ± 57 a.u.) growing more prominent and a third peak forming with three times the monomer intensity (1502 ± 89 vs. 492 ± 23 a.u.). In addition to the quantized increase in particle intensity induced by FGF23-WT stimulation (Fig. 4E), the diffusion coefficients (Fig. 4F) of the particles calculated from their MSD indicated that their diffusion coefficient (mean ± SE = 1.99 ± 0.057 × 10−9 cm2·s−1) was similarly reduced (by 22–23%, P < 0.0001) by FGF23-WT binding (1.53 ± 0.050 × 10−9 cm2·s−1) as well as by binding of dimeric anti-KLA nanobody Nb85-Fc (1.49 ± 0.049 × 10−9 cm2·s−1), but not by the monovalent FGF23-R1 or FGF23-R2 variants (1.95 ± 0.059 and 1.91 ± 0.065 × 10−9 cm2·s−1, respectively). These results directly demonstrate that FGF23-WT acts as a bivalent ligand of KLA molecules on the surface of living cells.
Finally, we performed single-molecule imaging analysis of cells expressing fluorescently labeled Halo-tag-KLA to detect potential effects of disulfide bridge formation across R2 of FGF23 on the dynamic properties of KLA expressed in the cell membrane. The experiment presented in SI Appendix, Fig. S10 shows that FGF23-WT and FGF23-CS induce similar changes in the diffusion coefficient (SI Appendix, Fig. S10A) and in the fluorescence intensity distribution (SI Appendix, Fig. S10B) of fluorescently labeled Halo-tag-KLA.
Discussion
The current view of the mechanism of action of FGF23 and the other two members of the endocrine-FGF family of circulating hormones is that FGF23 binding promotes the assembly of a signaling complex on the cell membrane composed of FGFR and the KLA receptor. The FGF domain of FGF23 binds to the extracellular domain of FGFR and the CT binds to the extracellular domain of KLA. Biochemical and structural analyses revealed a similar mechanism of action of FGF19 and FGF21, i.e., the FGF moieties of FGF19 or FGF21 bind to the extracellular domains of FGFRs and their C-terminal regions bind to the extracellular domain of KLB. The prevailing thought was that Klotho proteins function as coreceptors of endocrine FGFs similar to the role played by HSPG in cell signaling by canonical FGFs. However, as the binding affinities of the CTs of endocrine FGFs toward Klotho receptors are 1,000–10,000 fold stronger than the binding affinities of their FGF moieties toward FGFRs, it became clear that Klotho proteins function as the primary surface receptors for endocrine FGFs whereas FGFRs function as a catalytic subunit of the assembled activated signaling complex (8). It was also proposed that HSPG molecules on the cell membrane may facilitate conversion of a ternary FGF23/KLA/FGFR complex into to a FGF23/KLA/FGFR hexamer with stimulated tyrosine kinase activity (17).
The schematic presented in Fig. 4G depicts interactions among endocrine-FGF molecules, FGFRs, and Klotho proteins. Three separate binding events were identified: dissociation constant K1 for heterodimerization of FGFR1c with KLB, dissociation constant K2 for binding of the FGF moiety of FGF21 to FGFR1c, and dissociation constant K3 for binding of the CT of FGF21 to KLB. Binding measurements of each separate association revealed that K1 is ∼1 μM, K2 is ∼100 μM, and K3 is in the ∼20-nM range. In this report we demonstrate that the CT of FGF23 contains in addition to the previously identified KLA binding site (R1), a second distinct binding site toward KLA designated R2. Engineered FGF23 containing a single KLA binding site, FGF23-R1 or FGF23-R2 as well as FGF23-WT (with both R1 and R2) bind to sKLA with similar dissociation constants and stimulate similar tyrosine phosphorylation of FRS2α and MAPK response in cells expressing FGFR1c together with KLA. We also show that a FGF23 variant with an inactive R1 in the context of FL CT utilizes R2 for KLA binding and stimulation of cell signaling by FGFR activation.
An important conclusion of this study is that FGF23-WT may act as a bivalent ligand of KLA, based on the dimerization and activation of a chimeric receptor composed of the extracellular domain of KLA fused to the cytoplasmic domain of FGFR1 and visualization of individual KLA molecules on the cell surface using TIRF microscopy before and after FGF23 stimulation. Previous studies reported that the soluble extracellular domain of KLA forms dimers (42, 43), but we were not able to detect dimerization of our own preparations of the sKLA. While the experiments presented in this report demonstrate that FGF23 is capable of stimulating KLA dimerization, it is not clear whether KLA dimerization will take place in cells coexpressing FGFRs. The schematic presented in Fig. 4G depicts interactions taking place among FGF23, FGFR1c, and KLA. A major difference between the interactions mediated by FGF23 to the interactions mediated by FGF21 (or FGF19) is that the CT of FGF23 contains tandem repeats designated R1 and R2 which function as distinct high-affinity ligands for KLA. Yet, since FGF23-R1, FGF23-R2, and FGF23-WT bind to sKLA with similar dissociation constants and are capable of inducing similar FGFR1c activation and cell signaling, it appears that a single R1 or R2 ligand is sufficient for KLA binding and cell stimulation. Moreover, even the bivalent FGF23-WT utilizes a single R1 or R2 for KLA binding and for cell activation. Furthermore, treating mice with either FGF23-WT or with a truncated FGF23-R1-like variant resulted in similar regulation of serum phosphate concentration demonstrating that a FGF23 molecule with a single KLA binding site is capable of stimulating an in vivo physiological response (31). The bivalency of FGF23 toward KLA may facilitate an efficient assembly of signaling complexes on the cell membrane composed of KLA and FGFRs. With a dissociation constant of K1 of ∼1μM for KLA binding to FGFR, a bivalent FGF23 molecule may stimulate dimerization between a population of preexisting KLA/FGFR heterodimers with either a free KLA molecule or with another pair of preexisting KLA/FGFR heterodimers. While the binding affinities of R1 and R2 to free KLA are very similar to each other, it is possible that FGF23-WT binding to FGFR1c/KLA may be more constrained and limited to interactions with R1 and that the preference of the R2 ligand is to bring together a free KLA molecule to the signaling complex. Accordingly, the architecture of a FGFR1c/KLA heterodimer may preferentially permit interactions with R1 while R2 flanked by two cysteines connected by disulfide bridge may bind to free KLA molecules or vice versa. Detailed understanding of the mode of action of FGF23 awaits future structural insights and determination of the in situ stoichiometry of FGF23/FGFR1c/KLA interactions and complexes. Comparison of the physiological properties of FGF23-WT to those of monovalent FGF23 variants may reveal potential biological differences in the actions and activities of FGF23 variants under normal in vivo physiological concentrations. Finally, we describe in this report an engineered Fc-R2 protein that functions as a FGF23 antagonist by blocking KLA binding and cell signaling via FGFR activation, offering a potential new pharmacological approach for treating phosphate metabolism disorders caused by excessive activity of FGF23.
Materials and Methods
Plasmid Construction.
A complementary DNA (cDNA) encoding for FL human KLA with a C-terminal HA-tag was amplified by PCR and subcloned into the lentiviral transfer plasmids, pLenti CMV Hygro DEST. In order to generate GST fusion proteins, DNA fragments of the CT of human FGF23, FL (aa 180–251), R1 (aa 180–205), and R2 (aa 212–243) were amplified by PCR and cloned into the pGEX4T1 vector (GE Healthcare). DNA fragments encoding FL human FGF23 (FGF23-WT; aa 25–251) as well as FGF23-R1 (aa 25–205) and FGF23-R2 (aa 25–179 fused to aa 212–243) were amplified by PCR and cloned into the bacterial expression plasmids pET-28a. To reduce proteolytic cleavage, arginine 179 was substituted by glutamine (R179Q) in all plasmids. The mammalian expression vector of FGF23 composed of a cDNA encoding the signal peptide of human FGF23 followed by a FLAG tag (DYKDDDDK) and human FGF23 (aa 25–251) was subcloned into a modified pOptiVec vector (pCMV). Expression vectors of FGF23 variants harboring point mutations or deletions within the C-tail region were generated following standard site-directed mutagenesis protocol (quick change). All FGF23 encoding plasmids harbor four mutations (R140A/R143A/R176Q/R179Q) to increase ligand stability. Expression vectors encoding Fc-fusion proteins were generated using cDNA fragments encoding the signal peptide of mouse immunoglobulin G 1 (IgG1) (aa 1–20) and human IgG1 (aa 223–449) connected to the CT of FGF23, FL (aa 180–251), R1 (aa 180–205), or R2 (aa 212–243) cloned into pCMV. Expression vectors for the chimeric receptor composed of the extracellular region of human KLA (aa 1–981) fused to the transmembrane and intracellular regions of human FGFR1c (aa 377–822) were cloned into the pBabe-Puro system. Expression vector encoding the entire extracellular domain of human KLA (aa 1–980) fused to a 6xhistidine tag (sKLΑ) was cloned into the mammalian expression vector pCEP4 (Thermo Fisher Scientific).
Protein Expression and Purification.
Purification of sKLA.
sKLA (aa 1–980) fused to a 6xHis tag was expressed in HEK293 EBNA cells and purified from the cell culture medium. For protein purification, cells were maintained in Pro293 serum-free medium for 6 d, and the harvested medium was centrifugated at 300 × g, filtered through a 0.45-μM membrane, and incubated with Ni sepharose excel resin (GE Healthcare) for 1 h at 4 °C. The resin was then washed with 25-mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) (pH 7.5) containing 150-mM NaCl and 10-mM imidazole. sKLA was eluted from the resin with the same buffer containing 300-mM imidazole. The eluted protein was diluted with 20-fold of 25-mM 2-amino-2-hydroxymethyl-1,3-propanediol (Tris) pH 8.0 and subjected to anion exchange chromatography (MonoQ 5/50 GL, GE Healthcare) with a linear NaCl gradient (0–0.4 M). Fractions containing sKLΑ were concentrated and applied to a Superose 6 column (GE Healthcare) equilibrated with 25-mM Hepes (pH 7.5) containing 150-mM NaCl.
Purification of GST-fusion proteins.
Plasmids encoding for GST-fusion proteins (GST-FL, GST-R1, and GST-R2) were expressed in BL21-Gold (DE3) competent cells (Agilent), and protein purification was conducted as previously described (8).
Purification of FGF23 variants expressed in E. coli.
Plasmids expressing the various FGF23 variants were expressed in E. coli BL21(DE3) cells. The ligands were purified from inclusion bodies followed by refolding as previously described (19). Refolded FGF23 proteins were captured on a heparin affinity HiTrap column (GE Healthcare), eluted using a linear NaCl gradient (0–2.0 M), and subjected to size-exclusion chromatography using HiLoad 26/600 Superdex 200 (GE Healthcare) with a buffer containing 25-mM Hepes and 150-mM NaCl at pH 7.5.
Purification of FLAG-tagged FGF23.
For the expression of FLAG-tagged FGF23 or its variants, plasmids were transfected into Expi293F cells (Thermo Fisher Scientific) cultured in 125-mL flasks in Expi293 expression medium according to manufacturer’s protocol. Cells were maintained in expression medium for 6 d, and the medium was collected and incubated with anti-FLAG M2 agarose affinity gel (MilliporeSigma) for 1 h at 4 °C. The gel was washed with 20 column volumes of 25-mM Hepes (pH 7.5) containing 150-mM NaCl, and the ligands were eluted with 100-mM glycine pH 3.0. The eluted fractions were immediately mixed with 1/10 volume of 1-M Tris⋅HCl (pH 8.0). Fractions containing FGF23 were concentrated and applied to a Superdex S200 Increase column (GE Healthcare) equilibrated with 25-mM Hepes buffer (pH 7.5) containing 150-mM NaCl as a final purification step.
Purification of Fc-FGF23 C-tail fragments.
C-tail fragments of FGF23 fused to IgG1 Fc were expressed in Expi293F cells (using the same protocol described above for the expression of FLAG-tagged FGF23). Proteins were purified using protein A-sepharose (Thermo Fisher Scientific) followed by size-exclusion chromatography using Superdex S200.
Cell growth medium.
HEK293 cells stably coexpressing WT FGFR1c and KLA, were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100-U/mL penicillin–streptomycin, and 0.1-mg/mL hygromycin, and 1-μg/mL puromycin.HEK293 EBNA cells expressing sKLA were grown in DMEM containing 10% FBS, 100-U/mL penicillin–streptomycin, 250-μg/mL G-418, and 200-μg/mL of hygromycin B. After cell density reached 70–80% confluency, the medium was changed to Pro293a-CDM (Lonza) supplemented with 100-U/mL penicillin–streptomycin. Expi293 cells were grown in Expi293 expression medium (Thermo Fisher Scientific). These cells were used for transient expression of the Fc-fusion FGF23 C-tail proteins and the FLAG-FGF23 molecules. L6 cells stably expressing KLA-FGFR1c chimeric receptors were grown in DMEM supplemented with 10% FBS, 100-U/mL penicillin–streptomycin and 0.5-µg/mL puromycin.
BLI Measurements.
Kinetic parameters and dissociation constants of sKLΑ binding to the various forms of FL FGF23 or GST-fused C-terminal fragments of FGF23 were studied using BLI. An Octet RED96 system (Pall FortéBio) equipped with anti-mouse IgG Fc biosensors was used to study interactions between KLA and FLAG-tagged FGF23. Biosensor tips were loaded with anti-FLAG M2 antibody (MilliporeSigma) at 5 μg/mL for 2 min, washed in BLI buffer (25-mM Hepes, 150-mM NaCl, pH 7.5, 0.002% Tween-20, 1-mg/mL bovine serum albumin) for 60 s, and then loaded with FLAG-tagged FGF23 at 5 μg/mL for 4 min. Alternatively, anti-GST biosensor tips were loaded with 5-μg/mL GST fused with various FGF23 C-terminal fragments for 15 s. Subsequently, ligand-loaded sensor tips were dipped into microplate wells containing different sKLA concentrations, ranging from 6.25 to 200 nM in twofold dilutions of BLI buffer. After each binding cycle, the sensor tips were regenerated with 10-mM glycine (pH 1.5). The collected data were referenced using a parallel buffer control subtraction, and sensorgrams were fitted globally to a 1:1 Langmuir binding model using FortéBio Data Analysis 10.0 software provided by the manufacturer.
Deglycosylation of FGF23 Expressed in Mammalian Cells.
Purified FGF23 variants were treated with O-glycosidase and α-(2→3,6,8,9)-neuraminidase (New England BioLabs) for 4 h at 37 °C as directed by manufacturer’s protocol.
Shotgun Proteomic Identification of the Disulfide Bridge.
Disulfide-linked peptide mapping was performed by following a published method (34), and the pLink software was used for the identification of these peptides (34). Briefly, gel bands were processed by following a standard gel-based digestion protocol (44) with the modifications of digestion condition in which pH 6.5 with 10-mM N-ethylmaleimide was used to avoid disulfide scrambling (34). Trypsin (Promega) digestion was performed at a 10-ng/μL concentration for the gel bands overnight and Glu-C (New England Biolab) at 5 ng/μL for 8 h. Approximately 0.5-μg peptide digest was used for each liquid chromatography–MS measurement, using the shotgun mode on the Orbitrap Fusion Lumos Tribrid MS (Thermo Fisher Scientific) instrument that was described previously (45). For pLink (33) identification, the combination of GluC and trypsin was specified, and up to three miss cleavages were allowed with all of the other settings kept as defaults. The MS/MS spectrum was annotated by pLabel (46).
Parallel Reaction Monitoring Quantification of the Disulfide Bridge.
Peptide samples of E. coli produced FGF23 were injected to monitor the MS quantitative response by parallel reaction monitoring (PRM) mode for relative quantification for the standard nonmiss cleavage peptide containing Cys206–Cys244 (disulfide-linked MTAPAPSCQE and GCRPFAK). The theoretical MS1 and MS2 m/z values for the linked peptide were generated by Skyline (42) and imported into the PRM method. The isolation window was set to be 1.4 m/z. The Orbitrap resolution for PRM was set at 30,000, AGC target 1.0e5, and maximum injection time 150 ms. A stepped higher energy collisional dissociation collison energy of 2% (centered at 28%) was used. The resultant PRM data were imported (47) to Skyline for manual inspection.
Limited Proteolysis.
Limited proteolysis of FGF23-WT and FGF23-CS that were produced in mammalian cells were performed using the Proti-Ace Kit (Hampton Research) under manufacturer's recommendations. Digested samples were analyzed by SDS/PAGE followed by Coomassie blue staining.
TIRF Microscopy.
For single-molecule imaging experiments, L6 cells were plated on 35-mm glass-bottom dishes (MatTek Corporation) at a density of 2.5 × 105 cells per dish and transfected with 0.25-μg HaloTagKLA plasmid the next day using Lipofectamine 3000 reagent (Invitrogen), according to the manufacturer’s instructions. Cells were labeled with 0.25-μM Alexa488 HaloTag ligand (Promega) for 15 min at 37 °C and then washed three times with phenol-red–free DMEM (imaging media). After labeling, cells were immediately imaged at 37 °C and 5% CO2 in a cage incubator (OkoLab) housing a Nikon Eclipse Ti2 microscope (Nikon) equipped with a motorized Ti-LA-HTIRF module with a 15-mW LU-N4 488 laser, using a CFI Plan Apochromat Λ 100×/1.45 oil TIRF objective and a Prime95B complementary metal oxide semiconductor camera (110-nm pixel size; Teledyne Photometrics). Images were acquired using a 100-ms exposure time at 10 Hz with the laser power set at 100%. The penetration depth of the evanescent field was ∼118 nm.
Automated Single-Particle Tracking.
Particles were localized and tracked using the Matlab software GaussStorm (40, 41). Briefly, particles were automatically detected by application of a bandpass filter to remove noise, followed by convolution with a Gaussian kernel, and then the selection of above-threshold pixels. Particles were then fitted with elliptical 2D Gaussian functions, which yielded their intensities expressed as the volume under the curve, as well as their positions with subpixel accuracy. Particles were tracked frame to frame using a tracking algorithm with a tracking window of 8 pixels between consecutive frames. The distribution of the displacements of single particles was used to calculate a mean diffusion coefficient in a field of view encompassing an entire cell.
Supplementary Material
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018554117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Eswarakumar V. P., Lax I., Schlessinger J., Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 16, 139–149 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Turner N., Grose R., Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 10, 116–129 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Dorey K., Amaya E., FGF signalling: Diverse roles during early vertebrate embryogenesis. Development 137, 3731–3742 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sochacka M., et al. , FHF1 is a bona fide fibroblast growth factor that activates cellular signaling in FGFR-dependent manner. Cell Commun. Signal. 18, 69 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldfarb M., Fibroblast growth factor homologous factors: Evolution, structure, and function. Cytokine Growth Factor Rev. 16, 215–220 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C., Chung B. C., Yan H., Lee S.-Y., Pitt G. S., Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure 20, 1167–1176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S., et al. , Structures of β-klotho reveal a ‘zip code’-like mechanism for endocrine FGF signalling. Nature 553, 501–505 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen S. K., et al. , Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc. Natl. Acad. Sci. U.S.A. 101, 935–940 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spivak-Kroizman T., et al. , Heparin-induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation, and cell proliferation. Cell 79, 1015–1024 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Schlessinger J., et al. , Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Mohammadi M., Olsen S. K., Ibrahimi O. A., Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 16, 107–137 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Degirolamo C., Sabbà C., Moschetta A., Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nat. Rev. Drug Discov. 15, 51–69 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Urakawa I., et al. , Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 444, 770–774 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Ogawa Y., et al. , BetaKlotho is required for metabolic activity of fibroblast growth factor 21. Proc. Natl. Acad. Sci. U.S.A. 104, 7432–7437 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurosu H., et al. , Regulation of fibroblast growth factor-23 signaling by klotho. J. Biol. Chem. 281, 6120–6123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kharitonenkov A., et al. , FGF-21/FGF-21 receptor interaction and activation is determined by betaKlotho. J. Cell. Physiol. 215, 1–7 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Chen G., et al. , α-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature 553, 461–466 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzina E. S., et al. , Structures of ligand-occupied β-Klotho complexes reveal a molecular mechanism underlying endocrine FGF specificity and activity. Proc. Natl. Acad. Sci. U.S.A. 116, 7819–7824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin B. C., Wang M., Blackmore C., Desnoyers L. R., Liver-specific activities of FGF19 require Klotho beta. J. Biol. Chem. 282, 27277–27284 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Kurosu H., et al. , Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem. 282, 26687–26695 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuro-o M., et al. , Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390, 45–51 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Li S.-A., et al. , Immunohistochemical localization of Klotho protein in brain, kidney, and reproductive organs of mice. Cell Struct. Funct. 29, 91–99 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Goetz R., et al. , Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol. Cell. Biol. 27, 3417–3428 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimada T., et al. , Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J. Clin. Invest. 113, 561–568 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolek O. I., et al. , 1alpha,25-Dihydroxyvitamin D3 upregulates FGF23 gene expression in bone: The final link in a renal-gastrointestinal-skeletal axis that controls phosphate transport. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G1036–G1042 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Shimada T., et al. , Mutant FGF-23 responsible for autosomal dominant hypophosphatemic rickets is resistant to proteolytic cleavage and causes hypophosphatemia in vivo. Endocrinology 143, 3179–3182 (2002). [DOI] [PubMed] [Google Scholar]
- 27.White K. E., et al. , Autosomal-dominant hypophosphatemic rickets (ADHR) mutations stabilize FGF-23. Kidney Int. 60, 2079–2086 (2001). [DOI] [PubMed] [Google Scholar]
- 28.de Las Rivas M., et al. , Molecular basis for fibroblast growth factor 23 O-glycosylation by GalNAc-T3. Nat. Chem. Biol. 16, 351–360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato K., et al. , Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J. Biol. Chem. 281, 18370–18377 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Tagliabracci V. S., et al. , Dynamic regulation of FGF23 by Fam20C phosphorylation, GalNAc-T3 glycosylation, and furin proteolysis. Proc. Natl. Acad. Sci. U.S.A. 111, 5520–5525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goetz R., et al. , Isolated C-terminal tail of FGF23 alleviates hypophosphatemia by inhibiting FGF23-FGFR-Klotho complex formation. Proc. Natl. Acad. Sci. U.S.A. 107, 407–412 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agoro R., et al. , Inhibition of fibroblast growth factor 23 (FGF23) signaling rescues renal anemia. FASEB J. 32, 3752–3764 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z.-L., et al. , A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 10, 3404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu S., et al. , Mapping native disulfide bonds at a proteome scale. Nat. Methods 12, 329–331 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Hern J. A., et al. , Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc. Natl. Acad. Sci. U.S.A. 107, 2693–2698 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasai R. S., et al. , Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J. Cell Biol. 192, 463–480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Los G. V., et al. , HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Calebiro D., et al. , Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc. Natl. Acad. Sci. U.S.A. 110, 743–748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilmes S., et al. , Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science 367, 643–652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holden S. J., et al. , Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys. J. 99, 3102–3111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontana M., Fijen C., Lemay S. G., Mathwig K., Hohlbein J., High-throughput, non-equilibrium studies of single biomolecules using glass-made nanofluidic devices. Lab Chip 19, 79–86 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Imura A., et al. , Secreted klotho protein in sera and CSF: Implication for post-translational cleavage in release of klotho protein from cell membrane. FEBS Lett. 565, 143–147 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Tucker Zhou T. B., King G. D., Chen C., Abraham C. R., Biochemical and functional characterization of the klotho-VS polymorphism implicated in aging and disease risk. J. Biol. Chem. 288, 36302–36311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M., In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Li W., et al. , Assessing the relationship between mass window width and retention time scheduling on protein coverage for data-independent acquisition. J. Am. Soc. Mass Spectrom. 30, 1396–1405 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Lu S., et al. , Mapping disulfide bonds from sub-micrograms of purified proteins or micrograms of complex protein mixtures. Biophys. Rep. 4, 68–81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacLean B., et al. , Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.