Highlights
-
•
Angiogenesis is critical to the development and survival of solid tumors.
-
•
Vanucizumab inhibits both vascular endothelial growth factor and angiopoietin-2.
-
•
Vanucizumab exhibits anti-tumor, anti-angiogenic and anti-metastatic effects.
-
•
It markedly reduced blood-vessel markers in tumor/skin samples in cancer patients.
-
•
Skin biopsies are a valuable surrogate for studying angiogenesis-related mechanisms.
Keywords: Vanucizumab, Mode of action, Biomarkers, Phase I clinical trial, Micro vessel density, Angiopoietin-2
Abbreviations: ANG, angiopoietin; CD, cluster of differentiation; CNS, central nervous system; FFPET, formalin-fixed paraffin-embedded tissues; IHC, immunohistochemistry; IF, immunofluorescence; IQR, interquartile range; miRNA, micro RNA; MVD, microvessel density; VEGF, vascular endothelial growth factor; VGS, vanucizumab gene signature
Abstract
Vanucizumab is a novel bispecific antibody inhibiting vascular endothelial growth factor (VEGF-A) and angiopoietin-2 (Ang-2) that demonstrated safety and anti-tumor activity in part I of a phase I study of 42 patients with advanced solid tumors. Part II evaluated the pharmacodynamic effects of vanucizumab 30 or 15 mg/kg every 2 weeks in 32 patients. Serial plasma samples, paired tumor, and skin-wound-healing biopsies were taken over 29 days to evaluate angiogenic markers. Vanucizumab was associated with marked post-infusion reductions in circulating unbound VEGF-A and Ang-2. By day 29, tumor samples revealed mean reductions in density of microvessels (−32.2%), proliferating vessels (−47.9%) and Ang-2 positive vessels (−62.5%). Skin biopsies showed a mean reduction in density of microvessels (−49.0%) and proliferating vessels (−25.7%). Gene expression profiling of tumor samples implied recruitment and potential activation of lymphocytes. Biopsies were safely conducted. Vanucizumab demonstrated a consistent biological effect on vascular-related biomarkers, confirming proof of concept. Skin-wound-healing biopsies were a valuable surrogate for studying angiogenesis-related mechanisms.
Introduction
Angiogenesis is a key process in tumor growth and metastasis, as adequate vasculature and blood supply ensures tumor growth and development [1]. In normal tissues, blood vessel growth is usually kept to a minimum, and angiogenesis is regulated by a balance of pro-angiogenic (e.g. vascular endothelial growth factor; VEGF) and anti-angiogenic (e.g. thrombospondin-1) factors [2]. In the tumor environment, the balance between angiogenic and anti-angiogenic processes is disturbed, favoring angiogenic stimulation, blood vessel growth, and tumor perfusion [2]. This knowledge has allowed targeting of angiogenesis in the development of effective anticancer agents [3]. Indeed, several agents that target the pro-angiogenic VEGF ligands and the VEGF receptor are now approved, including the anti-VEGF-A monoclonal antibody (mAb) bevacizumab, the anti-VEGF-2 mAb ramucirumab, the dual inhibitor of VEGF-A and placental growth factor aflibercept, and the small molecule tyrosine kinase inhibitors sunitinib, sorafenib and axitinib [4,5]. Despite the introduction of these agents and the associated benefits for patients [4,5], it appears that given the complexity of the angiogenic control mechanisms that allow the tumor to compensate and continue the angiogenic drive [6], further control of angiogenesis is desirable [3]. One approach to this problem is the development of a bispecific antibody with two targets, potentially allowing for improved efficacy by addressing anti-angiogenic escape and resistance mechanisms [7]. Targeting VEGF-A and angiopoietin-2 (Ang-2) may provide an opportunity in this respect.
VEGF-A has a major role in tumor development, angiogenesis and vascular permeability [8], and the Tie2 receptor ligands angiopoietin-1 (Ang-1) and Ang-2 have also been implicated in remodeling of the tumor vasculature [9], [10], [11]. Ang-1 has been shown to have a role in regulating vascular maturation and stability, and Ang-2 has a role in promoting angiogenesis and tumor growth by priming the vasculature to angiogenic stimuli via Tie2 and causing endothelial cell migration and sprouting via stimulation of integrins [9], [10], [11]. Furthermore, Ang-2 abrogates the positive effects of anti-VEGF therapy on the tumor vasculature [12,13] and has been shown to be upregulated in several tumor types and associated with poor prognosis [14], [15], [16]. As Ang-2 acts mainly as a counterpart to Ang-1, a shift in the Ang-1:Ang-2 ratio in favor of Ang-2 therefore represents a change to a more pro-angiogenic state.
Vanucizumab (RO5520985) is a novel bispecific humanized immunoglobulin G-1 (IgG-1)-like antibody that acts as a dual targeting inhibitor of VEGF-A and Ang-2 [17]. In preclinical studies, vanucizumab was shown to possess notable anti-tumor, anti-angiogenic, and anti-metastatic effects [17], providing the rationale for clinical development and evaluation of the agent in a phase I study of patients with advanced solid tumors (NCT01688206). In the first part of this investigation, vanucizumab was associated with an acceptable safety profile and favorable pharmacokinetic and pharmacodynamic effects [18]. Here, we report the biomarker data demonstrating the mode of action for vanucizumab for a proportion of patients recruited to this study.
Materials and methods
Study design
This was a four-part, phase I, open-label, dose-escalation study to evaluate the safety, tolerability, pharmacokinetics, and pharmacodynamics of vanucizumab in patients with advanced solid tumors (NCT01688206); parts I–IV included 42, 32, 41, and 17 patients, respectively. Part I of this investigation involved single-agent dose escalation in 42 patients, with the aim of determining the safety profile and investigating pharmacokinetic and pharmacodynamic parameters (including circulating VEGF-A and Ang-2, vascular permeability, leakiness, and perfusion) as reported previously [18]. In part II of the investigation, reported here, a cohort of 32 patients treated with vanucizumab 30 mg/kg or 15 mg/kg every 2 weeks (the recommended dosages for phase II evaluation) provided blood and/or tumor/skin-wound-healing biopsy samples for biomarker analysis to confirm the mode of action for vanucizumab. This report is restricted to the cohort of patients in part II of the investigation and these specific measures (biomarkers and mode of action). Clinical methodology relating specifically to part I of this investigation has been previously reported [18]. The study (NCT01688206) was conducted in accordance with the principles of the Declaration of Helsinki and good clinical practice, with institutional review board/ethics committee approval and written informed consent from all patients.
Patients
Patients aged ≥18 years with histologically or cytologically confirmed locally advanced or metastatic non-resectable solid tumors being sensitive to anti-angiogenic treatment (colorectal cancer, non-squamous non-small-cell lung cancer, breast cancer, gastric cancer, pancreatic cancer or melanoma) and who had progressed on standard therapy or had no standard therapeutic option were eligible for participation. Patients were required to have measurable disease (RECIST V1.1), an Eastern Cooperative Oncology Group/World Health Organization performance status of 0–1 and adequate hematological, liver, renal, and cardiovascular function. Moreover, patients were obliged to have tumor tissue from which a biopsy can safely be obtained for pharmacodynamics assessments (lesion size ≥2 cm, in regions of motion ≥3 cm). Patients were excluded from participation if they had unstable central nervous system (CNS) tumors or CNS tumor involvement, non-squamous non-small cell lung cancer, significant cardiovascular/cerebrovascular disease or abdominal fistula or gastrointestinal perforation in the previous 6 months, hemoptysis grade ≥2 within 4 weeks, bleeding diathesis or coagulopathy, severe non-healing wound, active ulcer, untreated bone fracture, known HIV infection, or known active hepatitis B virus or hepatitis C virus infection. Exclusion criteria were also applied to those who had been treated with an anti-angiogenic agent 6 months before the start of the study, had received cyclical chemotherapy within a time that was shorter than the standard cycle length or a biological agent/targeted therapy within 4 weeks or five times the elimination half-life of that agent. Patients treated with an investigational agent up to 4 weeks (or the cycle duration) before study start or oral anticoagulants within 7 days, and those who had undergone major surgery up to 4 weeks before study enrolment or who had not adequately recovered were also excluded. Female and male patients were required to use appropriate contraception if applicable, and pregnant or nursing females were excluded.
Biomarker sampling and analysis
Biomarker sampling and analyses are reported in accordance with Biospecimen Reporting for Improved Study Quality guidelines [19].
Soluble biomarkers
Blood (approximately 6 mL) samples were drawn from the antecubital area of the arm at baseline and prior to vanucizumab infusion on days 2 and 5 of cycle 1, and prior to infusion on day 1 of cycles 2 and 3 for determination of plasma-free (unbound biologically active) VEGF-A, total (drug-bound inactive and unbound biologically active) VEGF-A, and free and total Ang-2 circulating levels. Samples for Ang2- analysis were stabilized in K3-EDTA, and those for VEGF-A in citrate, theophylline, adenosine and dipyridamole anticoagulant. Samples were frozen after preparation and centrally collated before analysis. Free VEGF-A levels were determined using the Human VEGF QuantikineⓇ enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems UK). Total VEGF-A was determined using the ElecsysⓇ assay (Roche Professional Diagnostics) comprising a biotinylated murine anti-VEGF-A antibody (clone 3C5) for detection and ruthenylated murine anti-VEGF-A antibody (clone A4.6.1) for capture. Free Ang-2 levels were determined by a two-step process, first using a novel magnetic bead-based depletion procedure developed in-house that employed an anti-idiotypic mAb specific for the VEGF-A-binding site of vanucizumab to capture the drug along with drug-bound Ang-2 [20], and then using the Ang-2 QuantikineⓇ ELISA kit.
Tissue biomarkers
Paired tumor punch needle (≥18 gauge) biopsies were obtained at baseline and on day 1 of cycle 3 from easily accessible representative tumor metastasis (e.g. liver, lymph node, lung) by needle drawing guided by ultrasound or computed tomography scan. Due to the inclusion in this study of patients with various cancer indications, biopsy site varied. However, baseline and on-treatment tumor biopsies for each patient were collected from the same lesion.
Given potential problems with performing sequential tumor biopsies in general, the potential of skin-wound-healing biopsies as a surrogate for tumor samples as described in the literature [21,22] was assessed. Paired skin biopsies were performed at baseline and on cycle 3 day 1 concurrent with tumor biopsies and were taken after local anesthesia from areas of normal skin. Skin wound-healing biopsies were collected from the same anatomical region of the right shoulder in each patient. An initial 2 mm punch biopsy was conducted to create a site of injury. The biopsy was guided by computed tomography scan or ultrasound and was performed using a ≥ 18 gauge needle under aseptic conditions and local anesthesia. This was followed by an overlapping 4 mm biopsy 7 days later to collect the wound-healing material as surrogate tissue of neoangiogenesis.
Tumor and skin biopsies were collected into 10% neutral buffered formalin-filled vials, fixed for 24 ± 2 h, transferred into 70% ethanol and shipped within 24–48 h to a central pathology laboratory (HistoGeneX or Penzberg) for paraffin embedding. Paraffin blocks were stored at −15 to −20 °C
Tissue processing and IHC
Eight sections (2–4 µm thick), per tumor block and skin biopsy, respectively, were processed according to routine histology and IHC protocols. Sections were stained with hematoxylin and eosin. Each sample containing at least 50 cells of interest was subjected to additional immunofluorescence (IF) and chromogenic assays. Formalin-fixed paraffin-embedded tissues (FFPET) were stained using IF multiplex assays. Ang-2/Tie-2, CD31/Ki67 (only assay used in skin) and CD31+CD34/aSMA were analyzed to determine ligands/receptors (e.g. Ang-2) as well as vessel-related parameters (MVD, proliferation and maturity of blood vessels). Single and duplex standard chromogenic assays were performed to assess CD3, CD4, CD8, CD16, and CD68/CD163 to assess the level of immune cells. Staining for Ki67 and CD31, Ang2/Tie2 and CD31+CD34/aSMA, CD4, CD8, and CD16 was performed at HistoGeneX. CD3 and CD68/CD163 samples were stained in the Good Clinical Practice Pathology Laboratory in Penzberg. Staining details are provided in Table 1.
Table 1.
Staining details Ki67, CD31, Ang2/Tie2, and CD31+CD34/aSMA, CD4, CD8, and CD16.
| Staining performed at HistogeneX | ||
|---|---|---|
| Antibody clone | Detection system | Staining instrument |
| CD4 – clone 1F6 monoclonal mouse Ab (Novocastra - NCL-CD4-1F6) |
Optiview DAB | Ventana Benchmark XT |
| CD8 – clone C8/144B (Dako, M7103) monoclonal mouse Ab | Ultraview DAB | Ventana Benchmark XT |
| CD16 – clone SP175 monoclonal rabbit Ab (Ventana – 760-4863) | Ultraview DAB | Ventana Benchmark XT |
| CD31 – clone 1A10 monoclonal mouse Ab (Novocastra – NCL-CD31-1A10) Ki67 – clone 30.9 monoclonal rabbit Ab (Ventana – 790-4286) |
Cy3 – goat anti-mouse (Jackson ImmunoResearch 115–165–166) Alexa488 – goat anti-rabbit (Invitrogen, A11070) |
Ventana Discovery XT |
| CD31 – clone 1A10 monoclonal mouse Ab (Novocastra – NCL-CD31-1A10) CD34 clone QBEnd/10 monoclonal mouse Ab (Ventana, 790-2927) aSMA – polyclonal rabbit Ab (Abcam, ab5694) |
Alexa647 – goat anti-mouse (Invitrogen A21236) Alexa647 – goat anti-mouse (Invitrogen A21236) Alexa488 – goat anti-rabbit (Molecular probes A11070) |
Ventana Discovery XT |
| Ang2 – clone F-1 monoclonal mouse Ab (Santa Cruz Technology, sc-74,403) Tie2 – polyclonal goat Ab (R&D Systems, AF313) |
Cy3 – rabbit anti-mouse (Invitrogen A11078) Alexa488 – rabbit anti-goat (Jackson Immunoresearch 315-165-003) |
Ventana Discovery XT |
| Staining performed in the Good Clinical Practice Pathology Laboratory in Penzberg | ||
| CD3 – clone 2GV6 monoclonal rabbit Ab (Ventana – 790-4341) |
Ultraview AP Red | Ventana Benchmark Ultra |
| CD68 – clone KP1 monoclonal mouse Ab (Ventana – 760-2931) CD163 – clone MRQ-26 monoclonal mouse Ab (Cell Marque), ready to use Dispenser (Ventana – 760-4437) |
Ultraview AP Red (Ventana – 760-501) Optiview DAB (Ventana – 760-700) |
Ventana Benchmark XT Ventana Benchmark XT |
Ab, antibody; CD, cluster of differentiation.
Slide analysis
Automated image analyses (IRIS – in-house platform)
Tissue slides were scanned in brightfield at 20 × magnification using the high-throughput whole-slide scanner Ventana iScan HT (Ventana Medical Systems, Inc.). Visual slide assessments of the samples were performed in the Roche Innovation Centre, Munich, with staining results checked for consistency. Tumor area was manually annotated by a certified pathologist according to internal guidelines before subjecting the digital whole-slide scans to automated image analysis in IRIS, an in-house-developed, next-generation, digital standardized oncology platform. Artefacts and necrotic areas were manually excluded. The different algorithm detection results were checked for accuracy. Tissue digital images were scored using whole-slide automated image analysis algorithms for perforin/CD3 and CD163/CD68. The image analysis included a color deconvolution step as described by Ruifrok and Johnson [23], followed by segmentation using morphological operations that separated the objects of interests from background and/or machine learning steps to classify different phenotypes based on its features and to remove non-specific staining.
Classification methods used included random forest, logistic regression with L1 regularization and support vector machine. The x and y coordinates of the detected objects were recorded in the form of seeds (dots over each cell), typically for nuclear and membrane stains. The performance of the algorithms was assessed by means of a verification step on a representative set of fields of view, before its release in our in-house developed R&D digital pathology platform, IRIS, and its use in studies (see following section). Notwithstanding, algorithm results obtained during the study execution were displayed over the tissue images for detection accuracy check by a pathologist. Artefacts were manually annotated and excluded. Tissue annotations and algorithm results (x,y coordinates and respective label) were stored in a spatial database for further data analysis.
Slide analysis – histogenex
Whole-slide images of the IHC-stained slides were generated at room temperature with a whole-slide scan device Pannoramic SCAN (3D-Histech, Budapest, Hungary). A 20 × Plan Apo objective (0.80 numerical apertures) and a Hitachi (HV-F22L) charge-coupled device progressive scan color camera with a resulting image resolution of 0.23 µm/pixel were used. Joint Photographic Experts Group (JPEG) image encoding with quality factor 80 and an interpolated focus distance of 15 with stitching in the scan options were chosen. Scanned images were examined in Pannoramic Viewer (3DHISTECH, Budapest, Hungary) to check for image quality and to confirm that the whole tissue section was captured.
The whole-slide images were analyzed with Tissue Studio 3.5 image analysis software from Definiens (Munich, Germany). The percentage marker area of the CD4+, CD8+, and CD16+ stained cells was determined in the center of the tumor and in the invasive margin. The invasive margin represents an area along the edge of the tumor and normal tissue that reaches 500 μm in the normal tissue and 500 μm in the tumor.
Image acquisition of the IF-stained slides was performed at room temperature with an AxioImager Z2 microscope (Carl Zeiss AG, Oberkochen, Germany), and the ZEN 2.0 (blue edition) software package. A 20 × Plan Apo objective (0.80 numerical apertures) and a Hamamatsu (OrcaFlash4.0) complementary metal-oxide semiconductor monochromatic camera with a resulting image resolution of 0.325 µm/pixel were used. The digital images were analyzed according to stereological principles: areas of interest were selected via systematically uniform random sampling and combined with a digital counting grid in the AxioVision 4.8 software package (Carl Zeiss AG, Oberkochen, Germany). For each sample in which at least three areas of interest could be identified, all vasculature structures were classified (positive for both markers of interest, positive for marker 1, positive for marker 2, negative for both markers) in a semiautomatic manner with the AxioVision 4.8 software. The average vessel diameter, the number of vessels per area and the area occupied by the vessels was calculated for each class.
Data analysis
All biomarker analyses were based on the full analysis population. For the analysis of pharmacodynamic biomarkers, the primary evaluation was based on the observed change from baseline. Both actual values and estimated parameters were collated and analyzed descriptively. To assess predictability of a biomarker, the association between clinical outcome (e.g. responders vs. non-responders; RECIST criteria V1.1) and the biomarker level was explored. Data were analyzed with SAS JMP software version 13.0.0 (SAS Institute Inc.) or R statistical programming language version 3.4.0. The distributions of concentrations of circulating biomarkers were checked for log-normality. If the data were well described with log-normal distribution, logarithmization was done. Relative changes of the biomarker levels for each patient were calculated and group medians as well as 25% and 75% quantiles were calculated and reported. Statistical analysis was performed using a two-tailed matched pairs t-test or ANOVA. All results are reported on the original (back-transformed scale).
Gene expression profiling
Randomization of tumor samples
At accessioning, tumor samples were randomized to avoid potential confounding effects with tumor entity, visit, dosing group, and response.
Protocol summary: RNA isolation from formalin-fixed paraffin-embedded tissue (FFPET)
Total RNA, including microRNA (miRNA), was isolated from FFPET using the Qiagen miRNeasy FFPE kit (Qiagen 217504, 50 reactions). Briefly, FFPE material was deparaffinized with xylene and lysed under denaturing conditions with proteinase K. The sample was heated to reverse formalin cross-linking and then treated with DNase to eliminate all genomic DNA. The lysate was then mixed with a buffer to optimize binding conditions and applied to an RNeasy MinElute column where the total RNA, including miRNA, binds to the membrane and contaminants are efficiently washed away. Total RNA, including miRNA, was then eluted with nuclease-free water.
Protocol summary: RNA sequencing utilizing illumina TruSeq RNA access library preparation and instrumentation
The library was generated using Illumina TruSeq RNA Access Library Preparation (recently renamed TruSeq RNA Exome, Illumina 20020189, 48 rxns). Cluster generation and sequencing of libraries was performed on the Illumina HiSeq.
Protocol summary
The TruSeq RNA Access method is a hybridization-based assay to enrich for coding RNAs from total RNA sequencing libraries. The assay consists of two major steps: 1) total RNA library preparation, and 2) coding RNA enrichment.
Preparation of total RNA library
First-strand complementary DNA (cDNA) synthesis was primed from total RNA using random primers, followed by the generation of second-strand cDNA with deoxyuridine triphosphate utilized in place of deoxythymidine triphosphate in the master mix. This facilitates the preservation of strand information, as amplification in subsequent steps stalls when it encounters uracil in the nucleotide strand. Double-stranded cDNA undergoes end-repair, A-tailing, and ligation of adapters that include index sequences. The resulting molecules were amplified via polymerase chain reaction (PCR), their yield and size distribution were determined, and their concentrations were normalized in preparation for the enrichment step.
Enrichment for coding RNA
Libraries were enriched for the mRNA fraction by positive selection using a cocktail of biotinylated oligos corresponding to coding regions of the genome. Targeted library molecules were then captured via the hybridized biotinylated oligo probe using streptavidin-conjugated beads. After two rounds of hybridization–capture reactions, the enriched library molecules were subjected to a second round of PCR amplification prior to paired-end 2 × 50 sequencing on the Illumina HiSeq.
Data quality-control metrics
Samples needed to have 100 ng of input RNA to move forward with library preparation. Samples with <30% of RNA fragments of >200 nucleotides in size (%DV200 < 30) are not recommended for analysis by this method. In total, 12 samples had DV200 values < 30 (one of which was < 20), two samples had yields <100 ng (one of which was also a DV200 value < 30). Since the material was limited, samples that did not qualify for the RNA quality control threshold proceeded to sequencing and sequencing quality control.
RNA sequence alignment
Raw reads data were aligned using an in-house pipeline against the human reference genome GRCh37/hg19.
Bioinformatic (gene expression) analysis
Statistical and bioinformatic analyses were performed with the computing environment R [24].
The RNA-seq RPKM matrix was log2-transformed. Differential gene expression analysis used linear mixed-effect models with the nlme package [25]. A set of 203 genes was obtained by selecting the most changing genes with a p-value < 0.05 and an absolute log 2-fold change of >0.5 (selected log-fold change [−2.01–4.94]). Signatures were obtained from the Molecular Signatures Database (MSigDB) [26,27] and peer-reviewed publications [28], [29], [30]. The Mann–Whitney U statistic was computed for those signatures in each sample. Differences between baseline and on-treatment samples were evaluated using a linear mixed-effect model with a p-value threshold of 0.05.
Spearman correlation coefficients were computed between gene expression levels and different biomarkers (CD8+ T cells, CD3+ T cells and macrophage levels, along with MVD and Ang-2). Spearman correlation coefficients were also calculated between the signature scores in each sample (Mann–Whitney U statistic) and the biomarkers.
Results
Patient population
A total of 32 patients participated in this analysis; all patients provided samples for soluble marker analysis, 26 gave evaluable paired skin biopsies and 19 gave evaluable paired tumor biopsies. Patient demographics showed that 15 (47%) were male, mean age (range) was 58.2 (31–75) years, and all recorded their ethnicity as White. The most common cancers were those of the colon/rectum (n = 11 patients), followed by cancer of the breast (n = 7), lung (n = 3), stomach/esophagus (n = 3), pancreas (n = 2), cervix, kidney, skin, thyroid, uterus (n = 1 each), or primary of unknown origin (n = 1).
Patients included in this analysis received vanucizumab for a median (range) of 4.0 (2.0–60.0) cycles at doses of 30 mg/kg (median 4.0 cycles; range 2.0–60.0; n = 23 patients) or 15 mg/kg (median 4.0 cycles; range 3.0–12.0; n = 9 patients). A documented best overall response (RECIST V1.1 criteria) of partial response was experienced by one patient (30 mg/kg), stable disease was experienced by 12 patients (15 mg/kg, n = 4; 30 mg/kg, n = 8) and progressive disease was experienced by 19 patients (15 mg/kg, n = 5; 30 mg/kg, n = 14). During the study, 28 patients withdrew because of progressive disease, one patient each withdrew because of gastrointestinal perforation or withdrawal of consent, and two patients were still receiving vanucizumab at data cut-off.
Mode of action based on soluble markers and immunohistochemistry (IHC)
Soluble markers
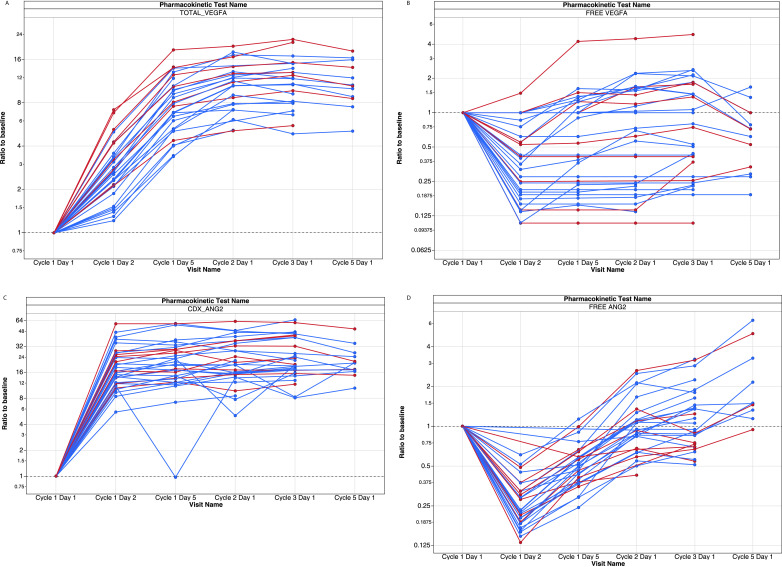
The mode of action of vanucizumab was confirmed by changes in circulating levels of VEGF-A and Ang-2 as soluble biomarkers of angiogenesis (Fig. 1). Although total (drug-bound and unbound) levels of both VEGF-A and Ang-2 increased during treatment (Fig. 1a and c), free (unbound biologically active) levels of both these biomarkers markedly decreased within 24 h after the first infusion of vanucizumab, returning to baseline prior to the next infusion (Fig. 1b and d). These effects were independent of dose or schedule.
Fig. 1.
Change from baseline in (A) total (bound and unbound) and (B) free (unbound) vascular endothelial growth factor-A (VEGF-A) and (C) total (bound and unbound) and (D) free (unbound) angiopoietin-2 (Ang-2) circulating levels during treatment with vanucizumab (n = 23 in the 30 mg/kg cohort [blue]; n = 9 in the 15 mg/kg cohort [red]). As VEGF-A and Ang-2 bind to vanucizumab, levels of free circulating VEGF-A and Ang-2 fall. Free biomarker levels return to baseline levels prior to the next infusion via mechanisms that may include antibody-binding kinetics, the de novo synthesis of these target molecules or, in the case of Ang-2, the release of this marker from Weibel-Palade bodies [31], [32], [33]. Total VEGF-A and Ang-2 levels increase over time given the prolonged half-life of vanucizumab and the de-novo synthesis of VEGF-A and Ang-2.
IHC
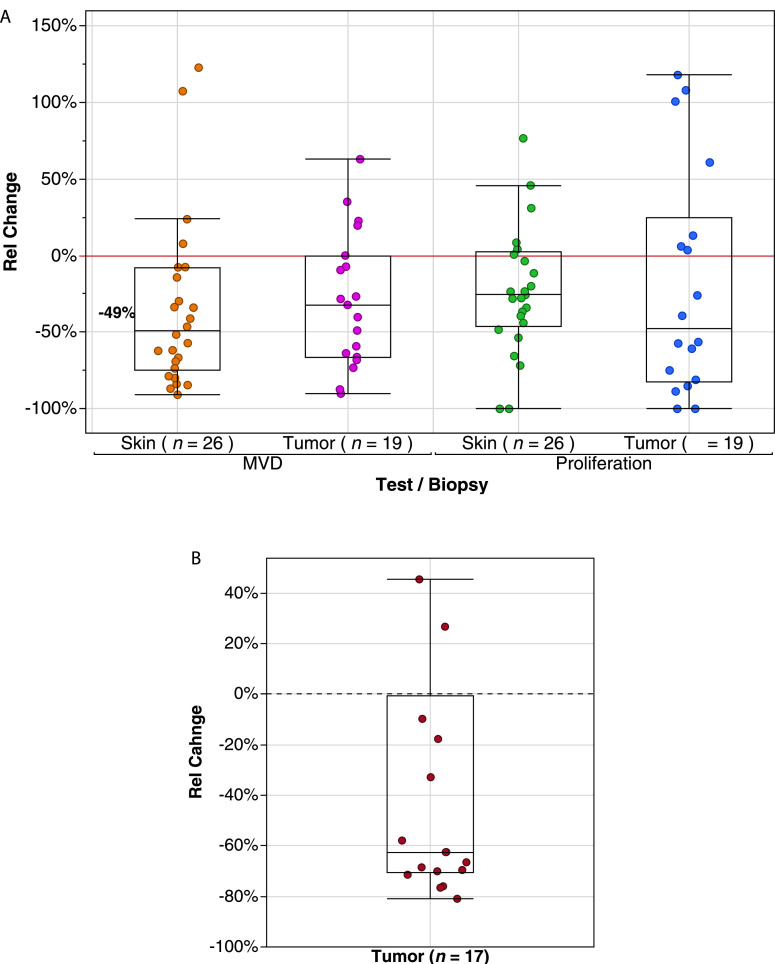
Consistent with the above finding relating to free circulating levels of angiogenesis markers, analysis of tumor biopsies revealed a significant median relative reduction during vanucizumab treatment (day 29) versus baseline for microvessel density (MVD) (cluster of differentiation [CD]-31 of −32.2% (interquartile range [IQR] −66.2%, 0.0%) and density of proliferating vessels (Ki67) of −47.9% (IQR −82.1%, +24.9%) (Fig. 2A). These findings were accompanied by a reduction in the median density of Ang-2 positive vessels of −62.5% (IQR −70.4%, −0.6%) at day 29 compared with baseline (Fig. 2B).
Fig. 2.
Box plot representing minimum, first quartile, median, third quartile, and maximum relative change (%) in (A) microvessel density (MVD) and density of proliferating vessels following vanucizumab treatment (day 29 vs. baseline) in tumor and skin-wound-healing biopsies, and (B) density of angiopoietin-2-positive (Ang2+) vessels following vanucizumab treatment (day 29 vs. baseline) in tumor biopsies (due to the small sample size, assessment was not possible in skin biopsy samples). Extreme values are located outside the limits of the box.
MVD and vessel proliferation (CD31 and Ki67) for skin-wound-healing biopsies followed a similar pattern as for tumor biopsies. At day 29, vanucizumab treatment was associated with a reduction in median MVD of −49.0% (IQR 74.9%, 7.7%) and a reduction in median proliferating vessel of −25.7% (IQR −46.2%, +2.4%) when compared with baseline (Fig. 2A).
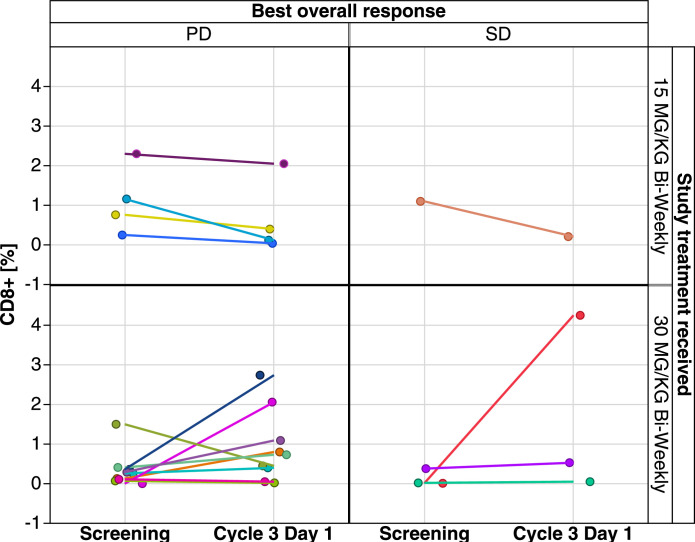
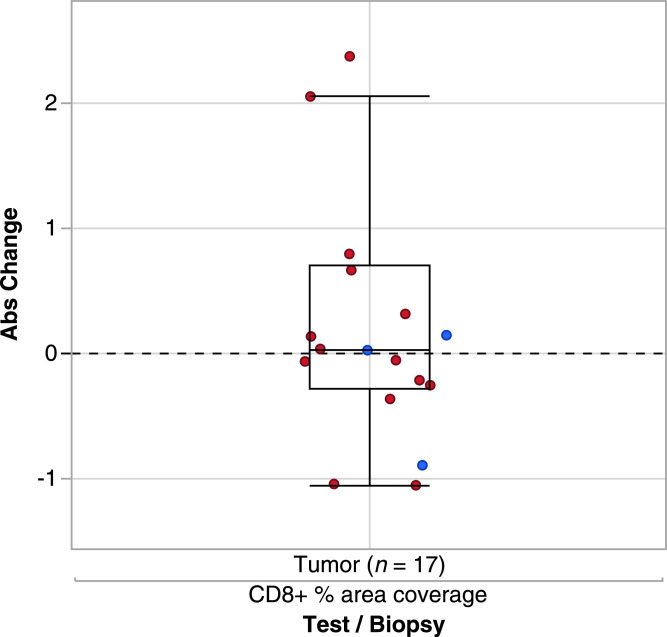
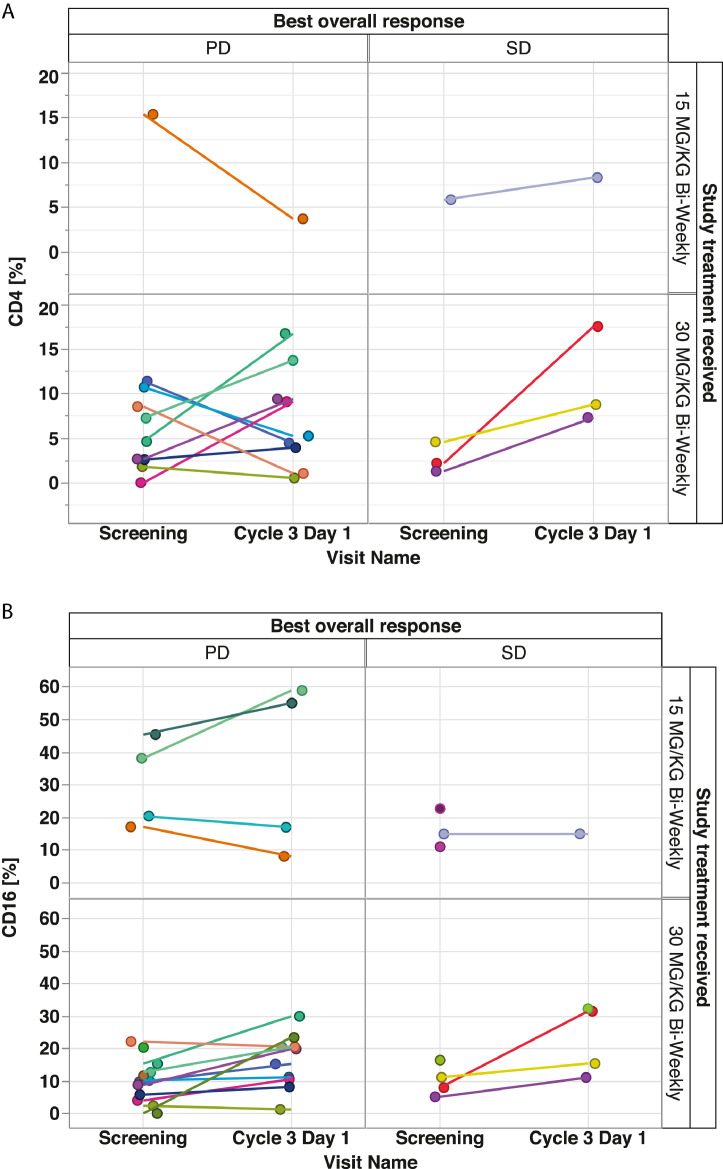
Evaluation of immune markers for patients with paired tumor samples classified by best overall response showed overall low levels of CD8+ T-cell infiltration (0–4.1% per central tumor area) at baseline and during treatment (see Fig. 3). There was a slight non-significant increase in CD8+ T cells on day 29 versus baseline for patients receiving vanucizumab 30 mg/kg (median change +0.04%; IQR −0.28%, 0.7%) (Fig. 3). However, the small sample size meant no reliable interpretation could be applied to CD8+ T-cell levels and best overall response (Figs. 3 and4). There was no discernible increase in CD8+ T cells at the lower dose. When the same evaluation was conducted for CD4+ T cells (Fig. 5A) and CD16+ T cells (Fig. 5B), no significant changes or trends were detected. Of the 20 patients, 11 showed an increase of >10% in CD3+ T cells. The patients with higher increase (>10%) represented the higher dosing group. Of 20 patients, 11 (all in the higher dose group) showed an increase in M2 (CD68+/CD163+) macrophage coverage of >10% (data not shown).
Fig. 3.
Extent of cell infiltration in tumor biopsies at baseline and during treatment (day 29) with vanucizumab for patients with a documented best overall response of progressive disease (PD) or stable disease (SD)a (n = 17) aNon-paired data points are shown for completeness where data points are missing. CD, cluster of differentiation.
Fig. 4.
Box plot representing minimum, first quartile, median, third quartile, and maximum change in percentage area coverage of cluster of differentiation (CD)−8+ cells in tumor samples from patients treated with vanucizumab (pooled data from both treatment groups) and a best overall response of progressive disease (red) or stable disease (blue). Extreme values are located outside the box.
Fig. 5.
Extent of (A) cluster of differentiation (CD)−4+ and (B) CD16+ T-cell infiltration in tumor biopsies at baseline and during treatment (day 29) with vanucizumab for patients with a documented best overall response of progressive disease (PD) or stable disease (SD)a. aNon-paired data points are shown for completeness where data points are missing.
Mode of action based on transcriptome data
Further transcriptome analysis of the paired tumor samples supports a potential vanucizumab-induced immune infiltration. A signature composed of 203 genes regulated by vanucizumab (the “vanucizumab gene signature” [VGS]), includes a large number of immune-related genes (e.g. T-cell receptors and IgGs).
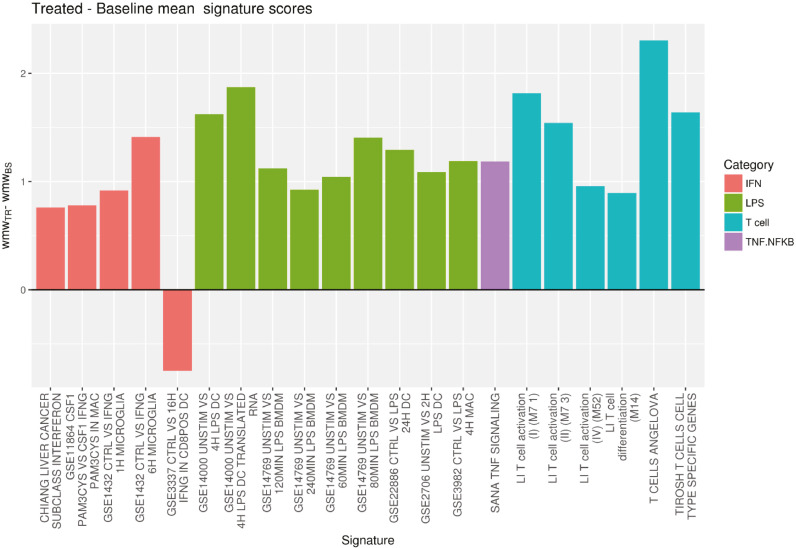
Signature score analysis showed an increased score for T-cell-specific signatures and for signatures related to inflammation processes after treatment (Fig. 6). Such analysis thus also supports the activation of the immune response after treatment.
Fig. 6.
Difference between baseline and end-of-treatment signature scores in tumor samples for interferon (IFN), lipopolysaccharide (LPS), T cell and tumor necrosis factor (TNF)/nuclear factor kappa-light-chain-enhancer of activated B (NF-κB) that achieved statistical significance (p < 0.05)a (n = 18). Signatures were divided into several categories that can be an indicator of inflammation (LPS, natural killer [NK], NF-κB and IFN) or of increased expression of genes related to T cells (T-cell signatures from scores from Angelova et al. [28], Li et al. [29] and Tirosh et al. [30]). Individual p-values obtained for the signatures represented above are shown below.
Signature; Category; P value
CHIANG LIVER CANCER SUBCLASS INTERFERON; IFN; 0.028
GSE11864 CSF1 PAM3CYS VS CSF1 IFNG PAM3CYS IN MAC; IFN; 0.034
GSE1432 CTRL VS IFNG 1H MICROGLIA; IFN; 0.039
GSE1432 CTRL VS IFNG 6H MICROGLIA; IFN; 0.046
GSE3337 CTRL VS 16H IFNG IN CD8POS DC; IFN; 0.019
GSE14000 UNSTIM VS 4H LPS DC; LPS; 0.021
GSE14000 UNSTIM VS 4H LPS DC TRANSLATED RNA; LPS; 0.029
GSE14769 UNSTIM VS 120MIN LPS BMDM; LPS; 0.003
GSE14769 UNSTIM VS 240MIN LPS BMDM; LPS; 0.041
GSE14769 UNSTIM VS 60MIN LPS BMDM; LPS; 0.024
GSE14769 UNSTIM VS 80MIN LPS BMDM; LPS; 0.004
GSE22886 CTRL VS LPS 24H DC; LPS; 0.042
GSE2706 UNSTIM VS 2H LPS DC; LPS; 0.013
GSE3982 CTRL VS LPS 4H MAC; LPS; 0.011
SANA TNF SIGNALING; TNF.NFKB; 0.022
LI T cell activation (I) (M7 1); Tcell; 0.031
LI T cell activation (II) (M7 3); Tcell; 0.032
LI T cell activation (IV) (M52); Tcell; 0.046
LI T cell differentiation (M14); Tcell; 0.032
T CELLS ANGELOVA; Tcell; 0.002
TIROSH T CELLS CELL TYPE SPECIFIC GENES; Tcell; 0.044.
Correlation between transcriptome and histological data
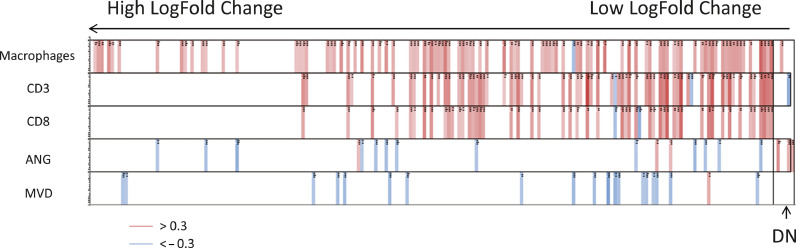
The expression of the VGS genes correlated moderately with CD8+ T cells (maximum correlation of 0.686), CD3+ T cells (maximum correlation of 0.819) and macrophage area (maximum correlation of 0.687) within tumor tissue. Furthermore, as expected, upregulation of some VGS genes upon treatment showed an inverse correlation with MVD and Ang-2 histological measurements (most negative correlation of −0.479 and −0.411, respectively) (Fig. 7). Moreover, T-cell-specific signatures correlated (maximum correlation of 0.741) with CD8+ T cells and CD3+ T cells as assessed by immunohistochemistry (IHC) displaying an agreement between histology and gene expression (Table 2). This is not the case for macrophage-specific signatures and macrophage coverage using IHC (Table 2).
Fig. 7.
Correlation of the biomarkers (Cluster of Differentiation [CD] 3, CD8; Angiopoietin [ANG] 2; microvessel density [MVD]) with the expression of the genes within the progressive disease (PD)-like signature for tumor samples. Each column is a gene selected among the most differentially expressed between treatment and baseline (p-value > 0.05 and log-fold change > 0.5). The genes are sorted from left to right, from the highest to the lowest log-fold change. The DN category indicates genes with a negative log-fold change (decreased expression in treated samples). Rows are the biomarker measurements. Cells are colored when there is a moderate to high correlation. A red cell indicates a Spearman correlation higher than 0.3, and a blue cell indicates a Spearman correlation inferior to 0.3 (n = 18).
Table 2.
Spearman correlation coefficients between the signature scores from Angelova et al., [28], Li et al., [29] and Tirosh et al., [30] and CD8+ and CD3+ biomarker quantification, and between the signature scores from Angelova et al., [28] and Tirosh et al., [30] and the measured macrophage areas.
| Signature | CD8+ cells | CD3+ cells |
|---|---|---|
| Li et al. [29] Enriched in T cells (I) (M7.0) |
0.665 | 0.741 |
| Li et al. [29] T-cell activation (IV) (M52) |
0.705 | 0.708 |
| Li et al. [29] T-cell activation (I) (M7.1) |
0.669 | 0.707 |
| Li et al. [29] T-cell surface signature (SO) |
0.595 | 0.702 |
| Li et al. [29] T-cell activation (II) (M7.3) |
0.650 | 0.701 |
| Angelova et al. [28] Treg cells |
0.600 | 0.646 |
| Angelova et al. [28] T cells |
0.511 | 0.633 |
| Tirosh et al. [30] T cells |
0.632 | 0.691 |
| Signature | All macrophages | |
| Angelova et al. [28] Macrophages |
0.229 | |
| Tirosh et al. [30] Macrophages |
0.090 | |
CD, cluster of differentiation; Treg, T regulatory cells.
Discussion
This is the first study exploring the intra-tumoral pharmacodynamic effect of an investigational new anti-angiogenic antibody by assessing paired tumor- and corresponding skin biopsies. Sequential skin- and core needle biopsies could be obtained safely and yielded in tissue material available for multiple translational applications.
This analysis of patients with a variety of advanced solid tumors treated with two doses of vanucizumab confirmed a biological effect on vascular-related biomarkers in plasma, tumor samples and surrogate wound-healing biopsies, thus demonstrating proof of mechanism for this bispecific anti-angiogenic antibody.
Soluble markers of angiogenesis were affected by administration of vanucizumab, with a reduction in circulating levels of free (unbound biologically active) VEGF-A and Ang-2 after infusion, confirming the mechanism of action of vanucizumab. Their return to baseline before the next infusion might reflect antibody-binding kinetics, a potential de novo synthesis of these target molecules or, in the case of Ang-2, the release of this marker from Weibel-Palade bodies [31], [32], [33]. Although total levels (bound and unbound) of VEGF-A and Ang-2 increased during treatment, this most likely reflects saturated vanucizumab-target complex formation [18] or, in the case of Ang-2, release from Weibel-Palade bodies [31,32].
In accord with the effects on soluble markers of angiogenesis, vanucizumab was also associated with notable reductions of angiogenesis markers in tissue. Indeed, the magnitude of the effect is worthy of note, with vanucizumab-associated reductions from baseline of −32.2% in MVD, −47.9% in density of proliferating vessels and −62.5% in the density of Ang-2-positive vessels in tumor biopsies. Moreover, the inverse correlations between some of the VGS genes and the MVD and Ang-2 histological markers reflect the anti-angiogenic mode of action of vanucizumab at the transcriptomic level. Finally, there was reasonable accordance (concordance) between markers of angiogenesis in tumor samples and surrogate skin-wound-healing biopsies, with MVD changes of −49.0% and density of proliferating vessels changes of −25.7% overall in the skin samples tested.
Over the treatment period of 1 month, the observed pharmacodynamic effects on angiogenic biomarkers, with a primary effect on circulating levels of VEGF receptor and Ang-2 were consistent during the observation period of 1 month and translated into secondary effects on tumor markers of angiogenesis, including tissue Ang-2, MVD, and vessel proliferation.
These observations are consistent with the moderate-to-significant sustained post-treatment reductions in tumor vascular permeability, vascular leakiness and permeability in patients undergoing magnetic resonance imaging in this study as reported previously [18] and overall reflects the anticipated inhibitory effect on the VEGF-A and Ang-2 axis. Our findings also suggest that wound-healing skin biopsies may be a useful surrogate marker and an alternative to tumor biopsies for the assessment of anti-angiogenic effects. Of note, paired tumor and skin biopsy samples were safely collected during this investigation without any bleeding events. Finally, the principal objective of this phase I study was to evaluate the safety, pharmacokinetics, and pharmacodynamics of vanucizumab. The low clinical activity of vanucizumab and the small numbers of responders versus non-responders precluded the investigation of any correlation between pharmacodynamic effects in plasma/tissue and treatment outcomes.
Immunological data suggest that the presence of tumor-infiltrating lymphocytes (including CD8+ T cells) is potentially associated with improved survival in patients with solid tumors [34]. Anti-angiogenic agents have been shown to improve the integrity of the tumor vasculature and to increase the associated T-cell infiltration [35], [36], [37]. Ad hoc analysis to evaluate the association between CD8+ T-cell infiltration in tumor samples by IHC and best response was inconclusive in this study. In the paired tumor samples analyzed, CD8+ T-cell infiltration was low at baseline and during treatment. A non-significant increase in CD8+ and CD3+ T cells and M2 macrophages for patients receiving vanucizumab at the higher dose was detected but was not replicated at the lower dose. Furthermore, a similar analysis of CD4+ and CD16+ T cells showed no significant changes or trends. Overall, the small sample size meant that no reliable interpretation could be applied to the relationship between clinical response and T-cell tumor infiltration. Besides, tumor biopsies can only reveal a snapshot of the tumor microenvironment, with an extrapolation of data for the whole tumor, potentially not entirely reflecting a heterogeneously activated and infiltrated immune contexture. However, signature score analysis after gene expression profiling of the paired tumor biopsies showed modest increase of inflammatory processes and T-cell-related signature scores under treatment with vanucizumab, implying recruitment and potential activation of lymphocytes (Fig. 6). This was not the case for macrophage-specific signatures and macrophage coverage using IHC. We hypothesize that two reasons are behind this lack of correlation. First, macrophage IHC readouts are based on the area covered by the macrophages, not the number of macrophages. The amount of messenger RNA (mRNA) is typically a function of the number of nuclei (cells) rather than the area covered by the cells, and the macrophage signatures were, of course, derived from gene expression (mRNA) data. Second, the subtype of macrophages (e.g. M1 vs. M2) represented by the macrophage signatures may not reflect the subtype detected by IHC in this study.
In conclusion, this analysis of patients with a variety of advanced solid tumors treated with vanucizumab in a phase I study confirmed an expected biological effect on vascular-related biomarkers in plasma, tumor samples, and wound-healing biopsies and demonstrated proof of concept for this bispecific antibody. These findings are consistent with previously reported improvements in tumor vascular permeability, leakiness and perfusion with anti-angiogenic therapy [18]. Our data also support the use of skin-wound-healing biopsies as a surrogate to explore angiogenesis. Furthermore, we demonstrated changes in immune-infiltrating cells induced by anti-angiogenic treatment deserving further investigation in a more homogenous and larger patient population to elaborate the relationship of angiogenesis and immune cell infiltration, as well as a potential association between the observed pharmacodynamic effects and the drug dose administered.
CRediT authorship contribution statement
Florian Heil: Conceptualization, Writing - review & editing, Data curation, Formal analysis. Galina Babitzki: Writing - review & editing, Data curation, Formal analysis. Alice Julien-Laferriere: Writing - review & editing, Data curation, Formal analysis. Chia-Huey Ooi: Writing - review & editing, Data curation, Writing - original draft. Manuel Hidalgo: Writing - review & editing. Christophe Massard: Writing - review & editing, Data curation, Writing - original draft. Maria Martinez-Garcia: Writing - review & editing, Data curation, Writing - original draft. Christophe Le Tourneau: Writing - review & editing, Data curation, Writing - original draft. Mark Kockx: Writing - review & editing, Data curation, Formal analysis. Peter Gerber: Writing - review & editing, Data curation, Formal analysis. Simona Rossomanno: Writing - review & editing, Data curation, Formal analysis. Oliver Krieter: Conceptualization, Writing - review & editing, Writing - original draft. Angelika Lahr: Conceptualization, Writing - review & editing, Writing - original draft. Norbert Wild: Writing - review & editing, Data curation, Formal analysis. Suzana Vega Harring: Writing - review & editing, Data curation, Formal analysis, Writing - original draft. Katharina Lechner: Conceptualization, Writing - review & editing, Data curation, Formal analysis, Writing - original draft.
Declaration of Competing Interest
The following authors are employees and shareholders of Roche Diagnostics GmbH: FH, GB, OK, SVH, KL; AJL is an employee of Soladis GmbH; CHO is an employee and shareholder of F. Hoffmann-La Roche AG; PG was an employee of Roche Innovation Center at the time of manuscript preparation, now retired; SR is an employee of F. Hoffmann-La Roche; AL and NW are employees of Roche Diagnostics GmbH; MH has received honoraria for acting as a speaker, consultant or advisory board member (Celgene, Pfizer, Novartis, MSD, EMD, Ipsem, Shire, SOBI, Champions Oncology, Agenus, Erytech, Pharmacyte, Bioline, BioOncotech, Oncomatrix, VCN, Bayer, BMS, Nelum, Eng T Cells), holds stock (Champions Oncology, Pharmacyte, BioOncotech, Nelum, Eng T Cell), and has received royalties (Myriad) and financial support paid to his institution for clinical trials or research grants (Berg, Bioline, Pfizer, EMD, Celgene, ASANA, Bycicle, Oncomatrix, BioExcell, Erytech); CM has acted as principal/sub-investigator of clinical trials (AbbVie, Aduro Biotech, Agios Pharmaceuticals, Amgen, Argen-X Bvba, Arno Therapeutics, Astex Pharmaceuticals, AstraZeneca, Aveo, Bayer Healthcare AG, Bbb Technologies Bv, Beigene, Bioalliance Pharma, Biontech AG, Blueprint Medicines, Boehringer Ingelheim, Bristol Myers Squibb, Ca, Celgene Corporation, Chugai Pharmaceutical Co., Clovis Oncology, Daiichi Sankyo, Debiopharm S.A., Eisai, Exelixis, Forma, Gamamabs, Genentech, Inc., Gilead Sciences, Inc, GlaxoSmithKline, Glenmark Pharmaceuticals, H3 Biomedicine, Inc, F. Hoffmann-La Roche AG, Incyte Corporation, Innate Pharma, Iris Servier, Janssen Cilag, Kura Oncology, Kyowa Kirin Pharm, Eli Lilly, Loxo Oncology, Lytix Biopharma AS, Medimmune, Menarini Ricerche, Merck Sharp & Dohme Chibret, Merrimack Pharmaceuticals, Merus, Millennium Pharmaceuticals, Nanobiotix, Nektar Therapeutics, Novartis Pharma, Octimet Oncology NV, Oncoethix, Oncomed, Oncopeptides, Onyx Therapeutics, Orion Pharma, Oryzon Genomics, Pfizer, Pharma Mar, Pierre Fabre, Rigontec GmbH, F. Hoffmann-La Roche, Sanofi Aventis, Sierra Oncology, Taiho Pharma, Tesaro, Inc., Tioma Therapeutics, Inc., Xencor), received research grants (AstraZeneca, BMS, Boehringer Ingelheim, Janssen Cilag, Merck, Novartis, Pfizer, F. Hoffmann-La Roche, Sanofi), received non-financial support (drug supplied) (AstraZeneca, Bayer, BMS, Boehringer Ingelheim, Johnson & Johnson, Eli Lilly, Medimmune, Merck, NH TherAGuiX, Pfizer, Roche) and received consultant/advisory fees (Amgen, Astellas, Astra Zeneca, Bayer, BeiGene, BMS, Celgene, Debiopharm, Genentech, Ipsen, Janssen, Eli Lilly, MedImmune, Novartis, Pfizer, F. Hoffmann-La Roche, Sanofi, Orion); MMG has acted as consultant/advisory board member (F. Hoffmann-La Roche) and received travel expenses (F. Hoffmann-La Roche, Pfizer); ClT has participated in advisory boards (AstraZeneca, F. Hoffmann-La Roche, MSD, BMS, Merck Serono, GSK, Nanobiotix, Amgen); MK is a co-founder and shareholder of HistoGeneX N.V.
Acknowledgments
Acknowledgments
The authors wish to thank their colleagues at HistoGeneX N.V., Antwerp, Belgium, for their contribution and expertise in immunohistochemical tissue analysis and Dr. Claudia Ferreira, pRED Roche Innovation Center Munich, for the digital algorithm used to compute the areas shown in Table 2. The authors also wish to thank David Peters and Mark O'Connor (Rx Communications, Mold, UK) for medical writing support.
Role of the Funding Source
The clinical trial was sponsored by F. Hoffmann-La Roche. The sponsor was involved in all stages of the study, conduct and analysis. F. Hoffmann-La Roche also paid all costs associated with the development and the publication of the present manuscript.
Ethical approval
This study was approved by the Institutional Ethical Committees at the following investigational sites: Centro Nacional de Investigaciones Oncológicas (CNIO), Madrid, Spain; START Madrid - Centro Integral Oncologico Clara Campal (CIOCC), HM Sanchinarro, Madrid, Spain; Medical Oncology Department, Hospital del Mar. Barcelona, Spain; Department of Medical Oncology, Institut Curie, Paris & Saint-Cloud, France; Department of Drug Development, Institut Gustave Roussy, Villejuif, France.
Footnotes
Summary: In this ex vivo study, a consistent biological effect of vanucizumab is demonstrated on vascular-related biomarkers in plasma, tumor and skin wound-healing biopsies. These surrogates provide valuable measures for studying angiogenesis-related mechanisms.
Contributor Information
Florian Heil, Email: florian.heil@roche.com.
Galina Babitzki, Email: galina.babitzki@roche.com.
Alice Julien-Laferriere, Email: alice.julien-laferriere@roche.com.
Chia-Huey Ooi, Email: chia-huey.ooi@roche.com.
Manuel Hidalgo, Email: mhidalgo@bidmc.harvard.edu.
Christophe Massard, Email: Christophe.MASSARD@gustaveroussy.fr.
Maria Martinez-Garcia, Email: MariaMartinezGarcia@parcdesalutmar.cat.
Christophe Le Tourneau, Email: christophe.letourneau@curie.fr.
Mark Kockx, Email: mark.kockx@histogenex.com.
Peter Gerber, Email: p.gerber1@gmx.de.
Simona Rossomanno, Email: simona.rossomanno@roche.com.
Oliver Krieter, Email: oliver.krieter@roche.com.
Angelika Lahr, Email: angelika.lahr@roche.com.
Norbert Wild, Email: norbert.wild@roche.com.
Suzana Vega Harring, Email: suzana.vega_harring@roche.com.
Katharina Lechner, Email: katharina.lechner@roche.com.
References
- 1.Folkman J. Role of angiogenesis in tumor growth and metastasis. Sem. Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D., Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet P., Jain R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuchs C.S., Tomasek J., Yong C.J., Dumitru F., Passalacqua R., Goswami C., Safran H., Dos Santos L.V., Aprile G., Ferry D.R., Melichar B., Tehfe M., Topuzov E., Zalcberg J.R., Chau I., Campbell W., Sivanandan C., Pikiel J., Koshiji M., Hsu Y., Liepa A.M., Gao L., Schwartz J.D., Tabernero J., REGARD Trial Investigators Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet. 2014;383:31–39. doi: 10.1016/S0140-6736(13)61719-5. [DOI] [PubMed] [Google Scholar]
- 5.Yadav L., Puri N., Rastogi V., Satpute P., Sharma V. Tumour angiogenesis and angiogenic inhibitors: a review. J. Clin. Diagn. Res. 2015;9:XE01–XE05. doi: 10.7860/JCDR/2015/12016.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergers G., Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weidle U.H., Tiefenthaler G., Weiss E.H., Georges G., Brinkmann U. The intriguing options of multispecific antibody formats for treatment of cancer. Cancer Genom. Proteom. 2013;10:1–18. [PubMed] [Google Scholar]
- 8.Ferrara N., Kerbel R.S. Angiogenesis as a therapeutic target. Nature. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 9.Felcht M., Luck R., Schering A., Seidel P., Srivastava K., Hu J., Bartol A., Kienast Y., Vettel C., Loos E.K., Kutschere S., Bartels S., Appak S., Besemfelder E., Terhardt D., Chavakis E., Wieland T., Klein C., Thomas M., Uemura A., Goerdt S., Augustin H.G. Angiopoietin-2 differentially regulates angiogenesis through TIE2 and integrin signalling. J. Clin. Investig. 2012;122:1991–2005. doi: 10.1172/JCI58832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scharpfenecker M., Fiedler U., Reiss Y., Augustin H.G. The Tie-2 ligand angiopoietin-2 destabilizes quiescent endothelium through an internal autocrine loop mechanism. J. Cell Sci. 2005;118:771–780. doi: 10.1242/jcs.01653. [DOI] [PubMed] [Google Scholar]
- 11.Thomas M., Felcht M., Kruse K., Kretschmer S., Deppermann C., Biesdorf A., Rohr K., Benest A.V., Fiedler U., Augustin H.G. Angiopoietin-2 stimulation of endothelial cells induces alphavbeta3 integrin internalization and degradation. J. Biol. Chem. 2010;285:23842–23849. doi: 10.1074/jbc.M109.097543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Bullock A.J., Zhang L., Wei L., Yu D., Mahagaoker K., Alsop D.C., Mier J.W., Atkins M.B., Coxon A., Oliner J., Bhatt R.S. The role of angiopoetins as potential therapeutic targets in renal cell carcinoma. Transl. Oncol. 2014;7:188–195. doi: 10.1016/j.tranon.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chae S.S., Kamoun S., Farrar C.T., Kirkpatrick N.D., Niemeyer E., de Graaf A.M., Sorensen A.G., Munn L.L., Jain R.K., Fukumura D. Angiopoietin-2 interferes with anti-VEGFR2-induced vessel normalization and survival benefit in mice bearing gliomas. Clin. Cancer Res. 2010;16:3618–3627. doi: 10.1158/1078-0432.CCR-09-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etoh T., Inoue H., Tanaka S., Barnard G.F., Kitano S., Mori M. Angiopoietin-2 is related to tumor angiogenesis in gastric carcinoma: possible in vivo regulation via induction of proteases. Cancer Res. 2001;61:2145–2153. [PubMed] [Google Scholar]
- 15.Ochiumi T., Tanaka S., Oka S., Hiyama T., Ito M., Kitadai Y., Haruma K., Chayama K. Clinical significance of angiopoietin-2 expression at the deepest invasive tumor site of advanced colorectal carcinoma. Int. J. Oncol. 2004;24:539–547. [PubMed] [Google Scholar]
- 16.Sfiligoi C., de Luca A., Cascone I., Sorbello V., Fuso L., Ponzone R., Biglia N., Audero E., Arisio R., Bussolino F., Sismondi P., De Bortoli M. Angiopoietin-2 expression in breast cancer correlates with lymph node invasion and short survival. Int. J. Cancer. 2003;103:466–474. doi: 10.1002/ijc.10851. [DOI] [PubMed] [Google Scholar]
- 17.Kienast Y., Klein C., Scheuer W., Raemsch R., Lorenzon E., Bernicke D., Herting F., Yu S., The H.H., Martarello L., Gassner C., Stubenrauch K.G., Munro K., Augustin H.G., Thomas M. Ang-2-VEGF-A crossMab, a novel bispecific human IgG1 antibody blocking VEGF-A and Ang-2 functions simultaneously, mediates potent antitumor antiangiogenic and antimetastatic efficacy. Clin. Cancer Res. 2013;19:6730–6740. doi: 10.1158/1078-0432.CCR-13-0081. [DOI] [PubMed] [Google Scholar]
- 18.Hidalgo M., Martinez-Garcia M., Le Tourneau C., Massard C., Garralda E., Boni V., Taus A., Albanell J., Sablin M.P., Alt M., Bahleda R., Varga A., Boetsch C., Franjkovic I., Heil F., Lahr A., Lechner K., Morel A., Nayak T., Rossomanno S., Smart K., Stubenrauch K., Krieter O. First-in-human phase I study of single-agent vanucizumab, a first-in-class bi-specific antiAng-2/anti-VEGF antibody, in adult patients with advanced solid tumors. Clin. Cancer Res. 2018;24:1536–1545. doi: 10.1158/1078-0432.CCR-17-1588. [DOI] [PubMed] [Google Scholar]
- 19.Moore H.M., Kelly A.B., Jewell S.D., McShane L.M., Clark D.P., Greenspan R., Hayes D.F., Hainaut P., Kim P., Mansfield E.A., Potapoya O., Riegman P., Rubinstein Y., Seijo E., Somiari S., Watson P., Weier H.U., Zhu C., Vaught J. Biospecimen reporting for improved study quality (BRISQ) Cancer Cytopathol. 2011;119:92–102. doi: 10.1002/cncy.20147. [DOI] [PubMed] [Google Scholar]
- 20.Stubenrauch K., Wessels U., Essig U., Vogel R., Waltenberger H., Hansbauer A., Koehler A., Heinrich J. An immunodepletion procedure advances free angiopoietin-2 determination in human plasma samples during anti-cancer therapy with bispecific anti-ANG-2/VEGF crossMAb. J. Pharm. Biomed. Anal. 2015;102:459–467. doi: 10.1016/j.jpba.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Mundhenke C., Thomas J.P., Wilding G., Lee F.T., Kelzc F., Chappell R., Neider R., Sebree L.A., Friedl A. Tissue examination to monitor anti-angiogenic therapy: a phase I trial with endostatin. Clin. Cancer Res. 2001;7:3366–3374. [PubMed] [Google Scholar]
- 22.Zhang D., Pier T., McNeel D.G., Wilding G., Friedl A. Effects of a monoclonal anti-αvβ3 integrin antibody on blood vessels – a pharmacodynamic study. Investig. New Drugs. 2006;25:49–55. doi: 10.1007/s10637-006-9013-8. [DOI] [PubMed] [Google Scholar]
- 23.Ruifrok A.C., Johnson D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001;23:291–299. [PubMed] [Google Scholar]
- 24.Core Team R. R Foundation for Statistical Computing; Vienna, Austria: 2017. R: A Language and Environment for Statistical Computing.https://www.R-project.org Available at: Accessed March 2018. [Google Scholar]
- 25.Pinheiro, J., D. Bates, S. DebRoy, D. Sarkar, and R. Core Team. 2017. Nlme: Linear and Nonlinear Mixed Effects Models. R Package Version 3.1–131. https://CRAN.R-project.org/package=nlme. Accessed December 2019.
- 26.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A., Tamayo P., Mootha V., Mukherjee S., Ebert E.L., GilLette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Nat. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angelova M., Charoentong P., Hackl H., Trajanoski Z. The colorectal cancer immune paradox revisited. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2015.1078058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S., Rouphael N., Duraisingham S., Romero-Steiner S., Presnell S., Davis C., Schmidt D.S., Johnson S.E., Milton A., Rajam G., Kasturi S., Carlone G.M., Quinn C., Chaussabel D., Palucka A.K., Mulligan M.J., Ahmed R., Stephens D.S., Nakaya H.I., Pulendran B. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tirosh I., Izar B., Prakadam S.M., Wadsworth M.H., II, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., Fallahi-Sichani M., Dutton-Regester K., Lin J.R., Cohen O., Shah P., Lu D., Genshaft A.S., Hughes T.K., Ziegler C.G., Kazer S.W., Gaillard A., Kolb K.E., Villani A.C., Johannessen C.M., Andreev A.Y., Van Allen E.M., Bertagnolli M., Sorger P.K., Sullivan R.J., Flaherty K.T., Frederick D.T., Jané-Valbuena J., Yoon C.H., Rozenblatt-Rosen O., Shalek A.K., Regev A., Garraway L.A. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiedler U., Augustin H.G. Angiopoietins: a link between angiogenesis and inflammation. Trends Immunol. 2006;27:552–558. doi: 10.1016/j.it.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Fiedler U., Scharpfenecker M., Koidl S., Hegen A., Grunow V., Schmidt J.M., Kriz W., Thurston G., Augustin H.G. The Tie-2 ligand angiopoietin-2 is stored in and rapidly released upon stimulation from endothelial cell Weibel-Palade bodies. Blood. 2004;103:4150–4156. doi: 10.1182/blood-2003-10-3685. [DOI] [PubMed] [Google Scholar]
- 33.Rondaij M.G., Bierings R., Kragt A., van Mourik J.A., Voorberg J. Dynamics and plasticity of Weibel-Palade bodies in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2006;26:1002–1007. doi: 10.1161/01.ATV.0000209501.56852.6c. [DOI] [PubMed] [Google Scholar]
- 34.Gooden M.J.M., de Bock G.H., Leffers N., Daemen T., Nijman H.W. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br. J. Cancer. 2011;105:93–103. doi: 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dirkx A.E., Oude Egbrink M.G., Kuijpers M.J., van der Niet S.T., Heijnen V.V., Bouma-ter Steege J.C., Wagstaff J., Griffoen A.W. Tumor angiogenesis modulates leukocyte-vessel wall interactions in vivo by reducing endothelial adhesion molecule expression. Cancer Res. 2003;63:2322–2329. [PubMed] [Google Scholar]
- 36.Dirkx A.E., Oude Egbrink M.G., Castermans K., van der Schaft D.W., Thijssen V.L., Dings R.P., Kwee L., Mayo K.H., Wagstaff J., Bouma-ter Steege J.C., Griffoen A.W. Anti-angiogenesis therapy can overcome endothelial cell anergy and promote leukocyte-endothelium interactions and infiltration in tumors. FASEB J. 2006;20:621–630. doi: 10.1096/fj.05-4493com. [DOI] [PubMed] [Google Scholar]
- 37.Shrimali R.K., Yu Z., Theoret M.R., Chinnasamy D., Restifo N.P., Rosenberg S.A. Antiangiogenic agents can increase lymphocyte infiltration into tumor and enhance the effectiveness of adoptive immunotherapy of cancer. Cancer Res. 2010;70:6171–6180. doi: 10.1158/0008-5472.CAN-10-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]