Summary
Amyloid precursor protein (APP) cleavage by the β-secretase produces the C99 transmembrane (TM) protein, which contains three dimerization-inducing Gly-x-x-x-Gly motifs. We demonstrate that dimeric C99 TM orientations regulate the precise cleavage lines by γ-secretase. Of all possible dimeric orientations imposed by a coiled-coil to the C99 TM domain, the dimer containing the 33Gly-x-x-x-Gly37 motif in the interface promoted the Aβ42 processing line and APP intracellular domain-dependent gene transcription, including the induction of BACE1 mRNA, enhancing amyloidogenic processing and signaling. Another orientation exhibiting the 25Gly-x-x-x-Gly29 motif in the interface favored processing to Aβ43/40. It induced significantly less gene transcription, while promoting formation of SDS-resistant “Aβ-like” oligomers, reminiscent of Aβ peptide oligomers. These required both Val24 of a pro-β motif and the 25Gly-x-x-x-Gly29 interface. Thus, crossing angles imposed by precise dimeric orientations control γ-secretase initial cleavage at Aβ48 or Aβ49, linking the former to enhanced signaling and Aβ42 production.
Subject Areas: Biological Sciences, Biochemistry, Molecular Biology, Neuroscience
Graphical Abstract
Highlights
-
•
C99 dimeric transmembrane orientations regulate γ-secretase processing line
-
•
Aβ42 production and signaling are linked to the 33Gly-x-x-x-Gly37 interface
-
•
SDS-resistant oligomers require the 25Gly-x-x-x-Gly29 interface and a pro-β motif
-
•
C99 dimeric orientations impact its localization and processing by PSEN1 or PSEN2
Biological Sciences; Biochemistry; Molecular Biology; Neuroscience
Introduction
The amyloid precursor protein (APP) is a type I transmembrane (TM) protein with a long extracellular domain, a single TM α-helix and a C-terminal cytosolic domain with no defined structure (Kang et al., 1987). Either α- or β-secretase first cleaves APP to generate two distinct fragments: a soluble N-terminal fragment (sAPPα or sAPPβ, respectively) and a membrane-anchored C-terminal fragment (α- or β-CTF, respectively) (Weidemann et al., 1989). Both α- or β-CTFs can be processed by γ-secretase to produce the APP intracellular domain (AICD). However, transcriptional activity associated with the release of AICD is preferentially correlates with the processing of the β-CTF or C99 (Belyaev et al., 2010) and only β-CTF can generate the amyloid β (Aβ) peptides. These are present in various lengths ranging predominantly from Aβ34 to Aβ46. It is considered that Aβ40 is predominant in terms of production and that Aβ42 and longer isoforms like Aβ43 are most toxic, due to their higher propensity to aggregate because of the extra hydrophobic residues at the C-terminus. Aβ42 forms the core of amyloid plaques in the brains of patients with Alzheimer's disease (AD) (Iwatsubo et al., 1994; Szaruga et al., 2017).
The first cleavage by γ-secretase on C99 occurs at the ε-site, corresponding to the residue 48 or 49 of the β-CTF, located at the cytoplasmic edge of the TM domain (Qi-Takahara et al., 2005; Takami et al., 2009). The ε-site is at a junction between the TM helix and the flexible cytosolic domain, where we showed a helical to coil transition to occur at the structural level (Sato et al., 2009). Support for such an unraveling close to the ε-site and progressive cleavage was recently provided by the single particle electron microscopy structure of a complex between a C83 fragment cross-linked to catalytically inactive presenillin 1 (PS1) (Zhou et al., 2019). The cleavage at ε-site leads to the formation of different AICD species, usually starting after residue 48 or after residue 49. The membrane-anchored fragment containing the β-amyloid sequence is further trimmed upstream by the γ-secretase carboxypeptidase activity. Two processing lines generate Aβ 46/43/40 and Aβ 45/42 fragments, respectively (Qi-Takahara et al., 2005; Takami et al., 2009). The AICD fragments have been reported to regulate gene transcription and require interaction with binding proteins in order to be translocated to the nucleus (Cao and Sudhof, 2001; Pardossi-Piquard et al., 2005). AICD-dependent signal transduction remains still controversial. Its regulatory properties on transcription have been a matter of debate, but experimental evidence associates AICD release to the transcriptional regulation of APP target genes (Pardossi-Piquard et al., 2005).
APP contains three Gly-x-x-x-Gly motifs or glycine zippers at the junction between the extracellular juxtamembrane (JM) and TM regions. It is known from the glycophorin A protein that Gly-x-x-x-Gly and Gly-x-x-x-Ala particular motifs can induce SDS-resistant dimerization of TM proteins by creating close apposition between the glycines in the interacting α-helices (Bormann et al., 1989; MacKenzie et al., 1997). Importantly, SDS-resistant dimerization also requires other residues besides the Gly-x-x-x-Gly motif, and the position of the motif relative to the thickness of the bilayer is also important (Bormann et al., 1989; MacKenzie et al., 1997). The Gly-x-x-x-Gly motifs were reported as key molecular determinants that stabilize the association of the α-helices of APP TM dimers, and that modulate APP processing by the γ-secretase complex (Kienlen-Campard et al., 2008; Munter et al., 2007). The Flemish mutation in which there is an Ala to Gly substitution at position 21, creates a fourth upstream in-register Gly-x-x-x-Gly motif and induces a conformational change by increasing the α-helix at position 21 (Tang et al., 2014). This promotes an increase in Aβ production, confirming the relation existing between APP TM dimerization and APP amyloidogenic processing. Still, the links existing between dimerization and processing remains elusive. For instance, the cryo-EM structure of γ-secretase in complex with C83 (Zhou et al., 2019) and other experimental data (Eggert et al., 2018) does not support the direct processing of C99 dimers. APP dimerization is known to affect its subcellular localization (Eggert et al., 2018; Ben Khalifa et al., 2012) but the precise mechanisms by which TM dimerization regulate the processing (and likely the function) of APP and, more specifically of C99, are not understood.
Here we asked whether a precise dimeric interface of C99 TM helices could influence processing by γ-secretase leading to AICD formation and signaling via ε-cleavage, as well as Aβ accumulation subsequent to processing at γ-sites. We used C99 fusion constructs with a coiled coil dimer (Put3) at different junction points within the TM domain. These Put3 dimeric constructs, referred to as cc-del(X) constructs, allowed us to individually dial-in all possible dimeric interfaces in the C99 TM domain. The effects of each of the fusion proteins, reproducing one at a time the individual dimeric interfaces, were tested in cell lines and primary neurons with respect to signaling, processing and oligomerization. We identified a precise dimeric interface where strong AICD release by γ-secretase is associated to signaling and likely favors the Aβ42 processing line. This orientation seems to be preferentially processed by a presenilin 2 or PS2-dependent γ-secretase. Another interface rotated clockwise from the previous one by 110° is associated with oligomerization of processed amyloid peptides and favors the Aβ43 processing line, preferentially through presenilin 1 or PSEN1-dependent γ-secretase. Strikingly, both of these interfaces are lined with glycines. This difference in TM packing appears to be translated into processing by different γ-secretases dependent on either of the two presenilin (PS1 and PS2) proteins, likely in different subcellular compartments, and into different signaling and oligomerization properties. We discuss how this knowledge can be translated in therapies aimed at preventing amyloid deposition, and we reveal that signaling and processing/oligomerization are supported by close dimeric interface.
Results
Design of the Fusion Proteins Put3 Coiled Coil C99
The left-handed leucine zipper coiled coil of the transcription factor Put3 (Put3-cc) from Saccharomyces cerevisiae was used to create fusion proteins at different junction points within the extracellular JM domain of C99. This strategy has previously been used to determine the effects of dimeric orientation on type II TM proteins (Mattoon et al., 2001), where the N-terminus is cytosolic, and on type I TM proteins where the N-terminus is extracellular (Seubert et al., 2003; Staerk et al., 2011). For the erythropoietin and thrombopoietin receptors, it has been shown by cysteine-mediated cross-linking that dimerization at different orientations imposed at the extracellular end of the TM domains is translated into the correct interfaces at the C-terminal ends of the TM domains (Staerk et al., 2011).
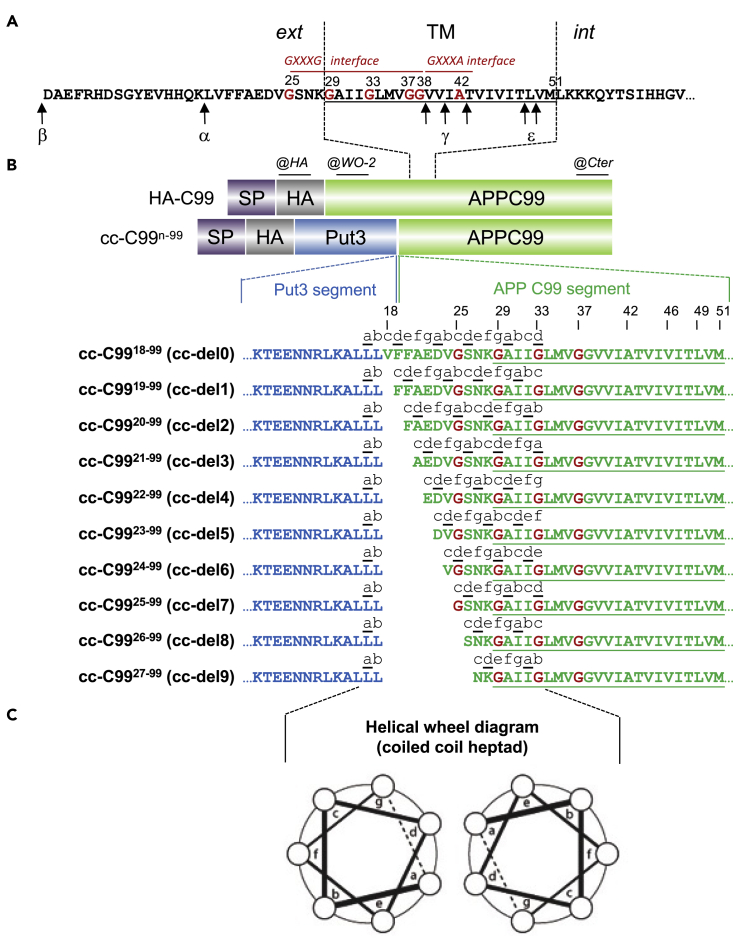
We replaced amino acids 1-17 of the C99 extracellular domain with the Put3-cc (Walters et al., 1997), which exhibits an overall neutral charge that makes it compatible with an extracellular topology. We engineered fusion proteins where the extracellular Put3-cc forces the C99 monomers to adopt each of the seven possible symmetric left-handed orientations downstream of the coiled coil, denoted cc-del0 to cc-del6 (cc-C9918−99 to cc-C9924−99, respectively). We also created three shorter constructs, denoted cc-del7 to cc-del9 (cc-C9925−99 to cc-C9927−99, respectively), where the interfaces are similar to the first three fusion proteins, but where significant residues of the JM region are deleted, as certain motifs such as 16LVFF20 have been reported to exert an inhibitory effect on processing by γ-secretase (Tian et al., 2010) (Figures 1A and 1B). All cc-C99 (cc-del(X)) fusion proteins and wild type C99 are HA-tagged and contain a signal peptide at the N-terminus. The positions of heptad repeats characteristic of left-handed coiled coils are denoted a-g with interface residues in “a” and “d” positions, with edge residues close to the interface in “e” and “g” positions and with residues outside of the interface in “b”, “c”, and “f” positions (Figure 1C). The cc-del0, cc-del1, and cc-del2 constructs exhibit the same orientations as the cc-del7, cc-del8, and cc-del9 constructs, respectively.
Figure 1.
Design of Coiled Coil APP C99 Fusion Proteins
(A) Representation of the juxtamembrane and transmembrane domains of human APP C99. The positions of the consecutive GxxxG motifs in Aβ numbering are in red. The cleavage sites of α-, β- and γ-(γ and ε) secretases are indicated by arrows. Ext, external; Int, internal; TM, transmembrane.
(B) Representation of HA-C99 and cc-del(X) constructs. The coiled-coil dimerization domain of Put3 was fused to consecutive residues of the juxtamembrane sequence of C99 starting at residue 17 (Aβ numbering) and where n is the first amino acid of the sequence. All seven possible registers of C99 TM dimers were imposed by successive N-terminal deletion. Constructs cc-C9918−99 to cc-C9927−99 are referred to as cc-del(X) with X from 0 to 9, respectively. cc-del7 to 9 have the same orientation as cc-del0 to 2, respectively, but without the 16LVFF20 motif. All fusion proteins contain a signal peptide (SP) and an HA tag upstream the Put3 sequence. Only HA-C99 harbors the WO-2 epitope. The Y188 antibody (Cter) is directed against the C-terminus of the C99 sequence.
(C) Helical wheel diagram showing the heptad repeats in cc-del(X). Position of heptad repeats are denoted a to g with the dimeric interface considered to be at positions a and d. See also Figures S1 and S2.
Validation of the TM Orientations of Fusion Proteins cc-del(X)
The dimer interface can be predicted for each fusion protein (Figures S1A and S1B). To test the model of the predicted orientation of the cc-del(X) constructs, a cysteine was substituted for Gly29 (G29C) in all the constructs (Figures S1A and S1B). Because this Cys residue is unique in the cc-del(X) fusion proteins, its position can be used as a readout for the dimerization interface by cross-linking studies. We used the Cys-dependent and irreversible cross-linking agent N, N′-1,2-phenylenedimaleimide (o-PDM) that forms dimers if two proteins are separated by a distance of 6–10 Å. We have predicted, based on α-helical wheel diagrams, which constructs can form dimers due to the presence of Cys residues within the interface itself (represented in red), with Cys residues in close enough proximity (represented in blue) or with Cys residues too far apart to dimerize (represented in green) (Figure S1B).
Cross-linking experiments were performed with living cells and validated the orientations imposed by the coiled coil structure in the JM domain. Western blot analysis using the C-terminal (Cter) APP antibody showed all cc-del(X) G29C constructs were expressed in CHO cells at the expected size of 14 kDa (Figure S1C). Cells treated with o-PDM are able to form intermolecular bonds maintaining stable dimers in denaturing and reducing conditions. In the presence of o-PDM, we observed the formation of cross-linked dimers for cc-del3 G29C and cc-del6 G29C, consistent with the hypothesis that these two constructs should have G29C in the dimeric interface (positions “d” and “a” of a heptad repeat). The cc-del0, cc-del2, cc-del7 and cc-del9 were also able to form dimers, the two cysteines being close enough to the interface via edge residues, placed at “e” or “g” positions of the heptad repeat. Specifically, G29C will be located in “g”, “e”, “g”, and “e” for cc-del0, cc-del2, cc-del7, and cc-del9, respectively. The other dimers that do not cross-link exhibit C29 at positions “f”, “c”, and “b”, namely cc-del1, cc-del4, cc-del5, and cc-del8, respectively. To validate that our constructs obeyed the predictions downstream of Gly29, we introduced a Cys mutation at position 37 (G37C) in cc-del4, cc-del6, cc-del7, and cc-del9. In the presence of o-PDM, we observed dimers in cc-del4 G37C and cc-del7 G37C but not in cc-del6 G37C and cc-del9 G37C (Figure S1C). This is in agreement with our predictions as G37C was expected to be in the dimeric interface for cc-del4 and cc-del7 (positions “d” and “a” of the heptad repeat, respectively) but outside the interface for cc-del6 and cc-del9 (position “b” and “f”, respectively).
While the TM domain of C99 adopts an α-helix secondary structure (Nadezhdin et al., 2012; Tang et al., 2014; Zhou et al., 2019), it has been demonstrated that unraveling occurs upstream of the ε-site, starting around residues 48-49 (Sato et al., 2009). This unraveling is followed by a short cytosolic β-strand downstream, which is induced by the contact with PS1 in the γ-secretase complex (Zhou et al., 2019). We asked whether our engineered dimers allow this level of flexibility or whether they are maintained in a rigid conformation at the C-terminus of the TM domain. For this purpose, we substituted L49 with cysteine (L49C) in all ten cc-del(X) fusion proteins. Cross-linking with o-PDM could be detected for all fusion proteins, consistent with unraveling at that position, similar to that described for wild-type C99 (Figure S1D) (Sato et al., 2009; Zhou et al., 2019).
The cryo-EM structure of γ-secretase in complex with C83 (Zhou et al., 2019) does not support processing of dimers. Our previous work also pointed to the need for unwinding of the α-helix around γ-sites for processing (Sato et al., 2009). The enzymatic cleavage of stable dimers is therefore very unlikely, although binding of CTF dimers to PS complexes may occur previous to entry in the catalytic site and actual cleavage. We used the cc-del6 G29C construct which form stable dimers upon crosslinking (Figure S1C) to assess its processing in dimeric form in a cell-free assay by measuring AICD release. We observed the release of AICD after incubation of isolated whole-cell membrane for 2 hr at 37°C when cc-del6 G29C was not crosslinked, and this release did was not detectable when cc-del6 G29C was previously crosslinked with o-PDM into stable dimers (Figure S2A). As an additional control, we verified that o-PDM did not by itself impede cleavage by γ-secretase, as AICD release could be observed with or without crosslinking upon processing of cc-del6 without the G29C mutation (Figure S2B). These results are in line with previous reports (Sato et al., 2009; Zhou et al., 2019) and suggest that neither C99 nor our constructs can be cleaved in dimeric form. Still, differences in addressing of C99 dimers and/or in initial docking of C99 dimers to γ-secretase are likely to affect their processing and functional properties.
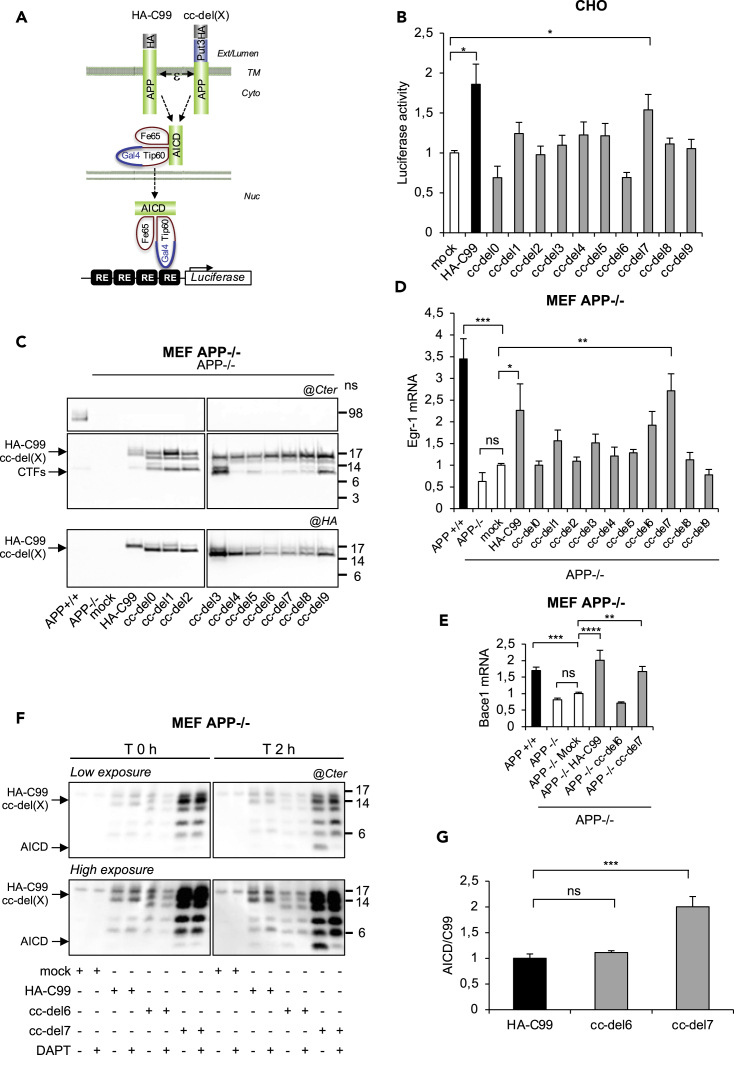
cc-del7 Favors Transcriptional Activity and AICD Production by γ-Secretase Activity
The release of AICD was measured in CHO cells which were co-transfected with the HA-C99 or cc-del(X) fusion proteins, Fe65, Tip60Gal4, Gal4RE-luciferase as a reporter gene (Cao and Sudhof, 2001) and the pRLTK (renilla luciferase) control reporter to normalize for transfection efficiency (Figure 2A). The level of transcription was significantly increased, as expected with the HA-C99 when compared to the mock control, but strikingly only with cc-del7 among the cc-del(X) constructs (Figure 2B). For the other constructs, the level of transcriptional activity was not significantly different when compared to the mock control. It appears therefore that only one precise TM dimeric orientation (cc-del7) promoted APP-dependent nuclear signaling.
Figure 2.
Signaling and Gene Expression via AICD Production of cc-del7 Fusion Proteins
(A) Transactivation assay to monitor the release of transcriptionally active AICD. Cleavage at the ε-site releases AICD, which translocates to the nucleus together with Fe65 and Tip60Gal4. The AICD/Fe65/Tip60Gal4 complex transactivates the Gal4RE reporter gene. Ext, exernal; Cyto, cytoplasm; TM, transmembrane; Nuc, nucleus.
(B) Transcriptional activity of the cc-del(X). Data are mean values ± SEM (n = 3, N = 3). Statistical analysis: non-parametric multiple comparisons Steel's test with control (mock). ∗p < 0.05.
(C) Western blots of HA-C99 and cc-del(X) constructs stably expressed in MEF APP−/−, revealed with the APP Cter (top) or the HA antibody (bottom).
(D) Egr-1 mRNA of MEF cells described above (C). Results (Means ± SEM; n = 3; N = 3) are given as fold induction compared to control (APP−/− mock).
(E) Bace1 mRNA of MEF cells described above (C). Results (Means ± SEM; n = 3; N = 3) are given as fold induction compared to control (APP−/− mock).
(F) AICD production in MEF APP−/− expressing cc-del(X) constructs. Total membrane extracts analyzed by Western blotting and revealed with APP Cter.
(G) Quantification of AICD. Results (Means ± SEM; n = 3; N = 2) are given as AICD:HA-C99 or AICD:cc-del(X) ratios, with AICD:HA-C99 ratio fixed at 1. Statistical analysis (D, E and G): one-way ANOVA followed by Dunnett's multiple comparison test. ns: non-significant, ∗: p < 0.05, ∗∗: p < 0.005, ∗∗∗: p < 0.001, ∗∗∗∗: p < 0.0001.
To determine whether this effect was truly due to the activity of cc-del(X) and the release AICD associated with transcriptional activity, we created stable cell lines using murine embryonic fibroblasts deficient for APP (MEF APP−/−). Western blotting with antibodies directed against the C-terminus of C99 (Cter) or the HA epitope indicated that cc-del(X) fusion proteins exhibited comparable levels of expression in MEF APP−/− cells (Figure 2C).
Within these stable cell lines, qPCR was used to analyze the expression of the Egr-1 gene known to be an APP downstream target (Hendrickx et al., 2013). Egr-1 is an immediate-early response gene regulating the transcription of late response genes important for synaptic plasticity processes, especially the maintenance of long-term potentiation (Jones et al., 2001). Exposure of mice to a novel environment induces Egr-1 expression in an APP-dependent manner (Hendrickx et al., 2013). We therefore chose Egr-1 as it is transiently induced via APP, and appears as a readout for the signaling function of APP upon cleavage by γ-secretase. We observed a decrease in the expression of Egr-1 in MEF APP−/− compared to MEF APP+/+ cells (Figure 2D). The expression of Egr-1 was restored in MEF APP−/− cells expressing HA-C99 or cc-del7 but not the other constructs. Importantly, Egr-1 is a transcriptional activator of the β-secretase (Bace1) and its inhibition decreases amyloidogenic processing in Alzheimer mouse models (Qin et al., 2016). We used qPCR to assess whether Bace1 expression was also increased in cells stably expressing cc-del7. The expression of Bace1 was decreased in MEF APP−/− compared to MEF APP +/+. Its expression was restored in MEF APP −/− cells stably expressing HA-C99 or cc-del7 but not in MEF APP−/− cells stably expressing a mock vector or cc-del6 (Figure 2E). In agreement with previous results (Qin et al., 2016), the expression profile of Bace1 mimicked the one observed for Egr-1.
Knowing that APP-dependent gene expression is associated with the release of AICD, we next assessed the production of AICD via a cell free experimental approach. Whole-cell membranes from MEF APP−/− cells expressing different cc-del(X) constructs were isolated with hypotonic buffer and ultracentrifugation. The membrane lysates were analyzed by Western blot using an antibody directed against APP C-terminus at two time points, 0 hr and 2 hr of incubation at 37°C in the absence or presence of a γ-secretase inhibitor (DAPT). The results indicated an increase in intensity of the band corresponding to AICD after 2 hr compared to the initial time point for all constructs tested. Strikingly, this increase was much stronger for cc-del7 than for cc-del6 or HA-C99 (Figure 2F). Quantification of the ratio of AICD produced confirmed a significant increase for cc-del7 when compared to the other constructs (Figure 2G). The bands considered (below 6 kDa) were strongly reduced in the presence of DAPT, indicating that they truly correspond to AICD produced by the γ-secretase.
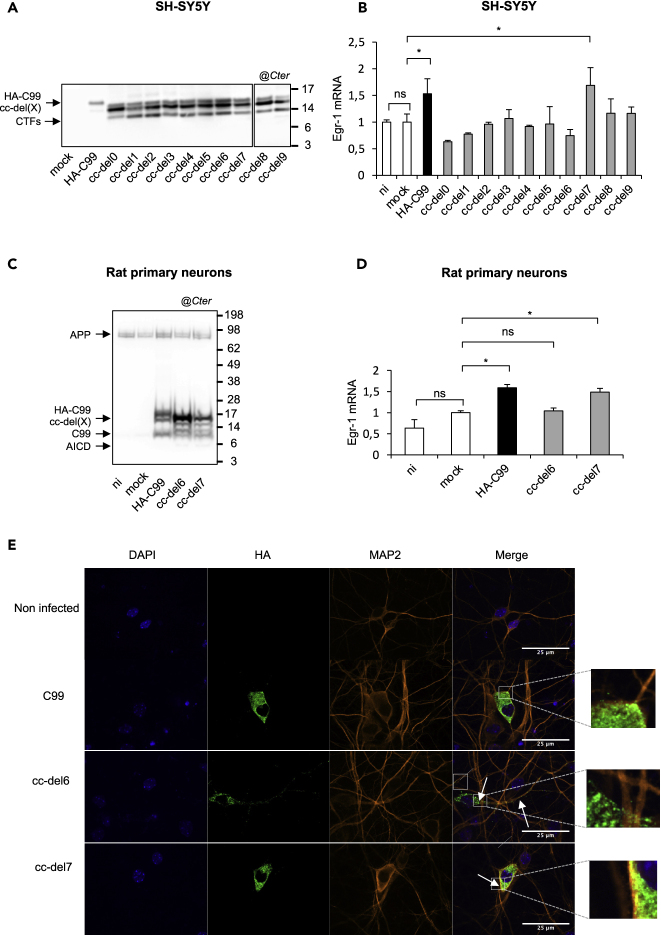
cc-del7 Orientation Favors AICD-Dependent Signaling in Human Neuronal Cell Lines and in Primary Neurons
To test whether the cc-del(X) fusion proteins exerted the same effects in human neuronal cells, we used the SH-SY5Y human neuroblastoma cell line, which exhibit neuronal properties upon differentiation with retinoic (Agholme et al., 2010). We generated SH-SY5Y cell lines stably expressing cc-del(X) by lentiviral transduction followed by puromycin selection. The expression profile of fusion proteins was tested 7 days after differentiation by Western blotting (Figure 3A). We next examined whether dimeric orientation also exerted an effect on gene expression in the neuronal cell line. Egr-1 expression increased for HA-C99 and for cc-del7, while the expression remained at a similar level to mock infected SH-SY5Y cells for the other constructs (Figure 3B).
Figure 3.
Regulation of the Transcriptional Activity by the cc-del7 Construct
(A) Cell lysates SH-SY5Y cells stably expressing HA-C99 or cc-del(X) constructs analyzed by Western blotting and revealed with the APP Cter antibody.
(B) Egr-1 mRNA levels in SH-SY5Ycells. Results (Means ± SEM; n = 3; N = 3) are given as fold induction compared to control (mock). Statistical analysis: one-way ANOVA followed by Dunnett's multiple comparison test. ∗p < 0.05.
(C) Rat cortical neurons were transduced with lentivirus expressing mock vector (GFP), HA-C99, cc-del6 and cc-del7 or non-infected (ni). Neuronal lysates were analyzed 4 days after infection by Western blotting and revealed APP Cter antibody (right).
(D) Egr-1 mRNA levels in primary neurons. Results (Means ± SEM; n = 3; N = 3) are given as fold induction compared to control the mock. Statistical analysis: one-way ANOVA followed by Dunnett's multiple comparison test. ns: non-significant, ∗p < 0.05.
(E) Mouse cortical neurons were transduced with lentivirus expressing HA-C99, cc-del6 and cc-del7 or non-infected (ni). Cells were stained with MAP2 neuronal marker (orange), HA antibody (green) and DAPI (blue) and analyzed by confocal microscopy.
See also Figure S3.
We further tested whether our model was also relevant for rat primary cortical neurons. Primary neurons were transduced with lentiviruses expressing mock vector, HA-C99, cc-del6 and cc-del7. The protein expression profile was examined by immunoblotting with the Cter APP antibody (Figure 3C). In line with our results in cell lines, Egr-1 gene expression was significantly increased in primary neurons transduced with HA-C99 and cc-del7, confirming their gene regulation activity (Figure 3D).
APP dimerization is known to affect its subcellular localization (Eggert et al., 2009) and processing (Ben Khalifa et al., 2012). Confocal microscopy on differentiated SH-SY5Y cells revealed that HA-C99 presented a diffuse expression. For the cc-del6 construct, the localization appeared mainly associated with the plasma membrane, while the cc-del7 seemed to exhibit predominantly intracellular localization (punctae) (Figure S3). We next examined whether these features were similar in primary neurons transduced with HA-C99, cc-del6 or cc-del7. HA-C99 and cc-del7 were essentially present in the soma with little membrane localization. In sharp contrast, cc-del6 seemed also present at the membrane and displayed important localization in neuronal processes (Figure 3E).
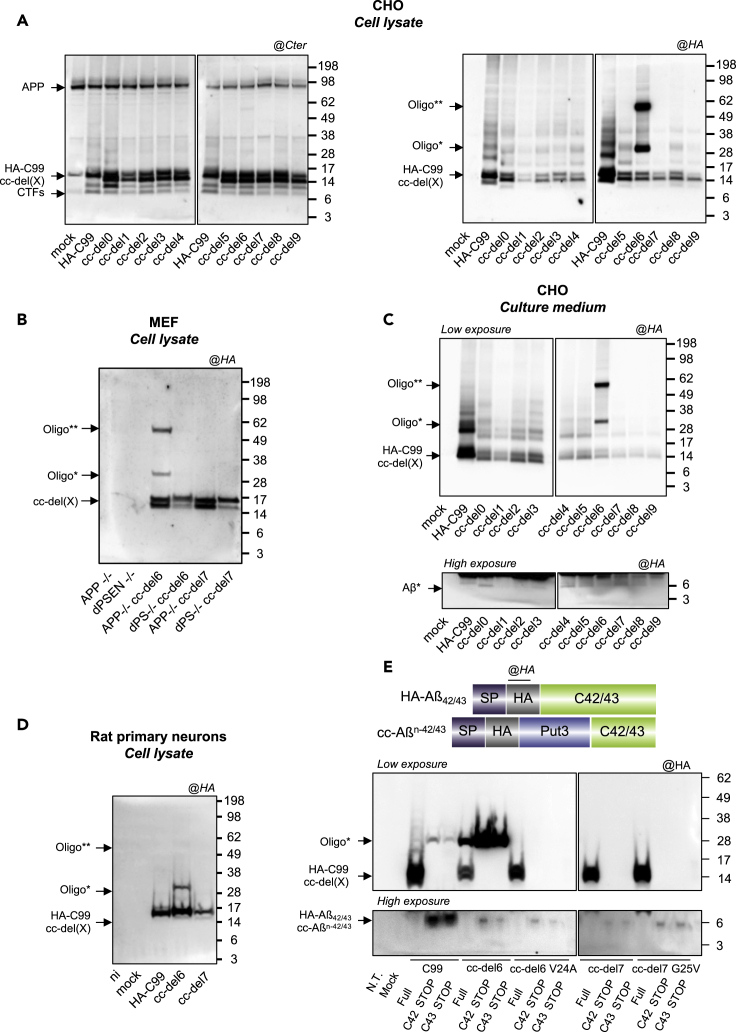
cc-del6 Favors the Production of SDS-Resistant Oligomers by γ-Secretase Processing
So far, the cc-del7 construct displayed unique features compared to other cc-del(X) proteins by promoting the release AICD associated with gene regulation. Apart from AICD, the processing of APP by γ-secretase produces a class of Aβ peptides directly involved in senile plaque formation. To assess in detail the amyloidogenic profile of all the cc-del(X) fusion proteins, CHO cells transiently expressing cc-del(X) constructs were lysed and separated by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis), then analyzed by Western blotting. The constructs were observed as monomers at the expected size (14-15 kDa) with the Cter antibody. Additional bands were detected at a lower size corresponding likely to a C83-like CTF (Figure 4A). Strikingly, the anti-HA antibody also detected a band around 28 kDa and 60 kDa uniquely for cc-del6. As we did not observe these bands with the Cter APP antibody, we infer the identities of these bands to be oligomeric “Aβ-like” species that we call “oligo∗” or “oligo∗∗” corresponding to the 28 kDa and 60 kDa, respectively. Importantly, expression of HA-C99 also led to the appearance of low levels of SDS-resistant 28 kDa oligomers, and those were also only recognized by the anti-HA antibodies, suggesting they lack C-term epitopes.
Figure 4.
cc-del6 Orientation in Conjunction with pro-β Motif Favors the Production of SDS-Resistant Oligomers by γ-Secretase Processing
(A) Analysis of the profile of cc-del(X) constructs in denaturing conditions. Cells lysates were analyzed by Western blotting and revealed with APP Cter antibody (left) or HA antibody (right).
(B) Western blot of MEF APP+/+, MEF double knockout for presenilin 1 and 2 (dPSEN−/−). Detection was performed using the HA-tag antibody.
(C) Analysis of extracellular media. Immunoprecipitation was performed on the supernatant. CHO cells were expressing HA-C99 and cc-del(X) constructs, analyzed by Western blotting and revealed with HA antibody (top). At high exposure (bottom), monomeric Aβ-like (Aβ∗) was detected.
(D) Neuronal lysates were analyzed 4 days after infection by Western blotting and revealed with HA antibody.
(E) Top: Representation of HA-C42/43 and cc-Aβn−42/43 constructs. Bottom: Analysis of the profile of truncated forms (C42/C43) of cc-del(X) constructs and Aβ-like peptides (top) in denaturing conditions. Cell lysates were analyzed by Western blotting and revealed with HA antibody (bottom).
See also Figures S4–S6.
To analyze whether the abovementioned oligomeric bands were produced by γ-secretase activity, we generated cell lines stably expressing cc-del(X) in MEFs deficient for presenilin 1 and presenilin 2 (MEF dPSEN−/−), which are devoid of γ-secretase activity. The expression profile of cc-del(X) constructs was identical in MEF APP−/− cells (Figure 4B) and CHO cells (Figure 4A), with only MEF APP−/− cc-del6 exhibiting oligomeric bands around 28 kDa and 60 kDa. We compared the expression profiles of cc-del6 in MEF APP−/− and MEF dPS−/− cells. No oligomeric bands were observed in MEF dPSEN−/− cc-del6 cells, indicating that processing by the γ-secretase is mandatory for production of the SDS-resistant 28 kDa and 60 kDa oligomeric bands.
We further investigated whether oligomers detected in cell lysates are also released in the culture medium. The release of cc-del(X) fragments was measured in the supernatant of CHO cells by immunoprecipitation with anti-HA antibodies. We observed the presence of the two bands at 28 kDa and 60 kDa secreted in the supernatant for cc-del6 (Figure 4C) and also the presence of cc-del(X) monomers. Importantly, upon higher exposure, we noticed for all the cc-del(X) constructs, the presence of a band around 6 kDa. This band likely corresponds to the monomer of the Aβ-like protein (Aβ∗) produced by the cc-del(X) constructs.
We next assessed whether these Aβ-like oligomers could also be detected in primary neurons transduced with lentiviruses expressing HA-C99, cc-del6 and cc-del7. Immunoblotting with the HA antibody (Figure 4D) showed that cc-del6 is able to form oligomers in primary neuronal cultures, like in the other cell lines tested.
The Production of SDS-Resistant Oligomers Requires a Pro-β Motif and a Specific Orientation of Aβ-like Peptides
CHO cells were also transfected with the constructs without the Put3 coiled coil system, differing only by the precise deletions in C99 (Figure S4A). This was performed in order to investigate whether the oligomerization profile of C99 is impacted by the deletions in the absence of the Put3 coiled coil. All constructs up to del5 expressed SDS-resistant oligomers in cell lysates (Figure S4B). Further deletions (del6-9) impaired insertion in the membrane as the TM segment became too short (unlike the Put3 fusions which always provide extra Leu residues) and did not give rise to any oligomers. These results indicate that progressive deletions at the outset of the TM domain do not by themselves induce oligomerization and that Put3 by providing the required hydrophobicity allows membrane insertion, cell-surface localization, in addition to imposing precise orientations.
The expression of cc-del(X) in dimeric form was verified by transient transfection in CHO cells by blue native PAGE with Cter APP antibody and the presence of oligomers with HA antibody (Figure S4C). The presence of bands at the expected size around 20 kDa (C-ter antibody) confirmed that all cc-del(X) fusion proteins are able to form dimers. We also observed (with the HA antibody) the presence of bands around 66 kDa for most constructs with highest levels in cc-del3, cc-del5 and cc-del6. Those results, correlated to the previous results (Figure 4A), show that although all the constructs are detected as dimers in native conditions, only cc-del6 is able to form SDS-resistant oligomers. Importantly, oligomers induced by other constructs, notably cc-del3 and cc-del5, are not SDS-resistant.
Next, we used the Tango algorithm to assess whether the addition of Put3 could provide an environment favoring the production of the SDS-resistant oligomers observed upon cc-del6 processing. The Tango algorithm predicts the cross-β aggregation propensity of amino acids sequences (referred to as pro-β motif) based on physico-chemical principles. Its accuracy was previously validated on a set of 250 peptides including the amyloid beta peptide (Fernandez-Escamilla et al., 2004). As expected, the C-terminal segment of the different forms of Aβ-like peptides was predicted to have a high propensity for cross-β aggregation in all cc-del(X) constructs (Figure S6A). In cc-del0 to cc-del2, all or part of the 16LVFF20 motif (due to the single amino acid deletion) was also predicted to have a high propensity for cross-β aggregation (Figure S5A), in agreement with previous studies (Fernandez-Escamilla et al., 2004). Strikingly, cc-del6 was the only construct beyond cc-del2 to contain a pro-β motif, 20ALLLV24, according to the algorithm.
The difference between cc-del6 and cc-del7 was the Val at position 24 present in cc-del6 and coming from endogenous C99 sequence, but deleted in cc-del7. We mutated Val24 in cc-del6 to Ala and Gly25 to Val in cc-del7 (Figure S5B) to assess whether this motif (ALLLV) was key for the production of SDS-resistant oligomers. The mutations were predicted to reduce cross-β aggregation propensity in cc-del6 and increase cross-β aggregation in cc-del7 without changing their dimeric TM orientation (Figure S5C). Strikingly, the V24A mutation in cc-del6 resulted in total disappearance of SDS-resistant oligomers (Figure S6D). In contrast, the presence of this pro-β motif in cc-del7 did not result in the release of SDS-resistant oligomers (Figure S5D). To assess whether this motif could be sufficient to form SDS-resistant oligomers in other constructs, we inserted the ALLLV motif in all constructs from cc-del0 to cc-del7 while keeping their respective orientation. With the exception of cc-del6, no other orientation led to the accumulation of SDS-resistant oligomers upon processing, even in the presence of the pro-β ALLLV motif (Figure S6). This motif was thus required, but not sufficient for the production of SDS-resistant oligomers, and the precise dimeric orientation imposed by cc-del6 appeared to be essential as well.
To discriminate the contribution of different TM orientations of Aβ-like peptides from that of different lines of cleavage possibly followed by cc-del6 and cc-del7, we created truncated forms of HA-C99, cc-del6, and cc-del7 constructs (with and without the predicted mutations) by adding a stop codon at position 42 and 43 (amyloid beta numbering) (Figure 4E, top). These constructs will be equivalent to the products of cleavage via either the 42 or the 43 lines. As expected, direct transfection of WT C42 and C43 led to the production of SDS-resistant oligomers. The direct transfection of truncated forms of cc-del6 at position 42 and 43 both led to the production of a much larger amount of SDS-resistant oligomers (Figure 4E). In line with our previous results, the V24A mutation blocked the formation of SDS-resistant oligomers in the cc-del6 context (Figure 4E). Importantly, direct transfection of the truncated forms of cc-del7 with or without the G25V mutation did not result in the apparition of SDS-resistant oligomeric bands (Figure 4E). The cc-del6 orientation thus appeared required for the stability of these oligomers.
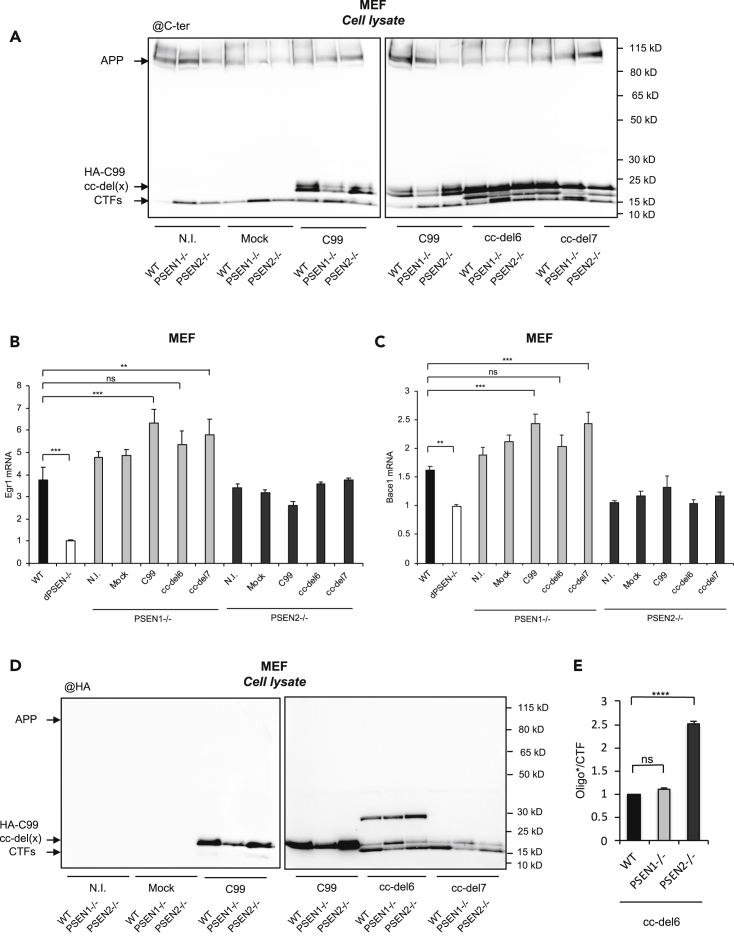
cc-del6 and cc-del7 Are Differentially Processed by PSEN1 and PSEN2-Dependent γ-Secretase
Presenilins (PSEN1 or PSEN2) form the catalytic subunits of γ-secretases. PSEN2-dependent γ-secretase is mainly localized in endosomes and lysosomes, whereas PSEN1-dependent γ-secretase is more present at the cell membrane (Sannerud et al., 2016). Our confocal images suggested that cc-del7 had a more intracellular localization, possibly in endo-lysosomal compartments, whereas cc-del6 was more present at the cell membrane and in neuronal processes in primary neurons (Figures 3E and S3). To assess whether cc-del6 and cc-del7 were differentially processed by PSEN1 and PSEN2-dependent γ-secretase, we infected MEF cells deficient for presenilin 1 (PSEN1−/−), presenilin 2 (PSEN2−/−), or both (dPSEN−/−), with C99, cc-del6, cc-del7 or a mock lentiviral vector (Figure 5A). Within these stable cell lines, qPCR was used to measure the expression of the two AICD target genes associated with cc-del7 processing. Strikingly, both Egr1 and Bace1 expression, previously shown to be increased upon cc-del7 processing (Figures 2D and 2E), was higher in PSEN1−/− cells in comparison to PSEN2−/− cells. A statistically significant increase in both Egr1 and Bace1 expression could only be observed in PSEN1−/− cells infected with either C99 or cc-del7 but not in PSEN1−/− infected with cc-del6 (Figures 5B and 5C). These results are in line with the idea that cc-del6 and cc-del7 are targeted to different subcellular compartments and suggest that cc-del7 orientation favors PSEN2-dependent γ-secretase processing.
Figure 5.
Differential Processing of cc-del6 and cc-del7 by PSEN1 and PSEN2
(A) Western blots of HA-C99, cc-el6, and cc-del7 constructs stably expressed in MEF WT, PSEN1−/−, or PSEN2−/−, revealed with the APP Cter antibody.
(B) Egr-1 mRNA of MEF cells described above (A). Results (Means ± SEM; n = 3; N = 3) are given as fold induction compared to control (dPSEN−/−).
(C) Bace1 mRNA of MEF cells described above (A). Results (Means ± SEM; n = 3; N = 3) are given as fold induction compared to control (dPSEN−/−).
(D) Western blots of HA-C99, cc-el6 and cc-del7 constructs stably expressed in MEF WT, PSEN1−/− or PSEN2−/−, revealed with the anti-HA antibody.
(E) Quantification of oligo∗. Results (Means ± SEM; n = 4; N = 2) are given as oligo∗:CTF-cc-del6 ratios, with oligo∗:CTF-cc-del6 WT ratio fixed at 1. Statistical analysis (B, C and E): one-way ANOVA followed by Dunnett's multiple comparison test. ns: non-significant, ∗: p < 0.05, ∗∗: p < 0.005, ∗∗∗: p < 0.001, ∗∗∗∗: p < 0.0001.
See also Figure S3.
We further tested whether the profile of SDS-resistant oligomers associated with cc-del6 was impacted by the knock-out of PSEN1 or PSEN2. Western blot analysis with an anti-HA antibody revealed more than 200% increased accumulation of SDS-resistant oligomers in PSEN2−/− cells infected with cc-del6 (Figure 5D). Still, oligomers were detected in PSEN1−/− cells indicating that they can be produced by either PS1- or PS2-dependent gamma secretase complexes. The higher accumulation of SDS-resistant oligomers in PSEN2−/− MEFs in comparison to WT MEFs infected with cc-del6 tends to indicate that oligomers are preferentially produced by PS1-dependent γ-secretases, PSEN1 being slightly upregulated in PSEN2-deficient cells (Stanga et al., 2018).
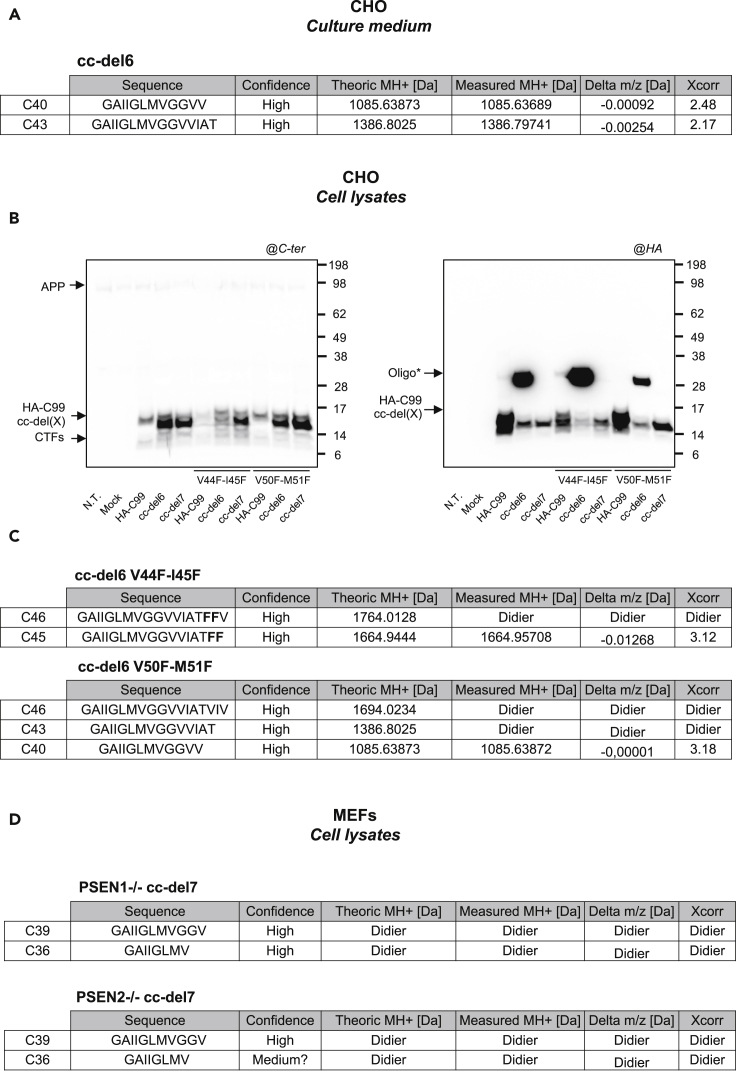
cc-del6 and cc-del7 Orientations Favors Distinct Processing Lines by Tripeptide Cleavage by γ-Secretase
A key goal was to assess whether different dimeric TM orientations of C99 favored distinct processing lines. We first created a specific protocol to identify the composition of the oligomers produced upon cc-del6 processing by mass spectrometry analysis (Figure S7A). The oligomeric bands observed from the medium after IP (Figure S7A arrow 1) can be disrupted with formic acid (Figure S7A arrow 2). The eluates obtained after IP with formic acid were analyzed by mass spectrometry (Figure S7A arrow 3). The oligomeric bands were composed of Aβ∗ that contained residues 40 and 43 of the Aβ sequence at the C-terminus (C40 and C43, respectively) (Figure 6A). Mass spectrometry data are shown in Figure S8. Next, we assessed whether our fusion proteins followed the tripeptide cleavage model of γ-secretase and that cc-del6 thus followed the processing line Aβ49>Aβ46>Aβ43>Aβ40. According to Bolduc et al. (2016), presenilins contain three putative S′ pockets that can each accommodate one amino acid at a time and occupancy of these three pockets precedes cleavage of a tripeptide. The S1′ and S3′ pockets are large and can easily accommodate any amino acids. The smaller S2′ pocket is less prone to accommodate large aromatic residues like phenylalanine. As a consequence, placing an aromatic residue at a given position is predicted to reduce overall cleavage beyond this point, although it was shown that γ-secretase was able to skip multiple residues to cleave at a further position (Bolduc et al., 2016). If cc-del6 follows the tripeptide cleavage model, mutating the amino acids at position 44 and 45 to phenylalanines would decrease further processing and result in accumulation of C46. Similarly, preventing initial ε-cleavage by mutating the residues at position 50-51 to phenylalanine should reduce overall processing.
Figure 6.
Processing Lines Favored by cc-del6 and cc-del7 Orientations
(A) Mass spectrometry analysis of oligomers from culture medium produced upon cc-del6 processing after disruption with formic acid. The results are summarized in the table. XCorr values are given for MS/MS spectra identified by Sequest. Spectra are available in Figure S9.
(B) Analysis of the profile of HA-C99, cc-del6, and cc-del7 constructs with or without the V44F-I45F and V50F-M51F mutations in denaturing conditions. Cells lysates were analyzed by Western blotting and revealed with APP Cter antibody (left) or HA antibody (right).
(C) Mass spectrometry analysis of oligomers from cell lysates produced upon cc-del6 processing with the V44F-I45F or V50F-M51F mutations after disruption with formic acid. The results are summarized in the table. XCorr values are given for MS/MS spectra identified by Sequest. Spectra or PRM traces are available in Figures S9 and S10.
(D) Mass spectrometry analysis of Commassie Blue positive bands detected in PSEN1 −/− or PSEN2 −/− MEFs infected with cc-del7 (Figure S11). The results are summarized in the table. PRM traces results are available in Figure S12.
See also Figures S7–S12.
In agreement with these predictions, we observed decreased production of SDS-resistant oligomers in presence of the V50F-M51F mutation in cc-del6 and absence of such oligomers in HA-C99 (Figure 6B). In contrast, the V44F-I45F mutation led to increased accumulation of SDS-resistant oligomers in HA-C99 and even more importantly in cc-del6 (Figure 6B). In order to identify the composition of these oligomers produced upon cc-del6 processing, we used the protocol described above for mass spectrometry analysis of cell lysates after IP (Figure S7B). Optimization of the detection enabled the identification of the C-terminus of C46, C43 and C40 in the lysates of cells expressing cc-del6 V50F-M51F (Figure 6C). In contrast, the band observed in cells expressing cc-del6 V44F-I45F was mainly composed of C46 containing the V44F-I45F mutation. We also detected a small amount of C45 with the V44F-I45F mutation (Figures 6C and S9), which can be interpreted as a side effect of the substitution to phenylalanines. Mass spectrometry data are given in Figures S9 and S10. Together, these results give strong evidence that cc-del6 obeys the tripeptide cleavage of γ-secretase and that it follows the Aβ49>Aβ46>Aβ43>Aβ40 processing line.
We also investigated the processing of cc-del7. We reasoned that if the cc-del6 orientation promoted the line of cleavage starting at position 49, a rotation of 110° counter-clockwise, like in cc-del7, bringing the more distal 33Gly-x-x-x-Gly37 in the interface, would lead to less tilt and thus an initial cleavage upstream at position 48 and thereby to the Aβ42 processing line. cc-del7 produced oligomers based on native gels, but the instability of Aβ-like peptide oligomers in SDS did not allow the acquisition of conclusive results by mass spectrometry analysis with a similar protocol as the one used for cc-del6, although suggestive data indicated the possibility of the Aβ48 line of cleavage.
To enhance the chance of purifying cc-del7–derived Aβ-like peptides and possibly aggregates/oligomers of cc-del7 products, we used MEFs knock-out for either PSEN1 or PSEN2 that we infected with cc-del7. It has been shown that in such MEFs, compensatory overexpression of the remaining PSEN occurs (Stanga et al., 2018), possibly triggering the processing of APP substrates. In addition, a recent study showed that longer Aβ forms (Aβ42) were preferentially produced by PSEN2 in endocytic compartments to generate an intracellular Aβ pool (Sannerud et al., 2016). This observation is fitting with our data indicating that Aβ-like peptides produced by cc-del(X) processing accumulate in cell lysates. In addition, PSEN1 and PSEN2 have been involved in autophagy and lysosomal proteolysis (Lee et al., 2010; Neely et al., 2011; Coen et al., 2012; Zhang et al., 2012); their absence thus favors the accumulation of cleavage products that would normally be degraded by these pathways. Confocal microscopy also showed large accumulation of HA positive intracellular vesicles in SH-SY5Y cells infected with cc-del7 (Figure S3), suggesting that cc-del7 associated oligomers could be present in stress granules that could tend to accumulate when phagocytosis is impaired.
We detected large accumulation of aggregated proteins by Ponceau red and by Coomasie Blue staining in MEFs PSEN1 or PSEN2 KO infected with cc-del7 (Figure S11). These oligomeric bands were relatively stable in NP-40 but sensitive to SDS unlike cc-del6 oligomers. We analyzed the Coomasie blue positive band, ranging from 49-62 kDa and recognized by HA antibodies, by mass spectrometry. We detected with high levels of confidence C36 and C39 peptides in these bands (Figures 6D and S12). More specifically, mass spectrometry showed the presence of C39 and C36 in PSEN1 −/− cc-del7 and C39 in PSEN2 −/− cc-del7. These fragments were not detected in a band of the corresponding size from non-infected MEFs. In line with our hypothesis, mass spectrometry analysis also detected the presence of p62, a marker of autophagosome, in Coomassie Blue positive bands in both PSEN1 and PSEN2 KO MEFs infected with cc-del7 but not in negative control. Together, these results indicated that products from the Aβ42>Aβ39>Aβ36 processing line are detectable in cells expressing the cc-del7 construct.
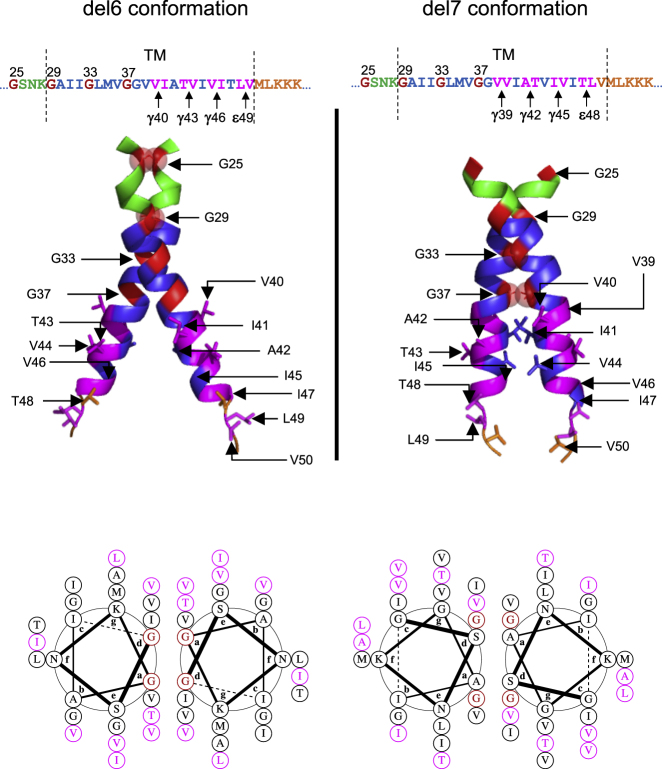
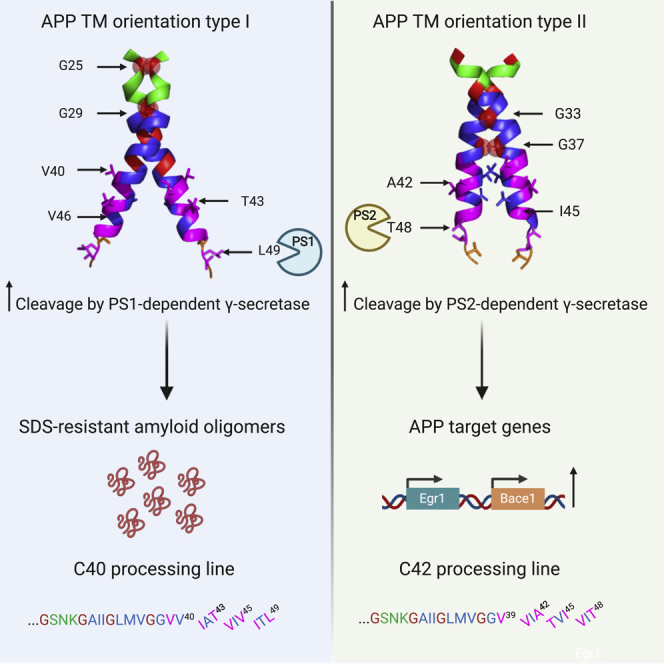
A Model for the Choice of γ-Secretase Cleavage Lines as a Function of Dimeric Orientation
A model of the dimeric orientations adopted by cc-del7 and cc-del6 based on the solved structure of the TM domain of APP (pdb: 2LOH) and modified according to (Sato et al., 2009; Zhou et al., 2019) is shown in Figure 7. We hypothesize that dimerization via the Gly-x-x-x-Gly motifs will affect the accessibility and configuration for initial binding of γ-secretase on dimeric C99 by exposing ε-sites and, subsequently, γ-sites for AICD and Aβ release, respectively. Specifically, as a function of which Gly-x-x-x-Gly motif is in the interface, the tilt of the helices will differ. Crossing at the N-terminus of the TM domains (for cc-del6) with Gly25 and Gly29 in interfacial “d” and “a” positions will result in a higher tilt at the C-terminus, when compared to crossing in the middle of the TM domain (for cc-del7). A higher tilt will lead to a lower position exposed up at the ε-cleavage site. After initial binding to the PS complex as a dimer, the dimeric substrate would be uncoupled to allow processing.
Figure 7.
Dimeric helical structure cc-del6 and C99 cc-del7
For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.isci.2020.101887.
Top: Dimeric model structures of cc-del7 and cc-del6 fusion proteins were adapted base on pdb: 2loh (Nadezhdin et al., 2012). The juxtamembane (JM) region is in green, transmembrane region (TM) in blue and AICD in orange. Glycine residues from GxxxG motifs are in red and the different positions of γ-secretase cleavage in pink. The arrows indicate the amino acids in the helix (Aβ numbering). Bottom: Helical wheel diagrams of the cc-del7 and cc-del6 fusion proteins. Glycine residues in the interface are in red and the residues accessible for γ-cleavage are in pink. The model does not take into account potential distortions/kinks induced by 33GlyxxxGly37Gly38.
In Figure 7 (left panel), with the helical lateral view, we show that the C99 cc-del6 dimer contains the 25Gly-x-x-x-Gly29 motif in the interface, which forms a more upstream crossing interface that requires a higher tilt of the helices. This exposes the amide bond of an amino acid further down the helix, at or after residue 49 within the ε-site. At those positions, the helix is already unraveled, as we have proven for our constructs as well with the L49C cross-linking (Figure S1). Cleavage at residue 49 would release AICD and process upstream at the γ43 and γ40 sites for releasing Aβ43 and Aβ40-like peptides, respectively (Qi-Takahara et al., 2005; Takami et al., 2009). The model that we propose fits very well with the experimental data in cell free assays for AICD production and mass spectrometry for the Aβ isoforms production, where we see for cc-del6 a preferential cleavage at C46, C43, and C40.
In Figure 7 (right panel), with the helical lateral view, we show that C99 cc-del7 contains 33Gly-x-x-x-Gly37 in the interface with Gly33 and Gly37 in interfacial “d” and “a” positions. Our data in MEFs deficient for PSEN1 or PSEN2 suggest that initial cleavage could occur at residue 48. This residue would be preferred in this more parallel dimer. Following tripeptide cleavage by γ-secretase, such initial cleavage at ε-site 48 would favor the Aβ42 processing line and an interesting overlap between signaling and generation of Aβ42 could be invoked.
Discussion
Our main observations are that C99 TM domain dimerization affects its cleavage by γ-secretase, and that different functional outcomes in terms of signaling and Aβ oligomerization depend on two of the seven possible symmetric dimeric orientations of the C99 TM domain, before and after processing. We have employed coiled coils to impose precise dimeric interfaces. We validated the predicted interfaces by Cys-cross-linking. We found that one dimeric interface (cc-del7) leads to enhanced release of AICD associated with Egr-1 gene expression as well as its downstream target Bace1. This orientation favors initial cleavage at position 48 and thereby the Aβ42 processing line. Another orientation, cc-del6 generates Aβ-like peptides that form SDS-resistant oligomers. Cleavage of cc-del6 at residue 49 leads to formation of SDS-resistant oligomers ending at positions 43 and 40 of Aβ. SDS-resistance of these oligomers requires the presence of a pro-β motif in addition to a precise dimeric orientation of Aβ-like peptides.
The two interfaces are related and rotated relative to each other by 110°. cc-del7 forms a dimer that crosses the TM helices at the middle 33Gly-x-x-x-Gly37 TM motif, while cc-del6 crosses the helices upstream at the 25Gly-x-x-x-Gly29. Our data and model argue that the initial cleavage at the ε-site differs because the tilt of helices differs exposing different sites to the γ-secretase, the higher the tilt in cc-del6 dimer, the lower start of cleavage (at 49).
Importantly, the cc-del7 fusion protein exhibits the same dimeric interface as cc-del0, but with a shorter extracellular region. It has been reported that the 16LVFF20 motif forms a β-sheet that prevents processing of C99 by γ-secretase (Hu et al., 2017; Tian et al., 2010). This particular motif is absent in the cc-del7, but present in cc-del0. Indeed, no effect has been observed in term of signaling for the cc-del0, consistent with decreased processing. Together, it indicates that the conformation of cc-del7 TM dimers is the active one for APP signaling.
The precise regulation of APP amyloidogenic processing depends on its subcellular localization and the nature of the γ-secretase present in the cellular compartments (presenilin 1- or presenilin 2-dependent). Recently, it has been reported that amyloidogenic processing by presenillin 2 occurs preferentially in recycling endosomes and late endosomes/lysosomes compartments (Sannerud et al., 2016). Our confocal immunofluorescence data from human neuroblastoma cell lines and mouse primary neurons (Figures 3E and S3) indicated that cc-del7 appears localized to intracytoplasmic structures, while cc-del6 seemed to harbor plasma membrane localization and was found in neuronal processes. In line with this, our data supports preferential processing of cc-del6 by PSEN1-dependent γ-secretase and of cc-del7 by PSEN2-dependent γ-secretase. The processing of cc-del6 and cc-del7 likely occurs in different cellular compartments and by different γ-secretases, with implications on the fate of produced Aβ or AICD.
The comprehensive model that we propose is C99 dimers with an orientation, where 33Gly-x-x-x-Gly37 is in the interface (cc-del7), allowing high levels of trimming by γ-secretase likely starting at position 48 (Figure 7). The structure of γ-secretase in complex with C83 (Zhou et al., 2019), the need for unwinding of the α-helix around γ-sites for processing and our data does not support cleavage of stable dimers. A mechanism has recently been described where stable substrate binding to γ-secretase is important for correct processing (Szaruga et al., 2017). This binding initially occurs just upstream of the region we described to shift from the TM helical structure to random coil (Sato et al., 2009). After initial binding, γ-secretase would induce unraveling of the helix for initial cleavage at the ε-site before cutting upstream in sections of three amino acids, to form gradually shorter Aβ species (Bolduc et al., 2016). Our mass spectrometry data in MEFs PSEN1−/− and PSEN2 −/− suggest that initial cleavage occurs at residue 48 for cc-del7, with tripeptide cleavage then releasing Aβ45, Aβ42, Aβ39 and, in some cases, Aβ36. The fact that Aβ-like peptides ending at position 42 were hardly detectable in our set up probably indicates that the carboxypeptidase (trimming) activity of the γ-secretase was particularly efficient to produce shorter peptides in the cc-del7 conformation.
In pathological conditions, it is possible that the generated Aβ peptides, like Aβ42, might then change orientation by rotating clockwise 110° to the same orientation where 25Glyx-x-x-Gly29 motif would come into the interface, corresponding to the orientation of cc-del6, and form stable Aβ oligomers. The shift from cc-del7 to cc-del6 would result in shifting the helix crossing point from Gly33x-x-x-G37 closer up to the extracellular JM region at 25Glyx-x-x-Gly29. Such an upward shift might alter interactions with membrane lipids (Zhou et al., 2019) and favor SDS resistance of the released Aβ peptides, just like in the case where the initial cleavage would occur at or after residue 49. The proportions of C99 that adopt one or the other dimeric interfaces might depend on their precise intracellular localization, membrane composition, pH, and temperature, as the precise interaction between the γ-secretase and its substrates, especially intermediates between Aβ49 and Aβ40 were shown to depend on temperature (Szaruga et al., 2017).
Because oligomers are nucleated by the coiled coil helices, we suggest the possibility that such oligomers might adopt transient α-helical structure during initial nucleation. We showed that SDS resistance is reminiscent of the similar resistance of the glycophorin A TM dimer (Bormann et al., 1989) and requires (i) close apposition of the mediated by close apposition of a specific 25Gly-x-x-x-Gly29 motif in the interface and (ii) a pro-β motif for secondary structure stabilization as suggested by the work of Misra et al. (2016). The fusion of Put3 coiled coil with the TM-cytosolic domain of C99 generates a novel sequence where Val24 of C99 is key for oligomerization, allowing the reconstitution of a pro-β motif. This is line with previous findings that substitution of Val17 (among others) in the 16LVFF20 motif similarly impedes SDS-resistance (Hilbich et al., 1992). Interestingly, SDS-resistant Aβ oligomers were detected in brains from Alzheimer's patients (Roher et al., 1996).
According to our model and to the cross-linking data in Figure S1, the cc-del6 construct exhibits a particular orientation in which two upstream glycines, Gly25 and 29 of the 25Gly-x-x-x-Gly29 motif are in the interface in “d” and “a” positions. Thus, we suggest that while these Gly residues mediate helix crossing before cleavage, they might also mediate initial nucleation of oligomers after cleavage. The observation that the other cc-del(X) fusion proteins exhibiting different orientations also dimerize (Figure S4C), produce Aβ-like monomers, but do not lead to the formation of SDS-resistant oligomers, even in presence of a pro-β motif (Figure S6), shows that the precise dimeric TM orientation with a particular Gly-x-x-x-Gly motif in the interface is key for the production of stable Aβ-like oligomers.
We show that cc-del6 obeys the tripeptide cleavage by γ-secretase, with processing likely starting at position 49, trimming in sections of three residues occurs (Bolduc et al., 2016), allowing the formation of Aβ46, -43 and −40. Several studies have shown that APP amyloidogenic processing leads to higher Aβ40 production compared to Aβ42 (10:1 ratio), but one study has demonstrated that Aβ43 is more common than Aβ40 in amyloid plaque cores found in AD brains (Welander et al., 2009). The aggregation propensity and potential pathogenic role of Aβ43 have been suggested (Saito et al., 2011; Veugelen et al., 2016) and recently documented (Szaruga et al., 2017). We suggest that different TM orientations can result in different lines of cleavage but that production of stable, SDS-resistant oligomers is linked with the specific TM orientation imposed in cc-del6 together with the presence of a central pro-β motif such as the 20ALLLV24 present in cc-del6 sequence or the 16LVFF20 motif of the human Aβ sequence.
The role of the precise dimerization of the C99 TM dimerization in signaling and processing might open novel therapeutic avenues to limit amyloidogenic processing. We posit that the identification of pathological conformations of C99 dimers could pave the way for the design of small molecules, nanobodies or monobodies which would intracellularly target specific conformation of C99 dimers associated with amyloidogenic processing. Sencanski et al. (2019) recently described the use of nanobodies to selectively target the active conformation of the β2-adrenergic receptor. Together with their high specificity, the small size of such biomolecular tools could provide interesting therapeutic avenues for intracellular targeting of pathological protein conformations, including in the context of Alzheimer's Disease. These results might also lead to the identification of the physiologic targets of APP signaling, which remain poorly known as does the physiological ligand for APP and related proteins APLP1 and APLP2. One provocative scenario is that in aged individuals a higher degree of signaling by APP is required to compensate for age-related hypoxia and senescence. Such overuse of the pathway modelized by cc-del7 would drive formation of pathogenic amyloidogenic peptides and possibly induce a self-perpetuating amyloidogenic loop by increasing the expression of Egr1 and its downstream target Bace1. Cholesterol is also known to affect the conformation and stability of C99 dimers (Tang et al., 2014; Dominguez et al., 2016). An alternative scenario is that the hypercholesterolemia observed in mouse models (Umeda et al., 2012) and in postmortem analysis of the brain of patients with AD (Lazar et al., 2012) could favor conformation of C99 dimers recapitulating cc-del6 and cc-del7. We suggest that our fusion protein cc-del7 could become a model of constitutive active APP or C99 useful for identification of physiologic APP signaling targets. Finally, this model would also explain pathologic signaling by Familial cases of Alzheimer's disease (FAD), where certain mutations favor dimerization via the Gly-x-x-x-Gly motifs like cc-del7 (Tang et al., 2014). Such mutations were recently also reported as somatic acquired mutations in sporadic Alzheimer's cases, thus increasing the relevance of understanding the mechanisms behind the pathogenicity of FAD mutations (Lee et al., 2018).
Limitations of the Study
Despite providing multiple lines of evidence for the role of APP TM dimeric orientation in the processing of C99 dimers and Aβ oligomerization properties, our study required the use of engineered coiled-coil constructs which reproduce all possible conformation of C99 TM dimers. Some of them might not be readily adopted in physiopathological conditions. The conclusions brought in this study should be explored in animal models using compound designed to modulate the natural C99 TM dimeric orientations. Finally, the precise localization of C99 dimers and their association with subcellular compartmentalization of PSEN1 and PSEN2 should be further investigated in subsequent studies.
Resources Availability
Lead Contact
Requests for further information and resources should be directed at the lead contact, Pr. Pascal Kienlen-Campard (pascal.kienlen-campar@uclouvain.be).
Materials Availability
Plasmids coding for our engineered coiled-coil constructs generated in this study will be made available on request, but we may require a payment and/or completed Materials Transfer Agreement if there is potential for commercial application.
Data and Code Availability
This study did not generate data set/code.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Dr. Jean-Philippe Defour for initial lentiviral cloning of coiled coil dimeric proteins, Joanne Van Hees and Lidvine Genet for expert technical assistance and Dr. Nicolas Dauguet and Dr. Xavier Cahu for Flow Cytometry support. SNC is Honorary Research Director at FRS-FNRS Belgium. Funding to SNC is acknowledged from Ludwig Institute for Cancer Research, Fondation contre le cancer, Salus Sanguinis and projects Action de recherche concertée (ARC) 16/21-073 and WelBio F 44/8/5 - MCF/UIG – 10955. Funding to PKC is acknowledged from SAO-FRA Alzheimer Research Foundation, Fondation Louvain and Queen Elisabeth Medical Research Foundation (FMRE). FP was supported by ARC 16/21-073 (to SNC) and ARC14/19-059 (to PKC). Support funds from FNRS grant PDRT.0177.18 to PKC, from NIH AG27317 to SOS and subcontract to NIH AG27317 to SNC are acknowledged. The graphical abstract was created with Biorender.com
Author Contributions
F.P., N.P., R.O., N.S., and C.V. designed and performed experiments and interpreted data; P.K.C., S.N.C., and S.O.S. designed experiments, defined the strategy of the study, and interpreted data; F.P., N.P., P.K.C., S.O.S., and S.N.C. wrote the paper; D.V. was responsible for mass spectrometry experiments; D.M.V. provided reagents and interpreted data.
Declaration of Interests
S.N.C. is co-founder of Alsatech – Boston, MA and close family is owning stock in A.C. Immune Lausanne. The work of this manuscript is not linked to any current project at Alsatech or A.C. Immune, which are companies that work in the field of Alzheimer's disease.
Published: December 18, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101887.
Contributor Information
Pascal Kienlen-Campard, Email: pascal.kienlen-campard@uclouvain.be.
Stefan N. Constantinescu, Email: Stefan.Constantinescu@bru.licr.org.
Supplemental Information
7
References
- Agholme L., Lindström T., Kågedal K., Marcusson J., Hallbeck M. An in vitro model for neuroscience: differentiation of SH-SY5Y cells into cells with morphological and biochemical characteristics of mature neurons. J. Alzheimers Dis. 2010;20:1069–1082. doi: 10.3233/JAD-2010-091363. [DOI] [PubMed] [Google Scholar]
- Belyaev N.D., Kellett K.A., Beckett C., Makova N.Z., Revett T.J., Nalivaeva N.N., Hooper N.M., Turner A.J. The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a {beta}-secretase-dependent pathway. J. Biol. Chem. 2010;285:41443–41454. doi: 10.1074/jbc.M110.141390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Khalifa N., Tyteca D., Marinangeli C., Depuydt M., Collet J.F., Courtoy P.J., Renauld J.C., Constantinescu S., Octave J.N., Kienlen-Campard P. Structural features of the KPI domain control APP dimerization, trafficking, and processing. FASEB J. 2012;26:855–867. doi: 10.1096/fj.11-190207. [DOI] [PubMed] [Google Scholar]
- Bolduc D.M., Montagna D.R., Seghers M.C., Wolfe M.S., Selkoe D.J. The amyloid-beta forming tripeptide cleavage mechanism of γ-secretase. Elife. 2016;5:e17578. doi: 10.7554/eLife.17578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann B.J., Knowles W.J., Marchesi V.T. Synthetic peptides mimic the assembly of transmembrane glycoproteins. J. Biol. Chem. 1989;264:4033–4037. [PubMed] [Google Scholar]
- Cao X., Sudhof T.C. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- Coen K., Flannagan R.S., Baron S., Carraro-Lacroix L.R., Wang D., Vermeire W., Michiels C., Munck S., Baert V., Sugita S. Lysosomal calcium homeostasis defects, not proton pump defects, cause endo-lysosomal dysfunction in PSEN-deficient cells. J. Cell Biol. 2012;198:23–35. doi: 10.1083/jcb.201201076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez L., Foster L., Straub J.E., Thirumalai D. Impact of membrane lipid composition on the structure and stability of the transmembrane domain of amyloid precursor protein. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E5281–E5287. doi: 10.1073/pnas.1606482113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert S., Midthune B., Cottrell B., Koo E.H. Induced dimerization of the amyloid precursor protein leads to decreased amyloid-beta protein production. J. Biol. Chem. 2009;284:28943–28952. doi: 10.1074/jbc.M109.038646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert S., Gonzalez A.C., Thomas C., Schilling S., Schwarz S.M., Tischer C., Adam V., Strecker P., Schmidt V., Willnow T.E. Dimerization leads to changes in APP (amyloid precursor protein) trafficking mediated by LRP1 and SorLA. Cell. Mol. Life Sci. 2018;75:301–322. doi: 10.1007/s00018-017-2625-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla A.-M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Hendrickx A., Pierrot N., Tasiaux B., Schakman O., Brion J.P., Kienlen-Campard P., De Smet C., Octave J.N. Epigenetic induction of EGR-1 expression by the amyloid precursor protein during exposure to novelty. PLoS One. 2013;8:e74305. doi: 10.1371/journal.pone.0074305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbich C., Kisters-Woike B., Reed J., Masters C.L., Beyreuther K. Substitutions of hydrophobic amino acids reduce the amyloidogenicity of Alzheimers disease βA4 peptides. J. Mol. Biol. 1992;228:460–473. doi: 10.1016/0022-2836(92)90835-8. [DOI] [PubMed] [Google Scholar]
- Hu Y., Kienlen-Campard P., Tang T.C., Perrin F., Opsomer R., Decock M., Pan X., Octave J.N., Constantinescu S.N., Smith S.O. Beta-sheet structure within the extracellular domain of C99 regulates amyloidogenic processing. Sci. Rep. 2017;7:17159. doi: 10.1038/s41598-017-17144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsubo T., Odaka A., Suzuki N., Mizusawa H., Nukina N., Ihara Y. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- Jones M.W., Errington M.L., French P.J., Fine A., Bliss T.V., Garel S., Charnay P., Bozon B., Laroche S., Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat. Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- Kang J., Lemaire H.G., Unterbeck A., Salbaum J.M., Masters C.L., Grzeschik K.H., Multhaup G., Beyreuther K., Muller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Kienlen-Campard P., Tasiaux B., Van Hees J., Li M., Huysseune S., Sato T., Fei J.Z., Aimoto S., Courtoy P.J., Smith S.O. Amyloidogenic processing but not amyloid precursor protein (APP) intracellular C-terminal domain production requires a precisely oriented APP dimer assembled by transmembrane GXXXG motifs. J. Biol. Chem. 2008;283:7733–7744. doi: 10.1074/jbc.M707142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar A.N., Bich C., Panchal M., Desbenoit N., Petit V.W., Touboul D., Dauphinot L., Marquer C., Laprévote O., Brunelle A., Duyckaerts C. Time-of-flight secondary ion mass spectrometry (TOF-SIMS) imaging reveals cholesterol overload in the cerebral cortex of Alzheimer disease patients. Acta Neuropathol. 2012;125:133–144. doi: 10.1007/s00401-012-1041-1. [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Yu W.H., Kumar A., Lee S., Mohan P.S., Peterhoff C.M., Wolfe D.A., Martinez-Vicente M., Massey A., Sovak G. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.H., Siddoway B., Kaeser G.E., Segota I., Rivera R., Romanow W.J., Liu C.S., Park C., Kennedy G., Long T., Chun J. Somatic APP gene recombination in Alzheimer's disease and normal neurons. Nature. 2018;563:639–645. doi: 10.1038/s41586-018-0718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie K.R., Prestegard J.H., Engelman D.M. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- Mattoon D., Gupta K., Doyon J., Loll P.J., DiMaio D. Identification of the transmembrane dimer interface of the bovine papillomavirus E5 protein. Oncogene. 2001;20:3824–3834. doi: 10.1038/sj.onc.1204523. [DOI] [PubMed] [Google Scholar]
- Misra P., Kodali R., Chemuru S., Kar K., Wetzel R. Rapid α-oligomer formation mediated by the Aβ C terminus initiates an amyloid assembly pathway. Nat. Commun. 2016;7:12419. doi: 10.1038/ncomms12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munter L.M., Voigt P., Harmeier A., Kaden D., Gottschalk K.E., Weise C., Pipkorn R., Schaefer M., Langosch D., Multhaup G. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadezhdin K.D., Bocharova O.V., Bocharov E.V., Arseniev A.S. Dimeric structure of transmembrane domain of amyloid precursor protein in micellar environment. FEBS Lett. 2012;586:1687–1692. doi: 10.1016/j.febslet.2012.04.062. [DOI] [PubMed] [Google Scholar]
- Neely K.M., Green K.N., Laferla F.M. Presenilin is necessary for efficient proteolysis through the autophagy-lysosome system in a γ-secretase-independent manner. J. Neurosci. 2011;31:2781–2791. doi: 10.1523/JNEUROSCI.5156-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardossi-Piquard R., Petit A., Kawarai T., Sunyach C., Alves da Costa C., Vincent B., Ring S., D'Adamio L., Shen J., Muller U. Presenilin-dependent transcriptional control of the Abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP. Neuron. 2005;46:541–554. doi: 10.1016/j.neuron.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Qi-Takahara Y., Morishima-Kawashima M., Tanimura Y., Dolios G., Hirotani N., Horikoshi Y., Kametani F., Maeda M., Saido T.C., Wang R., Ihara Y. Longer forms of amyloid beta protein: implications for the mechanism of intramembrane cleavage by gamma-secretase. J. Neurosci. 2005;25:436–445. doi: 10.1523/JNEUROSCI.1575-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Wang Y., Paudel H.K. Early growth response 1 (Egr-1) is a transcriptional activator of beta-Secretase 1 (BACE-1) in the brain. J. Biol. Chem. 2016;291:22276–22287. doi: 10.1074/jbc.M116.738849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roher A.E., Chaney M.O., Kuo Y.M., Webster S.D., Stine W.B., Haverkamp L.J., Woods A.S., Cotter R.J., Tuohy J.M., Krafft G.A. Morphology and toxicity of Aβ-(1-42) dimer derived from neuritic and vascular amyloid deposits of Alzheimers disease. J. Biol. Chem. 1996;271:20631–20635. doi: 10.1074/jbc.271.34.20631. [DOI] [PubMed] [Google Scholar]
- Saito T., Suemoto T., Brouwers N., Sleegers K., Funamoto S., Mihira N., Matsuba Y., Yamada K., Nilsson P., Takano J. Potent amyloidogenicity and pathogenicity of Aβ43. Nat. Neurosci. 2011;14:1023–1032. doi: 10.1038/nn.2858. [DOI] [PubMed] [Google Scholar]
- Sannerud R., Esselens C., Ejsmont P., Mattera R., Rochin L., Tharkeshwar A.K., De Baets G., De Wever V., Habets R., Baert V. Restricted location of PSEN2/gamma-secretase determines substrate specificity and generates an intracellular abeta pool. Cell. 2016;166:193–208. doi: 10.1016/j.cell.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Tang T.C., Reubins G., Fei J.Z., Fujimoto T., Kienlen-Campard P., Constantinescu S.N., Octave J.N., Aimoto S., Smith S.O. A helix-to-coil transition at the epsilon-cut site in the transmembrane dimer of the amyloid precursor protein is required for proteolysis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1421–1426. doi: 10.1073/pnas.0812261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sencanski M., Glisic S., Šnajder M., Veljkovic N., Ulrih N.P., Mavri J., Vrecl M. Computational design and characterization of nanobody-derived peptides that stabilize the active conformation of the β2-adrenergic receptor (β2-AR) Sci. Rep. 2019;9:16555. doi: 10.1038/s41598-019-52934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seubert N., Royer Y., Staerk J., Kubatzky K.F., Moucadel V., Krishnakumar S., Smith S.O., Constantinescu S.N. Active and inactive orientations of the transmembrane and cytosolic domains of the erythropoietin receptor dimer. Mol. Cell. 2003;12:1239–1250. doi: 10.1016/s1097-2765(03)00389-7. [DOI] [PubMed] [Google Scholar]
- Staerk J., Defour J.P., Pecquet C., Leroy E., Antoine-Poirel H., Brett I., Itaya M., Smith S.O., Vainchenker W., Constantinescu S.N. Orientation-specific signalling by thrombopoietin receptor dimers. EMBO J. 2011;30:4398–4413. doi: 10.1038/emboj.2011.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga S., Vrancx C., Tasiaux B., Marinangeli C., Karlström H., Kienlen-Campard P. Specificity of presenilin-1- and presenilin-2-dependent γ-secretases towards substrate processing. J. Cell. Mol. Med. 2018;22:823–833. doi: 10.1111/jcmm.13364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaruga M., Munteanu B., Lismont S., Veugelen S., Horre K., Mercken M., Saido T.C., Ryan N.S., De Vos T., Savvides S.N. Alzheimer's-causing mutations shift abeta length by destabilizing gamma-secretase-abetan interactions. Cell. 2017;170:443–456 e414. doi: 10.1016/j.cell.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Takami M., Nagashima Y., Sano Y., Ishihara S., Morishima-Kawashima M., Funamoto S., Ihara Y. Gamma-Secretase: successive tripeptide and tetrapeptide release from the transmembrane domain of beta-carboxyl terminal fragment. J. Neurosci. 2009;29:13042–13052. doi: 10.1523/JNEUROSCI.2362-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.C., Hu Y., Kienlen-Campard P., El Haylani L., Decock M., Van Hees J., Fu Z., Octave J.N., Constantinescu S.N., Smith S.O. Conformational changes induced by the A21G Flemish mutation in the amyloid precursor protein lead to increased Abeta production. Structure. 2014;22:387–396. doi: 10.1016/j.str.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Bassit B., Chau D., Li Y.M. An APP inhibitory domain containing the Flemish mutation residue modulates gamma-secretase activity for Abeta production. Nat. Struct. Mol. Biol. 2010;17:151–158. doi: 10.1038/nsmb.1743. [DOI] [PubMed] [Google Scholar]
- Umeda T., Tomiyama T., Kitajima E., Idomoto T., Nomura S., Lambert M.P., Klein W.L., Mori H. Hypercholesterolemia accelerates intraneuronal accumulation of Aβ oligomers resulting in memory impairment in Alzheimers disease model mice. Life Sci. 2012;91:1169–1176. doi: 10.1016/j.lfs.2011.12.022. [DOI] [PubMed] [Google Scholar]
- Veugelen S., Saito T., Saido T.C., Chavez-Gutierrez L., De Strooper B. Familial Alzheimer's disease mutations in presenilin generate amyloidogenic abeta peptide seeds. Neuron. 2016;90:410–416. doi: 10.1016/j.neuron.2016.03.010. [DOI] [PubMed] [Google Scholar]
- Walters K.J., Dayie K.T., Reece R.J., Ptashne M., Wagner G. Structure and mobility of the PUT3 dimer. Nat. Struct. Biol. 1997;4:744–750. doi: 10.1038/nsb0997-744. [DOI] [PubMed] [Google Scholar]
- Weidemann A., Konig G., Bunke D., Fischer P., Salbaum J.M., Masters C.L., Beyreuther K. Identification, biogenesis, and localization of precursors of Alzheimer's disease A4 amyloid protein. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- Welander H., Frånberg J., Graff C., Sundström E., Winblad B., Tjernberg L.O. Abeta43 is more frequent than Abeta40 in amyloid plaque cores from Alzheimer disease brains. J. Neurochem. 2009;110:697–706. doi: 10.1111/j.1471-4159.2009.06170.x. [DOI] [PubMed] [Google Scholar]
- Zhang X., Garbett K., Veeraraghavalu K., Wilburn B., Gilmore R., Mirnics K., Sisodia S.S. A role for presenilins in autophagy revisited: normal acidification of lysosomes in cells lacking PSEN1 and PSEN2. J. Neurosci. 2012;32:8633–8648. doi: 10.1523/JNEUROSCI.0556-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Yang G., Guo X., Zhou Q., Lei J., Shi Y. Recognition of the amyloid precursor protein by human gamma-secretase. Science. 2019;363:eaaw0930. doi: 10.1126/science.aaw0930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
7
Data Availability Statement
This study did not generate data set/code.