Abstract
We describe our collaborative efforts to increase representation of diverse populations in genomic research of kidney phenotypes to fill an unmet need to further understanding of the genetic contribution to chronic kidney disease (CKD) across the globe. These efforts led to the creation of the Continental Origins and Genetic Epidemiology Network Kidney (COGENT-Kidney) Consortium, focused on developing statistical methods for the analysis of genome-wide association studies (GWAS) across diverse populations and their application to renal traits and CKD. Resources generated within the consortium will provide a framework for future studies of genetic risk of CKD in worldwide populations. Our intent is to foster collaborations for studies of populations that are underrepresented in GWAS, increase awareness of the challenges and opportunities in studying these populations, and promote more genomic research across increasingly diverse populations.
Scope of the problem: CKD in the US and global populations.
The Global Burden of Disease study recently reported rising CKD incidence and prevalence worldwide, and increased deaths and disability due to CKD1. U.S. ethnic minorities have higher burden of CKD compared to those of European ancestry. Globally, CKD has an uneven distribution in its prevalence and causes1, 2. The limited knowledge on mechanisms underlying CKD development and progression, and the causes for regional and ethnic variation in CKD, have hampered efforts to prevent the disease globally and locally. It has also prevented the development of therapeutic tools for clinicians to effectively treat the disease.
Genetic studies in global populations: challenges and opportunities.
Genetic susceptibility to disease occurs in the context of lifetime exposures to lifestyle and environmental risk factors, some of which are amenable to intervention. To benefit individuals across the globe, irrespective of race/ethnicity, more studies are needed in populations that have shown a high risk of CKD. There have been relatively few GWAS undertaken across ancestral groups investigating the genetic susceptibility to other common diseases such as hypertension and diabetes3.. Understanding how genetic variation impacts on downstream molecular and biological processes across diverse populations provides greater insight into the genomic contributions to human health across the globe, and improved ability to apply this knowledge through clinical translation that will be relevant to everyone.
Populations vary in their DNA make-up, including the frequencies of alleles at genetic variants, and the correlation structure between these variants, referred to as linkage disequilibrium (LD BOX 1). Many genetic variants are shared across populations, and the power to identify genomic regions (“loci”) containing disease risk genes will be greatest in the population in which the causal genetic variants are most frequent. Differences in LD structure between populations can help to pinpoint causal variants among multiple variants at a locus through fine-mapping (BOX 1). The gain in knowledge and insights into disease mechanisms from gene discovery in one population benefits all groups, even those populations in which the causal variants at a locus are rare or absent.
BOX1. Definitions.
Genome-wide association study (GWAS).
Approach used to identify genetic variants associated with traits (i.e., eGFR) across the entire genome.
Trans-ethnic meta-analyses of GWAS. Approach that combines GWAS data from different population groups for a trait for variants that are shared across populations. The most powerful approaches allow for heterogeneity of effects of variants on the trait across ancestries.
Linkage disequilibrium (LD).
Correlation in alleles within individuals across genetic variants mapping to the same genomic region. LD patterns vary across ancestries: shorter range LD is found in African ancestry populations due to their longer population history.
Fine-mapping.
Approach that attempts to identify the causal variant at a GWAS locus from amongst the large numbers of all possible variants in the region using patterns of LD and association with the trait. Identified variants can then be prioritized for downstream experimental studies to understand the role the variant plays in disease pathogenesis.
Expression quantitative locus (eQTL).
Genetic variants that are associated with the expression of genes mapping to the same genomic region. eQTLS can be specific to one or a few tissues, or shared ubiquitously across tissues. Causal variants for a disease/trait that are also eQTLs provide insight into the gene through which the trait association is mediated.
Genetic risk scores.
Approach that aggregates multiple genetic variants associated with a disease through GWAS into a risk model for disease prediction.
Mendelian randomization (MR).
Approach to assess the causal associations of exposures with outcomes. Genetic variants (usually in aggregate) associated with a trait are proxies or “instrumental variables” for an exposure (or risk factor), with the advantage that they are not influenced by the confounding seen in observational studies or by reverse causality (because they are assigned at conception).
Genotype imputation.
Statistical methods to infer genotypes at untyped marker in a sample using population-specific reference panels. The process includes estimating the most likely genotype for each individual and provides an estimation of the quality of the imputation.
Challenges in studying populations that are underrepresented in GWAS include the less well-known genetic architecture, the lack of available public databases of genetic variants, and the small number of samples that can reduce power for gene discovery4. Reference panels for African, East Asian and South Asian ancestry populations, which describe allele frequencies and LD structure, have been greatly improved with increasing availability of large-scale whole genome sequencing resources 4, but knowledge on genetic variation for some worldwide populations is still lacking. There has also been a lack of statistical methods that are appropriate for combining GWAS data that adequately account for these genetic differences across populations. From the perspective of precision medicine, this underrepresentation in knowledge can exacerbate healthcare disparities and prevent an effective implementation of genomics in clinical care for all.
COGENT-Kidney Consortium description and goals
The COGENT-Kidney Consortium was established in 2014 to undertake trans-ethnic meta-analysis of GWAS of kidney traits in diverse populations. The Consortium recruitment targeted GWAS from four ancestral groups (African, Hispanic/Latino, European, and East Asian) composed of 71,638 individuals of whom 67% were of non-European ancestry (Table S1)5. The main goal of the Consortium is to increase representation of populations that have not been well characterized for genetic risk, particularly ethnic subgroups at high risk of CKD.
The Consortium is an open enrollment and equal partnership among investigators from different studies and it includes expertise in clinical nephrology, population health, human genetics, statistical genetics, bioinformatics and experimental research. Studies follow standardized protocols for trait definitions and statistical analyses, and they contribute GWAS results to a centralized repository, where quality control and meta-analyses are performed. Follow-up analyses include in silico and in vivo functional studies, and studies of clinical prediction of variants, and we welcome requests to contribute to any of our investigations The Consortium is modelled on the COGENT Blood Pressure Consortium, which is focused on genetics of blood pressure and hypertension in individuals of African ancestry6.
The COGENT-Kidney Consortium has made a considerable effort to increase the number of Hispanic/Latino GWAS included in genetic studies of CKD, and we are working towards recruitment of other diverse populations. Although sample sizes are not currently (and will not be) large when compared to studies of European ancestry, we have already demonstrated important gains in locus discovery and mapping of causal variants for kidney traits, in addition to detection of some population-specific risk variants5, 7. Our consortium GWAS on kidney traits in African Americans and Hispanics/Latinos is a resource to assess the generalization of findings to other clinical traits, including studies examining the impact of genetics in hypertension, metabolic abnormalities and CKD8, 9. As we expand the samples within each ethnic group, we expect that these data will become a resource for replication of genetic associations identified by multi-ethnic GWAS of kidney traits. We are continuing to develop trans-ethnic methods that account for the genomic differences across ancestries to map variants that are causal disease. These methods, when integrated with functional annotation obtained from CKD-relevant cells and tissues10, provide a reduced number of variants for characterization of causal genes in vitro and in experimental models to reveal biological mechanisms for CKD7.
What have we learned?
Trans-ethnic approaches identify novel loci where CKD associations are driven by causal variants with higher allele frequencies in non-European ancestries.
Our most recent trans-ethnic GWAS meta-analysis, undertaken in a total of 312,468 individuals of diverse ancestry, has identified 20 novel loci associated with estimated glomerular filtration rate (eGFR) that were replicated in two subsequent investigations from the (CKDGen Consortium and Million Veterans Project (Table S2). These include at least one locus, mapping to the region encompassing the genes PMF1 and BGLAP, that was not identified in European ancestry GWAS in a much larger sample size that is likely due to a larger effect on eGFR of the causal variant in other population groups.
Causal variants are shared across populations and amenable to risk prediction.
Genetic risk scores derived from aggregated GWAS variants can be integrated with traditional risk factors for disease prediction in clinical setting. However, studies have shown that genetic risk scores identified in European ancestry populations do not generalize to other populations11, thereby limiting the clinical utility of these GWAS findings for disease prediction in diverse populations. Our trans-ethnic GWAS meta-analysis has demonstrated, for the first time, that identified genetic associations with eGFR are shared across ancestries. Therefore, variants at these loci better represent genetic risk in multi-ethnic populations and can be used for disease prediction, irrespective of ancestry. We have used this genetic risk in Mendelian randomization studies to estimate the causal effects of eGFR on clinical outcomes7 (BOX 1, Figure 1).
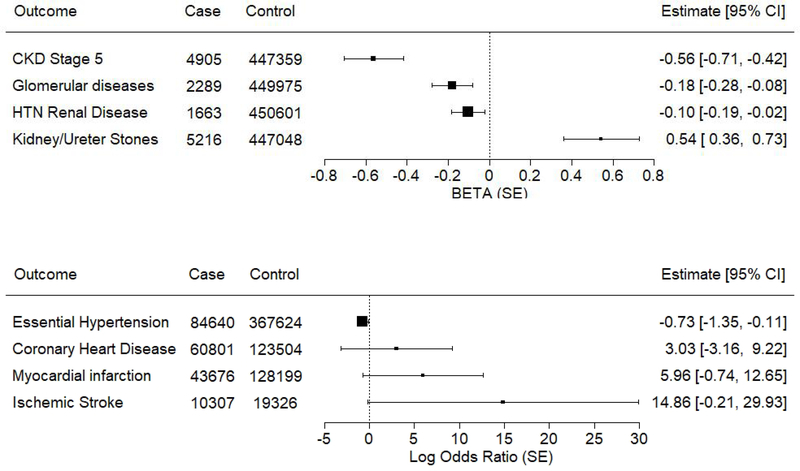
Figure 1. Mendelian randomization to assess causal effects of eGFR on CKD and cause-specific kidney disease.
We performed Mendelian randomization to assess the causal association between eGFR and clinical outcomes. We used genetic variants across 94 kidney function loci identified in the trans-ethnic meta-analyses of the COGENT-Kidney Consortium to construct an “instrumental variable” for eGFR. Clinical outcomes were obtained from ICD codes in the UK Biobank. The graph shows the Mendelian Randomization effect size (and standard deviation) of eGFR on outcomes for aggregated variants under inverse variance weighted regression. Note that low eGFR is casually associated with CKD stage 5, glomerular disease and hypertensive renal disease, but has an inverse causal association with kidney stones.
Improved localisation of causal variants at GWAS loci (i.e., fine-mapping).
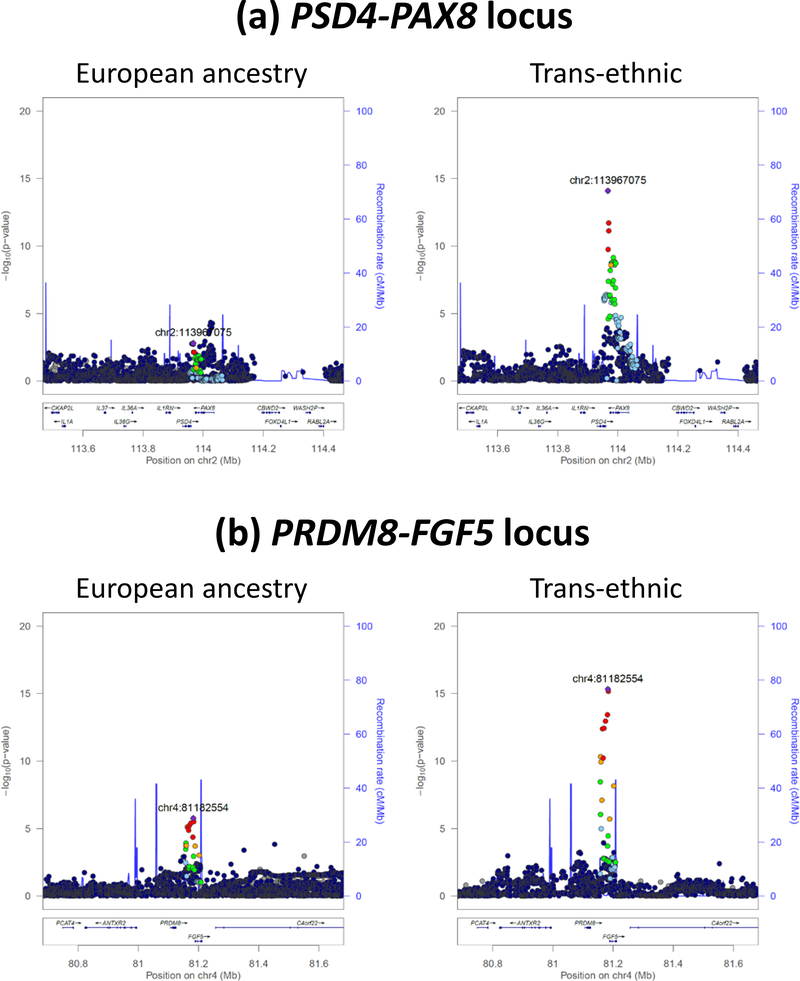
We have shown that trans-ethnic GWAS meta-analysis approaches that account for differences in LD patterns across populations and the heterogeneity in allelic effects improve fine-mapping resolution, reducing the number of likely causal variants associated with a trait to be queried for regulatory function to one or more nearby genes (Figure 2, Supplementary Figure 1). This is relevant as genetic associations identified in populations of European ancestry typically extend over large genomic regions containing multiple genes due to the extensive LD among variants.
Figure 2. Examples of improved fine-mapping resolution attained through trans-ethnic meta-analysis (312,468 individuals) over European ancestry meta-analysis (134,070 individuals) in the COGENT-Kidney Consortium.
Each point represents a variant, plotted with their p-value (on a -log10 scale) as a function of genomic position (NCBI build 37). In each plot, the index variant is represented by the purple symbol. The colour coding of all other variants indicates LD with the index variant in European ancestry haplotypes from the 1000 Genomes Project reference panel: red r2≥0.8; gold 0.6≤r2<0.8; green 0.4≤r2<0.6; cyan 0.2≤r2<0.4; blue r2<0.2; grey r2 unknown. Recombination rates are estimated from Phase II HapMap and gene annotations are taken from the University of California Santa Cruz genome browser.
Identified trans-ethnic variants for eGFR are enriched for functional annotation in kidney cells and improve understanding of disease biology.
Our studies have demonstrated significant enrichment of identified trans-ethnic variants associated with eGFR for regulatory sites in kidney-specific cells/tissues (for example, dnase I hypersensitivity sites in multiple kidney cell types, and kidney-specific histone modifications). Our recent trans-ethnic fine-mapping strategy incorporated the functional annotation from kidney cells and tissues to prioritize genomic variants. We further used kidney-specific gene expression data to link these variants to genes and to map genes to kidney cells in the nephron using single-nucleus RNAseq7. These studies have provided genes and variants for experimental follow-up studies, for example, genes related to salt sensitivity5.
What is needed?
We still need more information on genetic variants in diverse populations to construct reference panels for the design of efficient GWAS genotyping arrays and for imputation (BOX 1), and to uncover population-specific genetic risk variants. The National Human Genome Research Institute Human Heredity and Health in Africa (H3Africa) initiative is generating genomic data in Africans and have several projects related to CKD12. The National Heart Lung and Blood Institute Trans-Omic for Precision Medicine (TOPMed) program is performing deep whole genome sequencing in approximately 150,000 individuals across diverse populations2, including African Americans, Hispanics/Latinos, Caribbeans, East Asians, and Pacific Islanders. However, there are many global populations that are not represented in these efforts. Publication of high quality genetic studies in populations at high-risk of CKD is essential, even if they have small sample sizes, as they can provide insights into the genetic contribution to disease.
Recent studies have integrated high-dimensional multi-omics data and GWAS to uncover mechanisms by which genotypes influence a trait. However, multi-omics data are still limited in diverse populations. For example, expression quantitative trait loci (eQTL, BOX 1) can help prioritize gene(s) when the associated variant is located in a noncoding but regulatory regions of the genome. The landscape of genetic regulation of gene expression varies across populations, with some genes showing differential expression by ethnicity13, but eQTLs are mostly available only for European ancestry populations.
Finally, there is an increasing need for novel statistical methods that integrate omics and GWAS across diverse populations. Many existing approaches rely on knowledge of LD of the reference population, which may not be available or well-estimated in populations with ancestral admixture. In addition, approaches need to account for differences in allele frequencies and potential heterogeneity in allelic effects across populations when combining the data across ancestries.
In summary, studies of diverse populations have not been fully embraced by the research community, for reasons including the small available sample sizes, the complexity of genomic LD patterns, the presence of ancestry admixture, and a resistance to adopt methodology that is applicable to populations with or without admixture. We have established the COGENT-Kidney Consortium to overcome some of these challenges by targeting recruitment to diverse ancestries and fostering collaborations in research and methods development appropriate to these populations. By increasing awareness and representation of these populations, we have also shown important gains in translation of some of the loci to biologic mechanisms and through understanding the population impact in diseases associated with low eGFR.
Supplementary Material
Acknowledgments
Disclosures: The authors have no disclosures to report. This research is supported by the National Institutes of Health awards DK117445 and MD012765.
Footnotes
Supplementary Material
Supplementary information is available on Kidney International’s website
References
- 1.Xie Y, Bowe B, Mokdad AH, et al. Analysis of the Global Burden of Disease study highlights the global, regional, and national trends of chronic kidney disease epidemiology from 1990 to 2016. Kidney Int 2018; 94: 567–581. [DOI] [PubMed] [Google Scholar]
- 2.Jha V, Garcia-Garcia G, Iseki K, et al. Chronic kidney disease: global dimension and perspectives. Lancet 2013; 382: 260–272. [DOI] [PubMed] [Google Scholar]
- 3.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature 2016; 538: 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahajan A, Rodan AR, Le TH, et al. Trans-ethnic Fine Mapping Highlights Kidney-Function Genes Linked to Salt Sensitivity. Am J Hum Genet 2016; 99: 636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang J, Le TH, Edwards DRV, et al. Single-trait and multi-trait genome-wide association analyses identify novel loci for blood pressure in African-ancestry populations. PLoS Genet 2017; 13: e1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris AP, Le TH, Wu H, et al. Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat Commun 2019; 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sung YJ, Winkler TW, de Las Fuentes L, et al. A Large-Scale Multi-ancestry Genome-wide Study Accounting for Smoking Behavior Identifies Multiple Significant Loci for Blood Pressure. Am J Hum Genet 2018; 102: 375–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentley AR, Sung YJ, Brown MR, et al. Multi-ancestry genome-wide gene-smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nat Genet 2019; 51: 636–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magi R, Horikoshi M, Sofer T, et al. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum Mol Genet 2017; 26: 3639–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquez-Luna C, Loh PR, South Asian Type 2 Diabetes C, et al. Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet Epidemiol 2017; 41: 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium HA, Rotimi C, Abayomi A, et al. Research capacity. Enabling the genomic revolution in Africa. Science 2014; 344: 1346–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stranger BE, Montgomery SB, Dimas AS, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet 2012; 8: e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.