Abstract
Background
In the context of mass regulation, ‘muscle memory’ can be defined as long‐lasting cellular adaptations to hypertrophic exercise training that persist during detraining‐induced atrophy and may facilitate future adaptation. The cellular basis of muscle memory is not clearly defined but may be related to myonuclear number and/or epigenetic changes within muscle fibres.
Methods
Utilizing progressive weighted wheel running (PoWeR), a novel murine exercise training model, we explored myonuclear dynamics and skeletal muscle miRNA levels with training and detraining utilizing immunohistochemistry, single fibre myonuclear analysis, and quantitative analysis of miRNAs. We also used a genetically inducible mouse model of fluorescent myonuclear labelling to study myonuclear adaptations early during exercise.
Results
In the soleus, oxidative type 2a fibres were larger after 2 months of PoWeR (P = 0.02), but muscle fibre size and myonuclear number did not return to untrained levels after 6 months of detraining. Soleus type 1 fibres were not larger after PoWeR but had significantly more myonuclei, as well as central nuclei (P < 0.0001), the latter from satellite cell‐derived or resident myonuclei, appearing early during training and remaining with detraining. In the gastrocnemius muscle, oxidative type 2a fibres of the deep region were larger and contained more myonuclei after PoWeR (P < 0.003), both of which returned to untrained levels after detraining. In the gastrocnemius and plantaris, two muscles where myonuclear number was comparable with untrained levels after 6 months of detraining, myonuclei were significantly elongated with detraining (P < 0.0001). In the gastrocnemius, miR‐1 was lower with training and remained lower after detraining (P < 0.002).
Conclusions
This study found that (i) myonuclei gained during hypertrophy are lost with detraining across muscles, even in oxidative fibres; (ii) complete reversal of muscle adaptations, including myonuclear number, to untrained levels occurs within 6 months in the plantaris and gastrocnemius; (iii) the murine soleus is resistant to detraining; (iv) myonuclear accretion occurs early with wheel running and can be uncoupled from muscle fibre hypertrophy; (v) resident (non‐satellite cell‐derived) myonuclei can adopt a central location; (vi) myonuclei change shape with training and detraining; and (vii) miR‐1 levels may reflect a memory of previous adaptation that facilitates future growth.
Keywords: Hypertrophy, Atrophy, Muscle damage, Central nuclei, miR‐1, Tet‐On, H2B‐GFP
Introduction
It has been proposed that myonuclei gained during the hypertrophic process are permanent. 1 , 2 , 3 , 4 When hypertrophy is reversed with detraining, muscle fibres should be ‘hyper‐nucleated’ per unit size, theoretically constituting a ‘muscle memory’ of previous adaptation that may have consequences for retraining. 1 The majority of experimental data supporting the myonuclear number‐mediated muscle memory hypothesis were gathered using surgical or pharmacological manipulations to alter muscle fibre size, very young animals, or non‐mammalian model systems, 1 , 3 , 5 , 6 thereby limiting translatability to exercise training and detraining in adult mammals. Recently, using a novel progressive weighted wheel running (PoWeR) approach in adult mice (>4 month old), our laboratory showed that elevated myonuclear number observed during training returns towards untrained levels following 3 months of detraining in the primarily glycolytic plantaris muscle. 7 Although not completely reverted to untrained levels, myonuclear loss with detraining challenges the hypothesis that myonuclei acquired during hypertrophy are permanent. 1 , 3
The relationship between muscle fibre oxidative potential and the maintenance of myonuclear number during atrophy following growth is not well characterized. Myonuclei gained in oxidative muscle fibres during hypertrophy may be more stable than in glycolytic fibres because oxidative fibres are activated more frequently and have higher biosynethetic rates, 8 , 9 , 10 but this possibility has not been tested directly. Furthermore, information on the temporal nature of myonuclear loss following hypertrophy is somewhat limited. One study in quails reported that an apoptotic loss of myonuclei acquired during loading‐induced hypertrophy is initiated after 7 days of deloading, but myonuclear number was not quantified, nor was the time course of myonuclear removal. 11 Our finding of ~20% loss of myonuclei in the plantaris muscle after 3 months of detraining was noteworthy, 7 but whether myonuclear number returns completely to untrained levels across various muscle groups after PoWeR is unknown. Beyond myonuclear number, recent evidence suggests that DNA methylation changes in skeletal muscle induced by training could be long‐lasting and represent a memory of prior training exposure that facilitates future adaptation. 12 Our laboratory and others recently found that muscle‐specific miRNAs (myomiRs) persist for prolonged periods in the absence of the miRNA processing enzyme Dicer. 13 , 14 If prolonged miRNA stability is a quality specific to skeletal muscle, 15 myomiRs could be good candidates as long‐term epigenetic regulators of exercise adaptation, especially given their known function in regulating muscle mass. 16 , 17 , 18
In the current investigation, we provide evidence on myonuclear accretion and maintenance in skeletal muscles with different fibre type composition and function after 2 months of PoWeR training and prolonged 6‐month detraining. We hypothesized that oxidative muscle fibres in the soleus and gastrocnemius would be preferentially hyper‐nucleated per unit size following detraining and that myonuclear number in the plantaris would reflect untrained levels after detraining from PoWeR. Our data show that unlike the gastrocnemius and plantaris muscles, which lose training‐induced adaptations including fibre type distribution, fibre size, and myonuclear number following 6 months of detraining, the soleus muscle did not detrain according to these cellular parameters. Moreover, myosin heavy chain type 1 fibres of the soleus had elevated myonuclear number in response to PoWeR in the absence of hypertrophy. Contrary to our hypothesis, oxidative type 2a muscle fibres are not hyper‐nucleated per unit size after 6 months of detraining when hypertrophy is reversed in the gastrocnemius. We also report that myonuclear shape changes during training and detraining, with myonuclei being shorter following PoWeR training and elongated after detraining. Finally, we show that the myomiR miR‐1 is lower in PoWeR‐trained gastrocnemius and remains low after detraining, which may signify an epigenetic memory of prior growth that could facilitate future adaptation.
Materials and methods
Ethical approval
All animal procedures were approved by the IACUC of the University of Kentucky. C57BL/6J mice were obtained from the Jackson Laboratory (Bar Harbor, ME). The muscle‐specific Tet‐On (HSA‐rtTA) mouse, created by our laboratory, 19 was crossed with the tetracycline response element histone 2b green flurorescent protein (TRE‐H2B‐GFP) mouse, 20 obtained from the Jackson Laboratory, to generate the HSA‐GFP mouse, with pups genotyped as previously described by us. 19 Mice were housed in a temperature and humidity‐controlled room, maintained on a 14:10‐h light–dark cycle, and food and water were provided ad libitum. Animals were sacrificed via a lethal dosage of sodium pentobarbital injected intraperitoneally, followed by cervical dislocation.
Experimental design
An experimental design schematic is presented in Figure 1. Four month old female mice were assigned to a 2‐month PoWeR group that were 6 months old at the time of euthanasia (6M PoW; n = 14), a 2‐month PoWeR followed by 6 months of detraining group that were 12 months old at the time of euthanasia (12M PoW + DT; n = 8), a 6‐month‐old untrained control group age matched to the 6M PoW group (6M UT; n = 13), and a 12‐month‐old untrained control group age matched to the detraining group (12M UT; n = 9). Five mice from the PoWeR and UT groups were used for analyses outside the scope of the present report resulting in n = 8–9 in all groups for experiments reported here. We also analysed soleus muscles from a 2‐month PoWeR followed by 3‐month detrained group (9M PoW + DT, n = 8); the plantaris data were published previously. 7 The soleus muscle did not detrain after 3 months, so we focused our effort on the 6‐month detrained time point (see Supporting Information, Figure S1). Mice were singly housed during PoWeR to monitor distance run (km/day) for each mouse, which was recorded using ClockLab software (Actimetrics, Wilmette, IL). PoWeR was carried out as previously described. 7 Briefly, following 1 week of acclimation to an unweighted wheel, 2 months of PoWeR was 2 g of weight for the first week, 3 g for second week, 4 g for third week, 5 g for the fourth and fifth week, then 6 g for the final 3 weeks (Figure 1). Magnets weighing 1 g each were used to load the wheels (B661, K&J Magnetics, Pipersville, PA) and were affixed in one location along the outside circumference. Mice were moved to non‐wheel cages during detraining. In a separate experiment, HSA‐GFP mice (n = 3 sedentary and n = 4 running) were treated with 0.5 mg/mL doxycycline and 2% sucrose in drinking water, as described previously by our laboratory, 19 for 7 days followed by a 5‐day washout, and were then allowed free access to a running wheel for 6 days while consuming 0.5 mg/mL ethenyldeoxyuridine (EdU, Carbosynth, Berkshire, UK) with 2% glucose in drinking water, replenished on the third day; the wheel was locked for the final 24 h before euthanasia. Treatment with doxycycline induced stable GFP labelling of myonuclei in resting muscle, so only pre‐existing resident myonuclei were GFP+ for the remainder of the experiment (i.e. satellite cell‐derived myonuclei induced from running were not GFP labelled).
Figure 1.
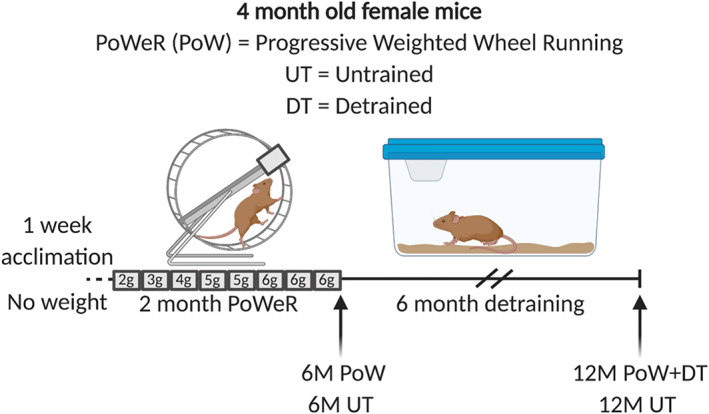
Study design schematic illustrating the four groups used for analysis: 6‐month‐old untrained (6M UT), 6‐month‐old PoWeR trained (6M PoW), 12‐month‐old untrained (12M UT), and 12‐month‐old PoWeR trained then 6‐month detrained (12M PoW + DT). All mice were 4 months old at the time of being assigned to experimenal groups.
Immunohistochemistry
Immunohistochemistry (IHC) analyses on entire soleus, gastrocnemius, and plantaris muscle cryosections was carried out following embedding in optimal cutting temperature compound (OCT compound) and snap freezing in isopentane, as previously described by our laboratory, using antibodies that are well established as specific. 21 For fibre type distribution, overall muscle fibre cross‐sectional area (CSA), fibre type‐specific CSA, and myonuclear density (analysed as myonuclei per fibre), frozen sections were incubated overnight in a cocktail of isotype‐specific anti‐mouse primary antibodies for types 1, 2a, and 2b, or embryonic MyHC (eMyHC) from Developmental Studies Hybridoma Bank (Iowa City, Iowa, USA), along with an antibody against dystrophin (1:100, ab15277, Abcam, St. Louis, MO, USA), followed by the appropriate secondary antibodies and/or 4′,6‐diamidino‐2‐phenylindole (DAPI), then mounted using a 50/50 solution of phosphate‐buffered saline and glycerol. EdU incorporation into DNA was detected in HSA‐GFP mice using the detection cocktail from Kirby et al., 22 which involves copper‐mediated Click‐iT chemistry and a TAMRA azide secondary (Click Chemistry Tools, Scottsdale, AZ, USA). The detection cocktail for EdU quenches GFP, so GFP in EdU‐labelled muscle sections was rescued using an anti‐rabbit GFP antibody (1:200 in phosphate‐buffered saline, ab6556, Abcam, Cambridge, United Kingdom) along with goat anti‐rabbit biotin secondary (1:500 in phosphate‐buffered saline, 111‐065‐144, Jackson Immunoresearch, West Grove, PA, USA) then streptavidin–horseradish peroxidase–tyramide signal amplification (TSA) amplification (Invitrogen, Waltham, MA, USA) of the fluorescent signal. Central nuclei were assessed manually using analysis tools in the Zen software, and a myonucleus was considered ‘central’ if there was discernable distance between DAPI and the dystrophin border (i.e. were not located peripherally, abutting dystrophin).
Single muscle fibre myonuclear analysis
In addition to IHC analysis, myonuclear counts were also conducted on isolated single muscle fibres and normalized to fibre length. This protocol for the plantaris was previously described by our laboratory and others, 7 , 21 , 23 , 24 and we adapted it without modification to the soleus and gastrocnemius muscles. Briefly, mouse hindlimbs were fixed in situ at resting length in 4% paraformaldehyde for 48 h to control for sarcomere spacing. Muscle fibres and adherent cells were liberated by NaOH incubation, pipetted onto glass slides, then mounted using VectaShield with DAPI (Vector). Thirty fibres per muscle per mouse (~300–500 μm random linear segments per individual fibre) were used for myonuclear number analysis of the soleus and plantaris, 21 whereas 50 fibres per muscle per mouse were analysed in the gastrocnemius given its large size and heterogenous mixture of all four primary skeletal muscle fibre types in mice. To measure myonuclear length, myonuclei from a random selection of 5 (soleus and plantaris) or 10 (gastrocnemius) muscle fibres per mouse were measured using tools in the Zeiss Zen software. In brief, a line tool was used to measure the length of individual nuclei along their longest axis (range: 116–240 per muscle in the soleus, 91–149 per muscle in the plantaris, and 174–320 per muscle in the gastrocnemius), and a weighted average myonuclear length value (i.e. accounting for the number of myonuclei measured in each fibre) for each muscle was used for statistics.
Image capture
For IHC, images were captured at ×20 magnification using a Zeiss upright fluorescent microscope (Zeiss AxioImager M1 Oberkochen, Germany). Whole muscle sections were obtained using the mosaic function in Zeiss Zen 2.3 for all muscles analysed. All measures were quantified on entire muscle cross sections using MyoVision semi‐automated analysis software, thus mimimizing bias during analysis. 25 For single fibre myonuclear analysis, fibres were imaged at ×20 magnification using the Z‐stack function within the Zen software. 21 All manual counting was performed by a blinded, trained technician.
RNA extraction and miRNA analysis
A piece of frozen gastrocnemius muscle was taken from the IHC mount and placed into RNAlater‐ICE Frozen Tissue Transition Solution (Invitrogen) overnight at −20°C. The following morning, residual OCT was gently removed from each piece of tissue. RNA was then isolated using the Qiagen miRNeasy Mini Kit (Qiagen, Germantown, MD, USA). Total RNA concentration was determined using a NanoDrop (ThermoFisher), and then, RNA quality was assessed using an Agilent 2100 Bioanalyzer Instrument (Agilent, Santa Clara, CA, USA). MiRNA abundance was then quantified using quantitative single molecule direct digital detection (mouse nCounter miRNA Expression Panel, NanoString, Seattle, WA, USA) and analysed using nSolver Analysis Software (NanoString). The nCounter panel multiplexes 577 mouse miRNAs as well as 6 positive controls, 8 negative controls, 3 ligation positive and 3 negative controls, 5 mRNA reference controls, and 5 spike‐in controls to ensure maximum accuracy. Raw read counts were normalized to the geomentric mean of the positive internal controls for each sample, then normalized to the geometric mean of the top 50 most abundant miRNAs (the geometric mean for the top 50 miRNAs was not significantly different between groups, analysed via one‐way ANOVA). The minimum threshold was set at 50 copies. All miRNAs that fell below this threshold were excluded from the analysis.
Statistics
Data were checked for normality and original or log‐transformed mean data were used for analyses. Differences between 6M UT, 6M PoW, 12M UT, and 12M PoW + DT were analysed with a one‐way ANOVA, in alignment with previous rodent studies of similar design, 2 , 4 , 6 and a Holm–Sidak post hoc analysis was used when differences were identified. Comparison of running volume between 6M PoW and 12M PoW + DT was evaluated using a Student's two‐tailed t‐test. To identify miRNAs that could be related to muscle memory, we overlapped the list of significantly different miRNAs in 6M UT versus 6M PoW with 12M UT versus 12M PoW + DT, which yielded a candidate list, and those candidates were then analysed via one‐way ANOVA to make all statistical comparisons. For myonuclear number and length analyses, values per fibre were averaged to generate one value per mouse, which was then used for statistics. Significance was set at P < 0.05, and all data are presented as mean ± standard error.
Results
Mouse characteristics
Female mice that PoWeR trained ran 12.5 ± 0.8 km per night on average (see Dungan et al. 7 ), and 12M PoW + DT mice ran 12.5 ± 0.6 km per night. Running volume was not different between 6M PoW and 12M PoW + DT. Body weights between 6M PoW and age‐matched untrained (6M UT) mice (6 month old; 23.9 g ± 1.0 vs. 24.0 g ± 2.1, respectively; P = 0.96), as well as 12M PoW + DT 12M DT mice (12 month old; 33.5 g ± 2.7 vs. 31.3 g ± 4.4, respectively; P = 0.20), were not different.
The soleus muscle adapted to PoWeR training but did not detrain after 6 months
We previously published that the plantaris detrained after a 3‐month cessation from PoWeR, which included a reduction in myonuclear number relative to PoWeR trained. 7 We similarly analysed the soleus muscle after 3 months of detraining, and muscle mass, fibre CSA, and myonuclear number measured on single fibres were the same as in PoWeR trained, indicating that the soleus did not detrain (Figure S1). As such, we focused our analyses on the 6‐month detraining time point to determine whether oxidative muscle fibres detrain and if this is associated with higher myonuclear number. Relative to 6M UT, soleus muscle wet weight was 35% greater in 6M PoW (Figure 2A, P < 0.0001). PoWeR‐trained mice had more type 1 fibres and fewer type 2a fibres relative to 6M UT (Figure 2B–F, P = 0.0003). Average soleus muscle fibre CSA, quantified using dystrophin IHC on entire cross sections, was not significantly larger in 6M PoW versus 6M UT (18%, Figure 2G, P = 0.14) because type 1 fibres were not larger after training (Figure 1H); however, type 2a average fibre size was significantly larger (Figure 2H, P = 0.018). Myonuclear number was 24% and 20% higher in 6M PoW compared with 6M UT using single fibre (Figure 3A, 3B, and 3E, P = 0.004) and IHC analyses (Figure 3F, P = 0.001), respectively. Fibre type‐specific analysis via IHC revealed that PoWeR‐trained type 1 fibres had greater myonuclei per fibre than 6M UT despite not being larger (Figure 3G, P = 0.007), while type 2a fibres had elevated myonuclei per fibre associated with larger fibre CSA (Figure 3G, P = 0.0005). In the soleus, 6.2% of muscle fibres had centrally located myonuclei after PoWeR (Figure 3H), with 4.7% of those being from type 1 muscle fibres and 1.5% being from type 2a fibres (Figure 3I). Myonuclear length was shorter in PoWeR‐trained muscles relative to 6M UT (Figure 3J, P = 0.03).
Figure 2.
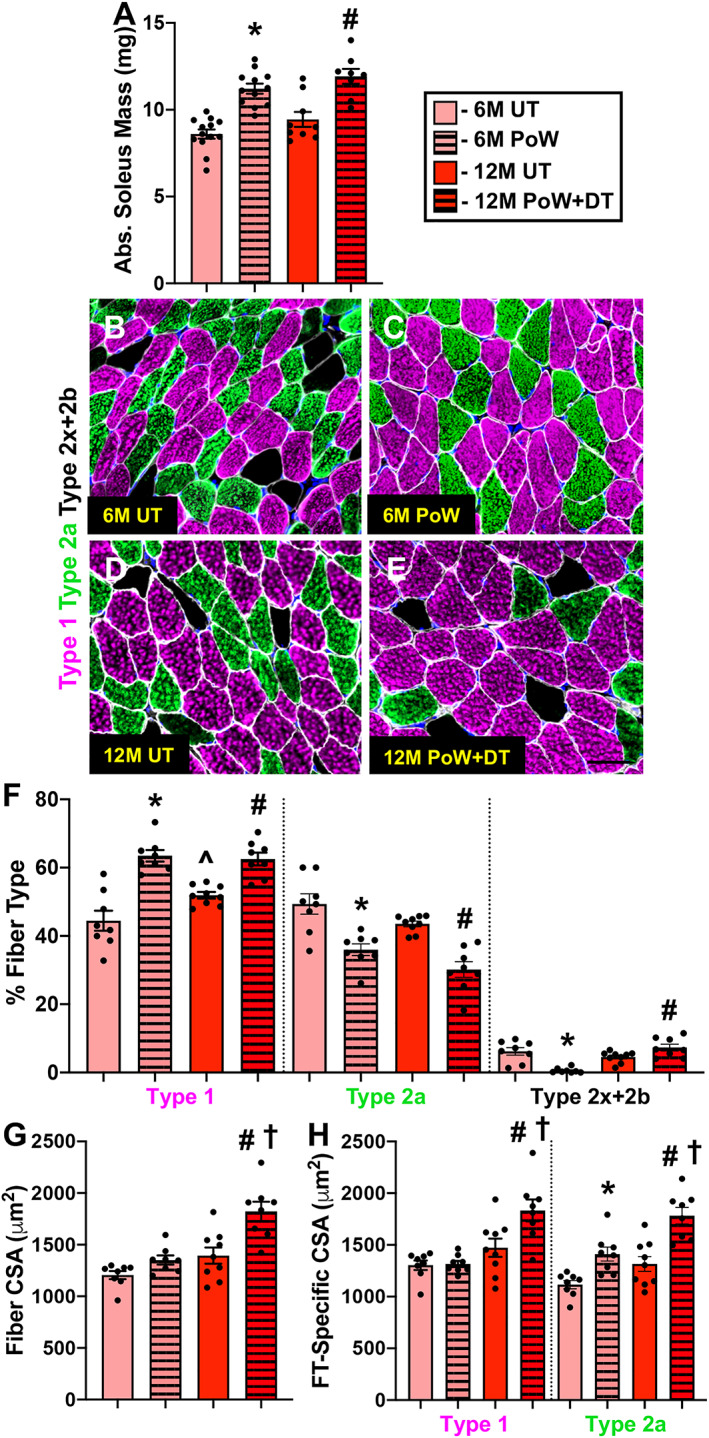
Soleus muscle size and fibre type with training and detraining. (A) Absolute soleus muscle wet weight in 6M UT, 6M PoW, 12M UT, and 12M PoW + DT. (B–E) Repesentative immunohistochemistry images from the four groups showing dystrophin (white), type 1 (pink), type 2a (green), unstained type 2x and/or 2b fibres (black), and DAPI (blue). (F) Fibre type (FT) distribution. (G) Average muscle fibre cross‐sectional area (CSA). (H) Fibre type‐specific CSA. *6M UT versus 6M PoW, ^6M UT versus 12M UT, #12M DT versus 12M PoW + DT, †6M PoW versus 12M PoW + DT. All P < 0.05, scale bars = 50 μm, n ≥ 8 per group for each analysis.
Figure 3.
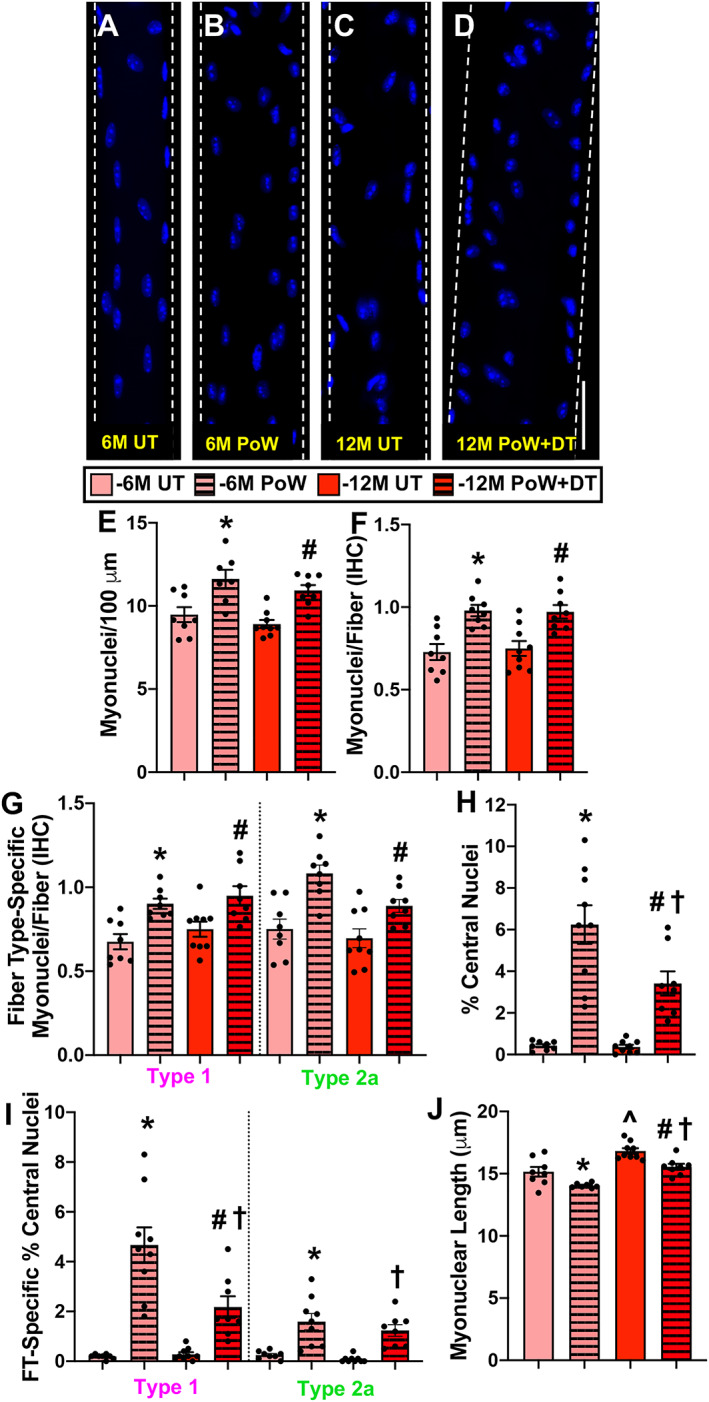
Soleus myonuclear analysis with training and detraining. (A–D) Representative images of longitudinal single muscle fibres from 6M UT, 6M PoW, 12M UT, and 12M PoW + DT. (E) Single fibre analysis of myonuclear number. (F) Immunohistochemistry (IHC) analysis of myonuclear number. (G) Fibre type‐specific myonuclear number via IHC. (H) Proportion of fibres with central nuclei (CN). (I) Myonuclear length. *6M UT versus 6M PoW, ^6M UT versus 12M UT, #12M DT versus 12M PoW + DT, †6M PoW versus 12M PoW + DT. All P < 0.05, scale bars = 50 μm, n ≥ 7 per group for each analysis.
Following 6 months of detraining, muscle mass (Figure 2A), muscle fibre size (Figure 2g), and myonuclear number (Figure 3E and 3F) were all larger than 12M UT (P < 0.01), indicating that the soleus did not detrain by 6 months after PoWeR training (consistent with observations at 3 months of detraining). Type 2x/2b proportion was modestly but significantly higher in 12M PoW + DT versus 12M UT (Figure 2F), while fibre size was larger versus 12M UT and 6M PoW (Figure 2G and 2H ). There were more central nuclei in 12M PoW + DT versus 6M and 12M UT, but fewer than in 6M PoW (Figure 3H and 3I, P = 0.003). Soleus myonuclear length was longer following detraining relative to PoWeR (P = 0.002), but was still shorter than age‐matched untrained (P = 0.007, Figure 3J).
One week of unweighted wheel running caused developmental myosin expression, myonuclear accretion, and resident myonuclear mispositioning in the soleus
We previously published that the proportion of central nuclei in the plantaris after PoWeR was <2%. 7 In light of there being over three times more fibres with central nuclei specifically in the soleus after PoWeR, we performed a study to determine whether unaccustomed activity early in training could explain these findings, as well as to determine the source of these central nuclei (Figure 4A). The HSA‐GFP mouse enables myonuclei to be fluorescently labelled only in the presence of doxycycline. 19 By labelling resident myonuclei prior to wheel running, and in combination with EdU treatment during running, we could for the first time test whether central nuclei in adult muscle fibres caused by unaccustomed activity (as opposed to toxin‐induced regeneration) were from fused‐in cells that underwent a round of division (e.g. proliferating satellite cells), from a cell that had fused in without proliferation, from a resident myonucleus that had migrated, or a resident myonucleus that had undergone DNA synthesis. In the soleus muscle of HSA‐GFP mice following 6 days of unweighted running, which is a normal component of initiating the PoWeR protocol, 7 there were eMyHC+ fibres that were large in calibre (likely pre‐existing muscle fibres that experienced significant satellite cell fusion, because eMyHC expression is satellite cell‐dependent in adult muscle 21 , 24 ) as well as smaller myofibers (likely de novo) (Figure 4B), many of which contained central nuclei (Figure 4C). Central nuclei after short‐term wheel running dovetails with previous reports. 26 , 27 The majority of central nuclei were GFP− (88%), likely derived from satellite cell fusion in the time after doxycycline treatment. Muscle fibres with central nuclei (8.2%) were eMyHC−/GFP+, eMyHC+/GFP+, or eMyHC+/GFP− (Figure 4C). Centrally nucleated eMyHC−/GFP− fibres were extremely rare; only two such events were identified across all sections analysed, indicating that eMyHC expression almost always occurs simultaneously with a central nucleus during short‐term exercise. To label nuclei that had undergone DNA replication, we delivered EdU in drinking water to sedentary and wheel running mice. Most central nuclei were GFP−/EdU+ in running mice, showing that myonuclear donors (i.e. satellite cells) divided prior to fusion; the remaining central nuclei were GFP+/EdU− (12%), suggesting that resident myonuclei can translocate to the centre of the fibre without DNA synthesis (Figure 4D–D”). GFP−/EdU− central nuclei were exceedingly rare (only three total identified across all muscle cross sections analysed). We also observed GFP−/EdU+ peripheral myonuclei (Figure 4E–E”). Nuclear dynamics and eMyHC data for running mice are reported in Figure 4F. The data for sedentary mice are not shown because there were no eMyHC+ fibres, negligible mispositioned myonuclei, and very few EdU+ nuclei. In context with the soleus PoWeR findings, and although we did not measure muscle membrane damage/disruption directly, these results support the hypotheses that myonuclear accretion could be driven by damage and not hypertrophy, that resident (non‐satellite cell‐derived) myonuclei are mobile and can participate in the damage response by relocating to the center of muscle fibres (without DNA synthesis), and that the majority of central nuclei found at the end of PoWeR may have resulted from unweighted wheel running at the beginning of training.
Figure 4.
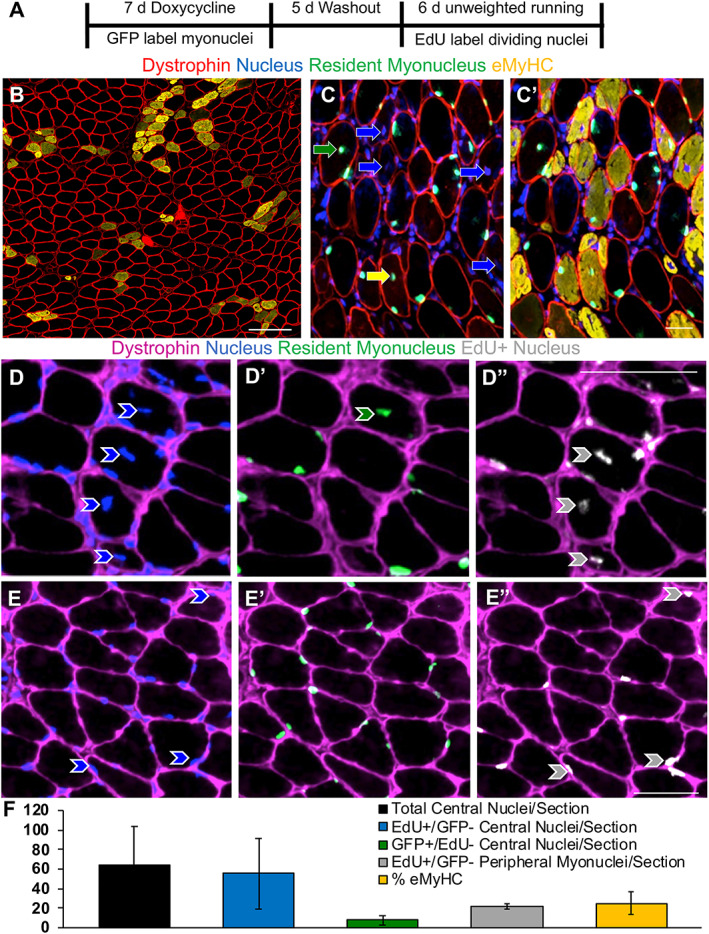
Soleus myonuclear dynamics during unweighted wheel running. (A) Study design illustrating resident myonuclear labelling in HSA‐GFP mice, ethenyldeoxyuridine (EdU) labelling for DNA synthesis, and unweighted wheel running. (B) Representative image of immunohistochemistry (IHC) for dystrophin (red) and embryonic myosin heavy chain (eMyHC, orange), scale bar = 100 μm. (C) Representative image of IHC for dystrophin (red), nuclei (DAPI), and green fluorescent protein (GFP)‐labelled non‐satellite cell‐derived resident myonuclei (green), with eMyHC (orange) added in (C’). In (C), blue arrows point to central nuclei found in eMyHC+ fibres, the green arrow points to a GFP+ central myonucleus in an eMyHC− fibre, and the yellow arrow points to a GFP+ myonucleus in an eMyHC+ fibre. (D–D”) Representative IHC images showing a GFP+/EdU− myonucleus (green arrow) and GFP−/EdU+ central myonuclei (grey arrows) within dystrophin (pink). (E–E”) Representative images showing EdU+ peripheral myonuclei (grey arrows). (F) Prevalence of total central nuclei, EdU+/GFP− central nuclei, GFP+/EdU− central nuclei, EdU+/GFP− peripheral myonuclei, and eMyHC expression. Scale bars = 50 μm, n = 4 for running and n = 3 for sedentary (data not shown for sedentary mice due to negligble EdU and eMyHC expression).
The gastrocnemius muscle undergoes fibre type‐specific adapations to PoWeR and detrains after 6 months
Relative to 6M UT, gastrocnemius wet weight was not different in 6M PoW mice (Figure 5A). PoWeR‐trained mice had more type 1 and type 2a fibres and fewer type 2x and 2b fibres relative to 6M UT (Figure 5B–F, P < 0.0001). It is important to note that even though there were more type 1 fibres following PoWeR, these fibres make up a small percentage of the total number of fibres in the gastrocnemius (~3–6%). Average gastrocnemius muscle fibre CSA was not different between 6M UT and 6M PoW mice (Figure 5G). No difference in average fibre CSA is likely attributable to the glycolytic (larger fibres) to oxidative (smaller) fibre type shift and lack of effect of PoWeR training on type 2b and 2x CSA; however, type 2a fibre CSA was larger in 6M PoW mice (Figure 5H, P < 0.0001). Type 1 fibres were not significantly larger following PoWeR (Figure 5H). Myonuclear number was 13% and 14% higher in 6M PoW compared with 6M UT gastrocnemius muscles via single fibre (Figure 6A, 6B, and 6E, P = 0.005) and IHC analyses (Figure 6F, P > 0.05), respectively. The latter analysis showed that only type 2a fibres had a higher myonuclear number in 6M PoW versus 6M UT (Figure 6G, P = 0.003). The less abundant type 1 fibres did not exibit myonuclear accretion (Figure 6G). Myonuclear length quantification showed that myonuclei from PoWeR‐trained mice were shorter than those in 6M UT mice (Figure 6H, P = 0.01).
Figure 5.
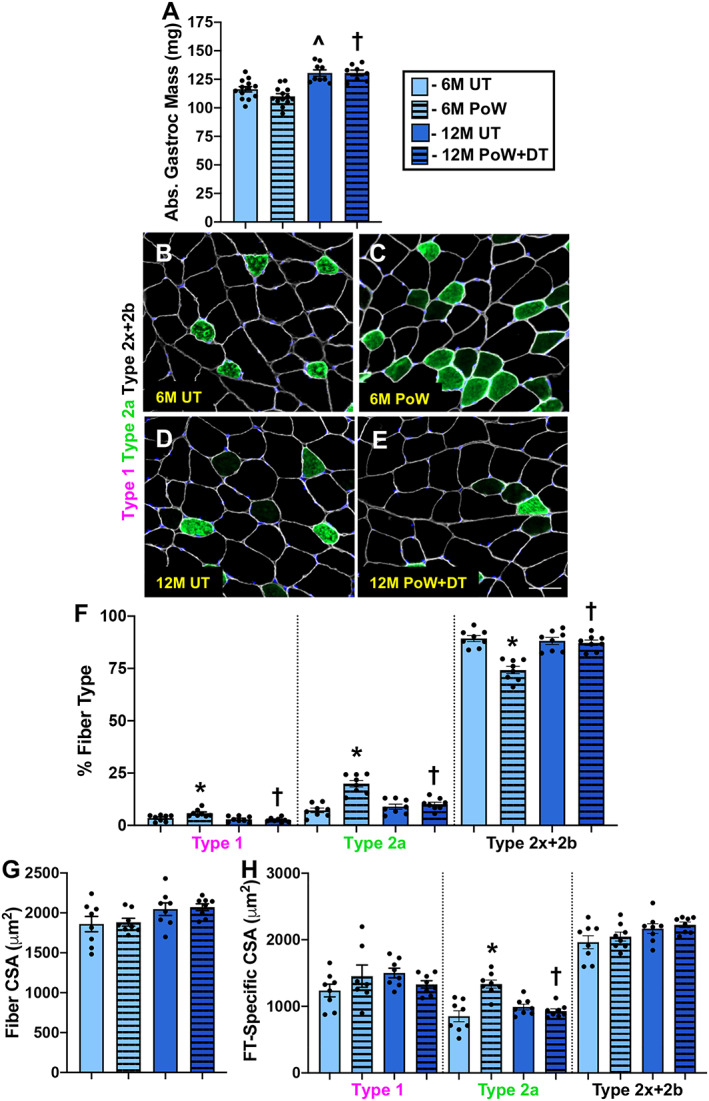
Gastrocnemius muscle size and fibre type with training and detraining. (A) Absolute gastrocnemius muscle wet weight in 6M UT, 6M PoW, 12M UT, and 12M PoW + DT. (B–E) Repesentative immunohistochemistry images from the four groups showing dystrophin (white), type 2a (green), unstained type 2x and/or 2b fibres (black), and DAPI (blue). (F) Fibre type (FT) distribution. (G) Average muscle fibre cross‐sectional area (CSA). (H) Fibre type‐specific CSA. *6M UT versus 6M PoW, ^6M UT versus 12M UT, #12M DT versus 12M PoW + DT, †6M PoW versus 12M PoW + DT. All P < 0.05, scale bars = 50 μm, n ≥ 8 per group for each analysis.
Figure 6.
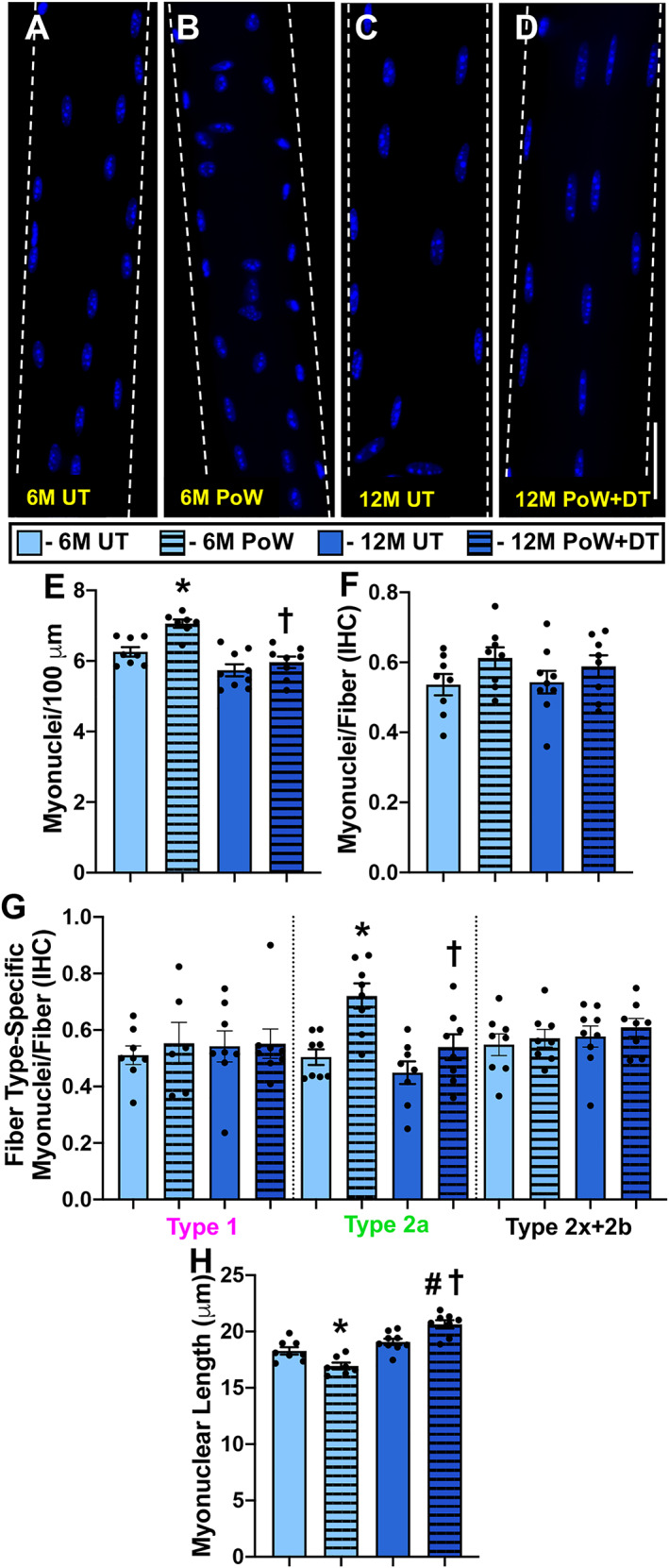
Gastrocnemius myonuclear analysis with training and detraining. (A–D) Representative images of longitudinal single muscle fibres from 6M UT, 6M PoW, 12M UT, and 12M PoW + DT. (E) Single fibre analysis of myonuclear number. (F) Immunohistochemistry (IHC) analysis of myonuclear number. (G) Fibre type‐specific myonuclear number via IHC. (H) Myonuclear length. *6M UT versus 6M PoW, ^6M UT versus 12M UT, #12M DT versus 12M PoW + DT, †6M PoW versus 12M PoW + DT. All P < 0.05, scale bars = 50 μm, n ≥ 7 per group for each analysis.
Fibre type composition (Figure 5F), fibre type‐specific CSA (Figure 5H), and myonuclear number (Figure 6E and 6F) in 12M DT were not different from 12M UT but were different from 6M PoW, suggesting that unlike the soleus, the gastrocnemius detrained from PoWeR. The deep region of the gastrocnemius is the most oxidative portion of any mouse hindlimb muscle. 28 We analysed fibre type‐specific myonuclear number via IHC in that region to address our hypothesis that oxidative muscle fibres would be hyper‐nucleated per unit size after detraining. Myonuclei per fibre was the same between 12M UT and 12M PoW + DT in highly oxidative type 2a fibres and lower than 6M PoW (Figure 6G), suggesting that myonuclei were lost in oxidative fibres with detraining. Myonuclei were significantly more elongated in 12M PoW + DT compared with UT and 6M PoW, quantified on isolated single fibres (Figure 6H, P < 0.01). [Correction added on 14 September 2020 after first online publication: The second value of “12M PoW” in the preceding sentence has been corrected to “6M PoW”.]
Myonuclear number is the same in detrained and untrained plantaris muscles after 6 months, but myonuclei are elongated in detrained muscle
We previously reported that muscle mass, fibre type distribution, and fibre CSA were identical to untrained mice following 3 months of detraining in the plantaris muscle. 7 Relative to 12M UT, the same was true following 6 months of detraining from PoWeR in the plantaris (Figure 7). Myonuclear number was signicantly lower in 3‐month detrained mice relative to PoWeR‐trained mice, but still 7% (single fibre) and 12% (IHC) higher than age‐matched controls. 7 Plantaris myonuclear number was essentially identical comparing 12M UT versus 12M PoW + DT via single fibre (Figure 7G–I) and IHC analyses (Figure 7J and 7K). Similar to the gastrocnemius, 12M PoW + DT mice had elongated myonuclei relative to 12M UT (Figure 7L, P = 0.003).
Figure 7.
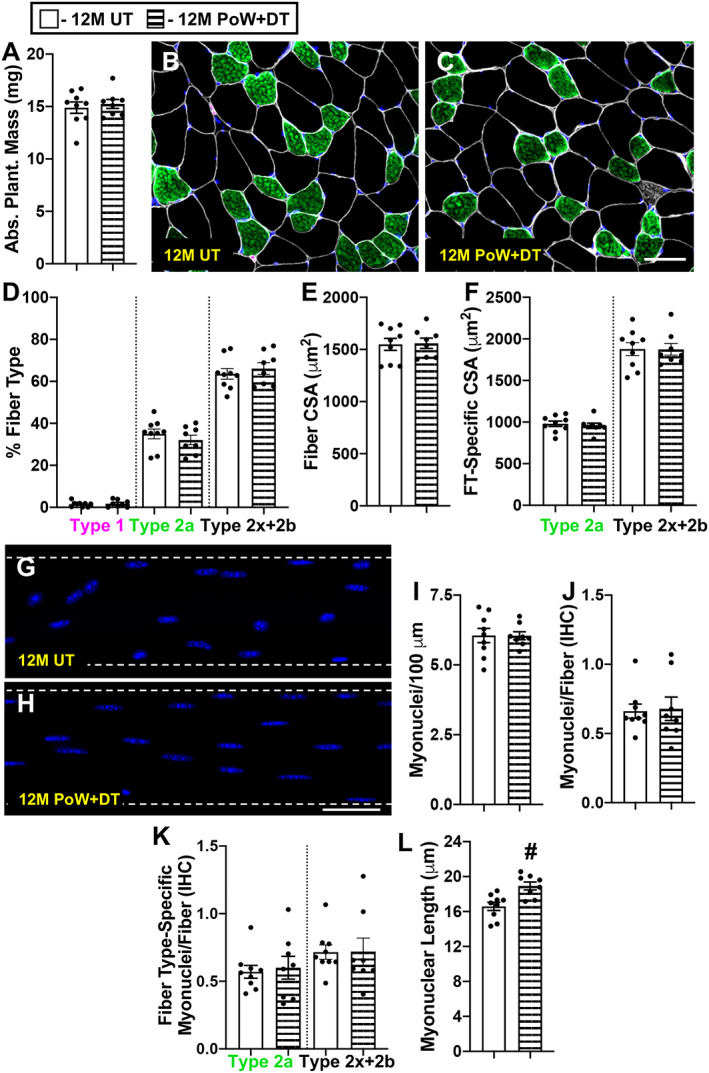
Plantaris adaptations to detraining (plantaris training data published in Dungan et al. 7 ). (A) Absolute plantaris muscle wet weight in 12M UT and 12M PoW + DT. (B–C) Repesentative immunohistochemistry (IHC) image from (B) 12M UT and (C) 12M PoW + DT mice showing dystrophin (white), type 2a (green), type 2x and/or 2b fibres (black), and DAPI (blue). (D) Fibre type (FT) distribution. (E) Average muscle fibre cross‐sectional area (CSA). (F) Fibre type‐specific muscle fibre CSA. (G–H) Representative images of longitudinal single muscle fibres from (G) 12M UT and (H) 12M PoW + DT. (I) Single fibre analysis of myonuclear number (J) IHC analysis of myonuclear number (K) fibre type‐specific myonuclear number via IHC. (L) Myonuclear length. # P < 0.05 12M DT versus 12M PoW + DT, scale bars = 50 μm, n ≥ 8 per group for each analysis.
Gastrocnemius miRNA levels change with training, and miR‐1 remained lower after 6 months of detraining
To gain insight into how PoWeR training affects muscle miRNA levels, and whether miRNA changes induced by PoWeR could persist throughout detraining, we conducted quantitative single molecule direct digital detection analysis (NanoString) in the gastrocnemius muscle that experienced fibre type‐specific hypertrophy and atrophy. After threshold filtering low‐abundance miRNAs, 139 miRNAs were analysed and 13 were uniquely differentially regulated in 6M PoW versus 6M UT (Figure 8A and 8B, P < 0.05). Relative to 12M UT mice, seven miRNAs were uniquely differentially regulated in 12M PoW + DT mice (Figure 8A and 8C, P < 0.05). Two miRNAs, miR‐1 and miR‐146a, were differentially regulated in both the PoWeR trained and detrained states (Figure 8A and 8D). Statistical analysis across all four groups revealed that miR‐1 was significantly lower in 6M PoW versus 6M UT (P = 0.002) and in 12M PoW + DT relative to 12M UT (P = 0.02). For miR‐146a, only the difference between 6M PoW versus 6M UT was significant via one‐way ANOVA (P = 0.007). Worth noting is that miR‐1 was the most abundant of all miRNAs analysed, more than twice as high as the next most abundant miRNAs measured (miR‐22 and miR‐133a). Raw data are presented in Data S1.
Figure 8.
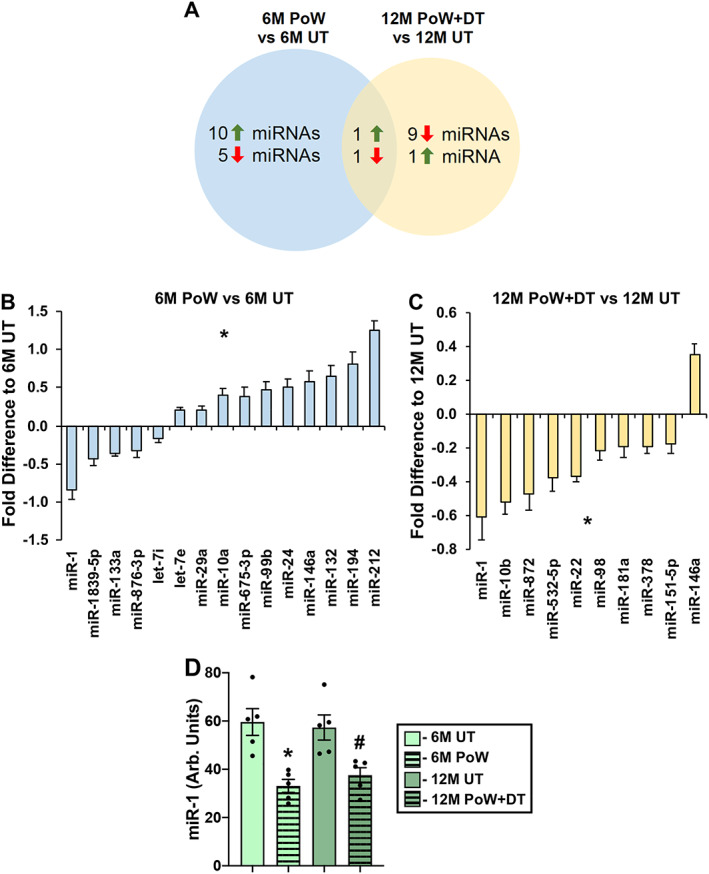
Quantitative analysis of miRNAs in gastrocnemius muscles with PoWeR training and detraining. (A) Venn diagram illustrating the number of miRNAs differentially regulated by PoWeR training (6M PoW versus untrained 6M UT), detraining (12M PoW + DT versus 12M UT), and differentially regulated miRNAs common to PoWeR trained and detrained. (B) Differentially regulated miRNAs in 6M PoW versus 6M UT, *P < 0.05. (C) Differentially regulated miRNAs in 12M PoW + DT versus 12M UT, *P < 0.05. (D) miR‐1 levels across all four groups, analysed via one‐way ANOVA. *P < 0.05 6M UT versus 6M PoW, # P < 0.05 12M DT versus 12M PoW + DT. n = 5 per time point.
Discussion
Utilizing PoWeR, a new model of voluntary murine exercise training, 7 we provide evidence on muscle and fibre type specificity in the training and detraining response. Although we did not measure muscle function, PoWeR results in classic cellular‐level changes associated with human resistance training, such as a faster‐to‐slower fibre type transition, 29 preferential hypertrophy of fast‐twitch muscle fibres (see review in Murach et al. 30 ), and myonuclear accretion (reviewed in Murach et al. 8 ), but is a voluntary ‘hybrid’ endurance/resistance stimulus that is much higher in volume and frequency than traditional resistance training, 31 so our results should be interpreted in this light. The current findings indicate that fibres of the gastrocnemius and plantaris muscles have larger CSA and myonuclear content with training and values similar to untrained mice following 6 months of detraining, regardless of fibre type. In our hands, the soleus did not detrain according to fibre type distribution, size, or myonuclear number after a 6‐month cessation from PoWeR. We speculate that the chronic activation pattern in this postural muscle, as well as the rapid increase in body weight that ensued during detraining, could explain why the soleus did not lose myonuclei, although there are likely other reasons that could account for this. Not all myonuclei are at their transcriptional maximum at rest, 22 and myonuclei are reportedly active in a pulsatile and potentially stochastic fashion. 32 , 33 If skeletal muscle maintains DNA dosage according to how frequently a nucleus is ‘turned on’, the recruitment pattern of soleus myonuclei during detraining was seemingly adequate to protect myonuclei from being targeted for removal. Furthermore, type 1 fibres of the soleus experienced myonuclear accretion in the absence of hypertrophy during PoWeR, which was likely a response to damage early in training. In muscles such as the gastrocnemius that demonstrated myonuclear loss with detraining, persistent repression of miR‐1 after training may serve as a muscle memory of previous adaptation that primes muscle for future hypertrophic growth.
To address our hypothesis that oxidative muscle fibres retain myonuclei following atrophy with detraining, we analysed type 2a muscle fibres of the gastrocnemius muscle. The gastrocnemius does not contain many type 1 fibres, but the deep ‘red’ region of the medial gastrocnemius is the most oxidative portion of any muscle in the mouse hindlimb and contains an abundance of highly oxidative type 2a fibres. 28 Contrary to our hypothesis, these oxidative (albeit fast‐twitch) muscle fibres in 12M PoW + DT mice had the same number of myonuclei as 12M UT mice; we therefore infer that the myonuclei gained during PoWeR in oxidative muscle fibres were lost upon detraining. Soleus type 2a muscle fibres did not show a detraining effect, with potential reasons discussed above, but it is also possible that type 2a fibres in the soleus are inherently different from gastrocnemius type 2a fibres. Even if detraining could be induced in the soleus, type 2a fibres may remain hyper‐nucleated per unit size long term; the same could be true for type 1 fibres. A study where post‐training cage activity and food consumption (and therefore body weight) are controlled to potentially facilitate soleus detraining and/or a longer detraining period may address these issues. Nevertheless, the current evidence suggests that oxidative muscle fibres contain fewer myonuclei when hypertrophy is reversed, comparable with glycolytic muscle fibres [Corrections added on 14 September 2020 after first online publication: The “6M DT” and “6M UT” mice in the preceding sentence have been corrected to “12M PoW + DT” and “12M UT” respectively.]. 7 , 34
Because there were more centrally located myonuclei in the soleus than in the plantaris muscle after 2 months of PoWeR training, 7 we hypothesized that muscle fibre damage in response to unaccustomed wheel activity may explain this observation. Following 6 days of unweighted wheel running, central myonuclei, as well as eMyHC+ fibres (suggestive of muscle damage), were apparent in the soleus. The HSA‐GFP mouse combined with EdU labelling revealed that central myonuclei are primarily GFP−/EdU+, so were derived from myonuclear donors, likely satellite cells. All centrally located GFP+ myonuclei were EdU−, so resident myonuclear repositioning also appears to contribute to the muscle fibre adaptive response to exercise. Type 1 fibres of the soleus experienced significant myonuclear accretion in the absence of hypertrophy after 2 months of PoWeR, raising the possibility that muscle fibre sarcolemmal damage and/or inflammation‐related signalling associated with unaccustomed exercise, as opposed to hypertrophy per se, creates a local cellular milieu that promotes satellite cell fusion to adult muscle fibres. 35 , 36 To this point, human investigations report increased myonuclear number after strenuous non‐hypertrophic activities such as multiday ultra‐endurance exercise, 37 intense endurance exercise training in athletes, 38 , 39 , 40 and traditional resistance training in older individuals, 41 the latter of which aligns with functional overload data in aged mice. 42 Rodent studies also show myonuclear accretion in the absence of hypertrophy with acute downhill running 36 and chronic voluntary wheel running, 43 as well as with induced genetic overexpression of Sirt1 specifically in muscle (which was not associated with increased muscle oxidative capacity) 44 and phosphatase and tensin homolog (PTEN) depletion/mTOR hyperactivation specifically in satellite cells. 45 It is possible that increased muscle oxidative potential triggers satellite cell fusion, 46 , 47 but the available evidence suggests that satellite cell fusion during running is not to support skeletal muscle metabolic adaptations because global oxidative shifts (e.g. fibre type transitions, succinate dehydrogenase and voltage‐dependent anion channels (VDAC) levels, muscle lipid content) occur to a similar extent with wheel running in the presence and absence of satellite cells. 48 , 49 , 50 Although type 1 fibres of the soleus were not larger after PoWeR, it is worth noting that they had the most central myonuclei and were significantly larger than untrained muscle fibres after 6 months of detraining; this delayed growth response could be related to the modest proportion of regenerating fibres and is worthy of further investigation.
Our laboratory and others report that depletion of satellite cells, the primary contributors of myonuclei, in adult sedentary mice generally does not result in a catastrophic nor appreciable loss of myonuclei across multiple muscles throughout the lifespan. 42 , 49 , 51 , 52 While alternative stem cell populations may contribute to adult muscle fibres in some capacity, 53 , 54 resident myonuclei may be permanent under non‐stressed conditions. By contrast, under conditions of dynamic remodelling such as exercise training, the myonuclei that are gained from satellite cells could be lost when hypertrophic adaptations are reversed. One avian study suggests that satellite cell‐derived myonuclei are the first to be targeted for removal with deloading. 11 Shortened myonuclei in PoWeR‐trained muscle fibres and elongated myonuclei in 6‐month detrained muscle fibres could be a response to a gain and loss of satellite cell‐derived myonuclei, and we captured the process of myonuclear migration. 55 , 56 Nuclear repositioning could be for the purpose of maintaining the ‘myonuclear domain’, 57 , 58 but could also be related to cytoskeletal rearrangement, 59 , 60 changes in innervation, 61 , 62 and/or blood vessel remodelling, 63 which may occur coincident with detraining. Nuclei are mechanosensors, 64 , 65 , 66 and myonuclear shape change could also be the consequence of alterations in transcriptional output. Alternatively, myonuclear shape change may be related to Pgc1α expression. 47 Regardless, the development of new genetic tools for specifically and conditionally labelling myonuclei in adult mice in vivo could provide further insight into how and which myonuclei migrate, as well as how and which myonuclei are removed during detraining. 19
MicroRNAs function epigenetically because they alter cellular transcript levels and/or protein abundance without affecting the genetic code. 67 In skeletal muscle, miR‐1 is highly abundant, represses pro‐growth processes, and is robustly and progressively down‐regulated in response to a hypertrophic stimulus 16 , 18 , 68 ; miR‐1 down‐regulation also occurs in PoWeR‐trained muscle as shown here. Others have found that atrophied muscles regrow more rapidly if previously having undergone hypertrophy. 4 , 12 The mechanism for persistently reduced miR‐1 after detraining is unclear, but if miR‐1 functions as a molecular ‘brake’ that tunes hypertrophic signalling, 16 , 17 , 68 it would be easier to overcome this attenuated brake in the detrained state when a second wave of hypertrophic signalling is encountered (i.e. retraining). Thus, lower miR‐1 with detraining could represent a ‘memory’ of previous adaptation that primes the muscle molecular machinery for efficient regrowth.
Anlaysis of human training/detraining data 69 indicates that myonuclei gained during hypertrophy are lost upon detraining, 70 which is congruent with recent training/detraining studies conducted in older individuals by independent laboratories, 34 , 71 as well as our current and previous data in mice. 7 With mounting evidence for myonuclear loss following atrophy, 8 , 72 we propose a model where an elevated number of myonuclei could potentially serve as one component of a muscle memory of previous hypertrophy in the short term (e.g. <3 months in mice), 2 , 4 but that alternative epigenetic mechanisms such as myonuclear DNA methylation, histone modifications, or miRNA expression could explain muscle memory in the long‐term after myonuclear number has stabilized 12 , 70 , 73 , 74 (Figure 9). Furthermore, myoblasts are shown to have a long‐term epigenetic memory of previous exposure to stress in vitro 75 , so satellite cells could theoretically contribute to muscle memory in myofibers via recently identified fusion‐independent mechanisms in vivo. 76 Future studies may employ newly developed voluntary 77 or involuntary resistance training paradigms (exercise dosage can be controlled for in the latter) 78 , 79 for mice that can complement PoWeR (recently reviewed here 31 ) and will aim to address how myonuclei are removed during detraining, the role of myonuclear translocation during exercise adaptation, whether epigenetic changes could explain myonuclear shape transformations with training and detraining, which exercise‐induced epigenetic alterations to myonuclei are permanent, and how miR‐1 could facilitate retraining adaptations.
Figure 9.
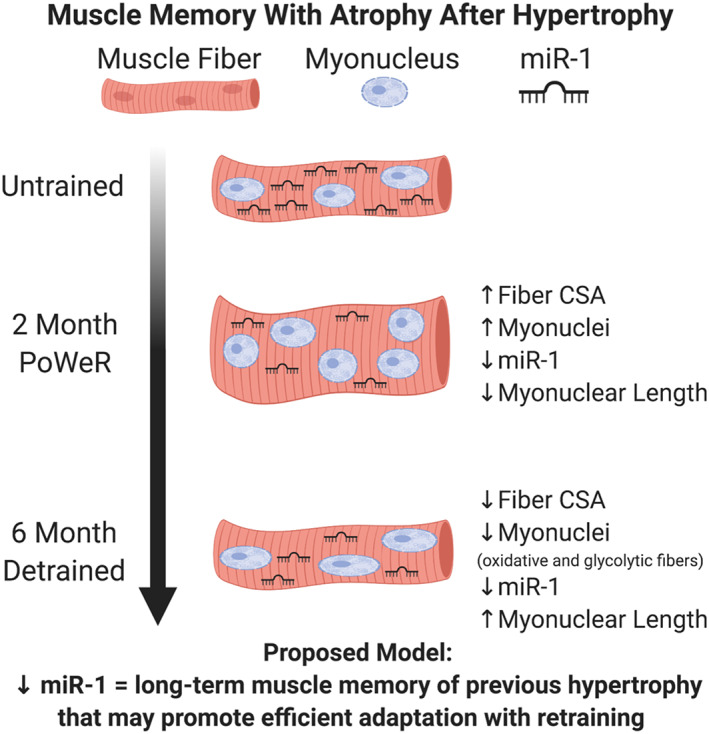
Summary figure: a proposed model for a cellular muscle memory of previous hypertrophy. With 2 months of PoWeR training in the plantaris (see Dungan et al. 2019) and gastrocnemius (type 2a fibres in the current investigation), myonuclear number increases simultaneous with hypertrophy, and miR‐1 levels are lower (measured in the gastrocnemius). Myonuclei are also shorter after PoWeR training in the gastrocnemius. Following 6 months of detraining after PoWeR, muscle fibre size and myonuclear number return to untrained levels in the plantaris and gastrocnemius, miR‐1 levels remain lower (measured in gastrocnemius), and myonuclei are longer (gastrocnemius and plantaris). We hypothesize that lower miR‐1 levels during PoWeR facilitate hypertrophy by promoting pro‐growth gene expression, while persistently lower levels with detraining could allow for more efficient regrowth with retraining.
Author contributions
K. A. M., J. J. M., C. A. P., and C. M. D. conceived the study. K. A. M., C. M. D., C. B. M., C. J. Z., K. K. F., and S. R. J. performed the experiments. K. A. M. and C. M. D. analysed the data and wrote the manuscript. All authors reviewed the manuscript.
Conflict of interest
The authors have no conflicts of interest to declare.
Supporting information
Figure S1 Muscle mass, fiber type, size, and myonuclear number after 3 months of detraining in the soleus. (A) Soleus muscle mass in PoWeR trained (6M PoW) versus 3 month detrained (9M PoW+DT). (B&C) Repesentative IHC images from 6M PoW and 9M PoW+DT showing dystrophin (white), Type 1 (pink), Type 2a (green), unstained Type 2x and/or 2b‐containing fibers (black), and DAPI (blue). (D) Fiber type (FT) distribution. (E) Fiber cross sectional area (CSA). (F&G) Representative images of longitudinal single muscle fibers from (F) 6M PoW and (G) 9M PoW+DT. (H) Single fiber analysis of myonuclear number.
Data S1 Raw data for quantitative single molecule direct digital detection (NanoString) analysis of miRNAs in the gastrocnemius muscle
Acknowledgements
Funding for this study was provided by National Institutes of Health grants AR071753 and AG063994 to K. A. M., AR060701 and AG049806 to C. A. P. and J. J. M., AG046920 to C. A. P., and T32 AG057461 to C. M. D. The study design figure and summary figure were generated using BioRender. The authors certify that they comply with the ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: update 2017. 80
Murach K. A., Mobley C. B., Zdunek C. J., Frick K. K., Jones S. R., McCarthy J. J., Peterson C. A., and Dungan C. M. (2020) Muscle memory: myonuclear accretion, maintenance, morphology, and miRNA levels with training and detraining in adult mice, Journal of Cachexia, Sarcopenia and Muscle, 11, 1705–1722, doi: 10.1002/jcsm.12617
References
- 1. Gundersen K. Muscle memory and a new cellular model for muscle atrophy and hypertrophy. J Exp Biol 2016;219:235–242. [DOI] [PubMed] [Google Scholar]
- 2. Bruusgaard JC, Johansen IB, Egner IM, Rana ZA, Gundersen K. Myonuclei acquired by overload exercise precede hypertrophy and are not lost on detraining. Proc Natl Acad Sci U S A 2010;107:15111–15116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwartz LM. Skeletal muscles do not undergo apoptosis during either atrophy or programmed cell death‐revisiting the myonuclear domain hypothesis. Front Physiol 2019;9:1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Egner IM, Bruusgaard JC, Eftestøl E, Gundersen K. A cellular memory mechanism aids overload hypertrophy in muscle long after an episodic exposure to anabolic steroids. J Physiol 2013;591:6221–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwartz LM, Brown C, McLaughlin K, Smith W, Bigelow C. The myonuclear domain is not maintained in skeletal muscle during either atrophy or programmed cell death. Ame J Physiology‐Cell Physi 2016;311:C607–C615. [DOI] [PubMed] [Google Scholar]
- 6. Lee H, Kim K, Kim B, Shin J, Rajan S, Wu J, et al. A cellular mechanism of muscle memory facilitates mitochondrial remodelling following resistance training. J Physiol 2018;596:4413–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dungan CM, Murach KA, Frick KK, Jones SR, Crow SE, Englund DA, et al. Elevated myonuclear density during skeletal muscle hypertrophy in response to training is reversed during detraining. Ame J Physiology‐Cell Physi 2019;316:C649–C654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murach KA, Fry CS, Kirby TJ, Jackson JR, Lee JD, White SH, et al. Starring or supporting role? Satellite cells and skeletal muscle fiber size regulation. Phys Ther 2018;33:26–38, PubMed PMID: 29212890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dickinson JM, Lee JD, Sullivan BE, Harber MP, Trappe SW, Trappe TA. A new method to study in vivo protein synthesis in slow‐ and fast‐twitch muscle fibers and initial measurements in humans. J Appl Physiol 2010;108:1410–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tseng BS, Kasper CE, Edgerton VR. Cytoplasm‐to‐myonucleus ratios and succinate dehydrogenase activities in adult rat slow and fast muscle fibers. Cell Tissue Res 1994;275:39–49. [DOI] [PubMed] [Google Scholar]
- 11. Siu PM, Pistilli EE, Butler DC, Alway SE. Aging influences cellular and molecular responses of apoptosis to skeletal muscle unloading. Ame J Physiology‐Cell Physi 2005;288:C338–C349. [DOI] [PubMed] [Google Scholar]
- 12. Seaborne RA, Strauss J, Cocks M, Shepherd S, O'Brien TD, van Someren KA, et al. Human skeletal muscle possesses an epigenetic memory of hypertrophy. Sci Rep 2018;8:1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vechetti IJ, Wen Y, Chaillou T, Murach KA, Alimov AP, Figueiredo VC, et al. Life‐long reduction in myomiR expression does not adversely affect skeletal muscle morphology. Sci Rep 2019;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oikawa S, Lee M, Akimoto T. Conditional deletion of dicer in adult mice impairs skeletal muscle regeneration. Int J Mol Sci 2019;20:5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res 2011;39:5692–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCarthy JJ, Esser KA. MicroRNA‐1 and microRNA‐133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol 2007;102:306–313. [DOI] [PubMed] [Google Scholar]
- 17. Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, Russo MA, et al. Reciprocal regulation of microRNA‐1 and IGF‐1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation 2009;120:2377–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaillou T, Lee JD, England JH, Esser KA, McCarthy JJ. Time course of gene expression during mouse skeletal muscle hypertrophy. J Appl Physiol 2013;115:1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iwata M, Englund DA, Wen Y, Dungan CM, Murach KA, Vechetti IJ, et al. A novel tetracycline‐responsive transgenic mouse strain for skeletal muscle‐specific gene expression. Skeletal muscle 2018;8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, et al. Defining the epithelial stem cell niche in skin. Science 2004;303:359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murach KA, White SH, Wen Y, Ho A, Dupont‐Versteegden EE, McCarthy JJ, et al. Differential requirement for satellite cells during overload‐induced muscle hypertrophy in growing versus mature mice. Skeletal Muscle 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirby TJ, Patel RM, McClintock TS, Dupont‐Versteegden EE, Peterson CA, McCarthy JJ. Myonuclear transcription is responsive to mechanical load and DNA content but uncoupled from cell size during hypertrophy. Mol Biol Cell 2016;27:788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brack AS, Bildsoe H, Hughes SM. Evidence that satellite cell decrement contributes to preferential decline in nuclear number from large fibres during murine age‐related muscle atrophy. J Cell Sci 2005;118:4813–4821. [DOI] [PubMed] [Google Scholar]
- 24. McCarthy JJ, Mula J, Miyazaki M, Erfani R, Garrison K, Farooqui AB, et al. Effective fiber hypertrophy in satellite cell‐depleted skeletal muscle. Development 2011;138:3657–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wen Y, Murach KA, Vechetti IJ Jr, Fry CS, Vickery CD, Peterson CA, et al. MyoVision: Software for automated high‐content analysis of skeletal muscle immunohistochemistry. J Appl Physiol 2018;124:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Soffe Z, Radley‐Crabb HG, McMahon C, Grounds MD, Shavlakadze T. Effects of loaded voluntary wheel exercise on performance and muscle hypertrophy in young and old male C57Bl/6J mice. Scand J Med Sci Sports 2016;26:172–188. [DOI] [PubMed] [Google Scholar]
- 27. Irintchev A, Wernig A. Muscle damage and repair in voluntarily running mice: strain and muscle differences. Cell Tissue Res 1987;249:509–521. [DOI] [PubMed] [Google Scholar]
- 28. Bloemberg D, Quadrilatero J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS ONE 2012;7:e35273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Williamson DL, Gallagher PM, Carroll CC, Raue U, Trappe SW. Reduction in hybrid single muscle fiber proportions with resistance training in humans. J Appl Physiol 2001;91:1955–1961. [DOI] [PubMed] [Google Scholar]
- 30. Murach KA, Raue U, Wilkerson BS, Minchev K, Jemiolo B, Bagley RJ, et al. Fiber type‐specific gene expression with taper in competitive distance runners. PLoS ONE 2014;9:e108547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murach KA, McCarthy JJ, Peterson CA, Dungan CM. Making mice mighty: Recent Advances in translational models of load‐induced muscle hypertrophy. Journal of Applied Physiology. 2020:In Press. PMID: 32673155. 10.1152/japplphysiol.00319.2020 [DOI] [PMC free article] [PubMed]
- 32. Newlands S, Levitt LK, Robinson CS, Karpf AC, Hodgson VR, Wade RP, et al. Transcription occurs in pulses in muscle fibers. Genes Dev 1998;12:2748–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cutler AA, Jackson JB, Corbett AH, Pavlath GK. Non‐equivalence of nuclear import among nuclei in multinucleated skeletal muscle cells. J Cell Sci 2018;131:jcs207670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Snijders T, Leenders M, de Groot L, van Loon LJ, Verdijk LB. Muscle mass and strength gains following 6 months of resistance type exercise training are only partly preserved within one year with autonomous exercise continuation in older adults. Exp Gerontol 2019;121:71–78. [DOI] [PubMed] [Google Scholar]
- 35. Murach KA, Englund DA, Dupont‐Versteegden EE, McCarthy JJ, Peterson CA. Myonuclear domain flexibility challenges rigid assumptions on satellite cell contribution to skeletal muscle fiber hypertrophy. Front Physiol 2018;9:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith HK, Maxwell L, Rodgers CD, McKee NH, Plyley MJ. Exercise‐enhanced satellite cell proliferation and new myonuclear accretion in rat skeletal muscle. J Appl Physiol 2001;90:1407–1414. [DOI] [PubMed] [Google Scholar]
- 37. Konopka AR, Castor WM, Wolff CA, Musci RV, Reid JJ, Laurin JL, et al. Skeletal muscle mitochondrial protein synthesis and respiration in response to the energetic stress of an ultra‐endurance race. J Appl Physiol 2017;123:1516–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frese S, Ruebner M, Suhr F, Konou TM, Tappe KA, Toigo M, et al. Long‐term endurance exercise in humans stimulates cell fusion of myoblasts along with fusogenic endogenous retroviral genes in vivo. PLoS ONE 2015;10:e0132099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frese S, Valdivieso P, Flück M, Jaecker V, Harms S, Konou T, et al. Expression of metabolic and myogenic factors during two competitive seasons in elite junior cyclists. Deutsche Zeitsc für Sportm 2016;67. [Google Scholar]
- 40. McKenzie AI, D'Lugos AC, Saunders MJ, Gworek KD, Luden ND. Fiber type‐specific satellite cell content in cyclists following heavy training with carbohydrate and carbohydrate‐protein supplementation. Front Physiol 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moro T, Brightwell CR, Volpi E, Rasmussen B, Fry CS. Resistance exercise training promotes fiber type‐specific myonuclear adaptations in older adults. J Appl Physiol 2020;128:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee JD, Fry CS, Mula J, Kirby TJ, Jackson JR, Liu F, et al. Aged muscle demonstrates fiber‐type adaptations in response to mechanical overload, in the absence of myofiber hypertrophy, independent of satellite cell abundance. J Gerontol A Biol Sci Med Sci 2016;71:461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kurosaka M, Naito H, Ogura Y, Kojima A, Goto K, Katamoto S. Effects of voluntary wheel running on satellite cells in the rat plantaris muscle. J Sports Sci Med 2009;8:51–57. [PMC free article] [PubMed] [Google Scholar]
- 44. Ross JA, Levy Y, Svensson K, Philp A, Schenk S, Ochala J. SIRT1 regulates nuclear number and domain size in skeletal muscle fibers. J Cell Physiol 2018;233:7157–7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yue F, Bi P, Wang C, Shan T, Nie Y, Ratliff TL, et al. Pten is necessary for the quiescence and maintenance of adult muscle stem cells. Nat Commun 2017;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Omairi S, Matsakas A, Degens H, Kretz O, Hansson KA, Solbra AV, et al. Enhanced exercise and regenerative capacity in a mouse model that violates size constraints of oxidative muscle fibres. Elife 2016;05:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ross JA, Pearson A, Levy Y, Cardel B, Handschin C, Ochala J. Exploring the role of PGC‐1α in defining nuclear organisation in skeletal muscle fibres. J Cell Physiol 2017;232:1270–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jackson JR, Kirby TJ, Fry CS, Cooper RL, McCarthy JJ, Peterson CA, et al. Reduced voluntary running performance is associated with impaired coordination as a result of muscle satellite cell depletion in adult mice. Skeletal Muscle 2015;5:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murach KA, Confides AL, Ho A, Jackson JR, Ghazala LS, Peterson CA, et al. Depletion of Pax7+ satellite cells does not affect diaphragm adaptations to running in young or aged mice. J Physiol 2017;595:6299–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Englund DA, Murach KA, Dungan CM, Figueiredo VC, Vechetti IJ Jr, Dupont‐Versteegden EE, et al. Depletion of resident muscle stem cells negatively impacts running volume, physical function and muscle hypertrophy in response to lifelong physical activity. Ame J Physiology‐Cell Physi 2020;318:C1178–C1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fry CS, Lee JD, Mula J, Kirby TJ, Jackson JR, Liu F, et al. Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regenerative capacity without affecting sarcopenia. Nat Med 2015;21:76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Keefe AC, Lawson JA, Flygare SD, Fox ZD, Colasanto MP, Mathew SJ, et al. Muscle stem cells contribute to myofibres in sedentary adult mice. Nat Commun 2015;6:7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Strömberg A, Jansson M, Fischer H, Rullman E, Hägglund H, Gustafsson T. Bone marrow derived cells in adult skeletal muscle tissue in humans. Skeletal Muscle 2013;3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu N, Garry GA, Li S, Bezprozvannaya S, Sanchez‐Ortiz E, Chen B, et al. A Twist2‐dependent progenitor cell contributes to adult skeletal muscle. Nat Cell Biol 2017;19:202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Folker ES, Schulman VK, Baylies MK. Translocating myonuclei have distinct leading and lagging edges that require kinesin and dynein. Development 2014;141:355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Azevedo M, Baylies MK. Getting into position: Nuclear movement in muscle cells. Trends Cell Biol 2020;30:303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pavlath GK, Rich K, Webster SG, Blau HM. Localization of muscle gene products in nuclear domains. Nature 1989;337:570–573. [DOI] [PubMed] [Google Scholar]
- 58. Taylor‐Weiner H, Grigsby CL, Ferreira DM, Dias JM, Stevens MM, Ruas JL, et al. Modeling the transport of nuclear proteins along single skeletal muscle cells. Proc Natl Acad Sci 2020;117:2978–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Capetanaki Y, Bloch RJ, Kouloumenta A, Mavroidis M, Psarras S. Muscle intermediate filaments and their links to membranes and membranous organelles. Exp Cell Res 2007;313:2063–2076. [DOI] [PubMed] [Google Scholar]
- 60. Staszewska I, Fischer I, Wiche G. Plectin isoform 1‐dependent nuclear docking of desmin networks affects myonuclear architecture and expression of mechanotransducers. Hum Mol Genet 2015;24:7373–7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc Natl Acad Sci 2005;102:4359–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Daou N, Hassani M, Matos E, de Castro GS, Costa RGF, Seelaender M, et al. Displaced myonuclei in cancer cachexia suggest altered innervation. Int J Mol Sci 2020;21:1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ralston E, Lu Z, Biscocho N, Soumaka E, Mavroidis M, Prats C, et al. Blood vessels and desmin control the positioning of nuclei in skeletal muscle fibers. J Cell Physiol 2006;209:874–882. [DOI] [PubMed] [Google Scholar]
- 64. Kirby TJ, Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol 2018;20:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, et al. Transcription upregulation via force‐induced direct stretching of chromatin. Nat Mater 2016;15:1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kirby TJ, Lammerding J. Cell mechanotransduction: stretch to express. Nat Mater 2016;15:1227–1229. [DOI] [PubMed] [Google Scholar]
- 67. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010;79:351–379. [DOI] [PubMed] [Google Scholar]
- 68. Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Ame J Physio‐Endocr Metab 2008;295:E1333–E1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Psilander N, Eftestøl E, Cumming KT, Juvkam I, Ekblom MM, Sunding K, et al. Effects of training, detraining, and retraining on strength, hypertrophy, and myonuclear number in human skeletal muscle. J Appl Physiol 2019;126:1636–1645. [DOI] [PubMed] [Google Scholar]
- 70. Murach KA, Dungan CM, Dupont‐Versteegden EE, McCarthy JJ, Peterson CA. “Muscle memory” not mediated by myonuclear number?: secondary analysis of human detraining data. J Appl Physiol 2019;127:1814–1816. [DOI] [PubMed] [Google Scholar]
- 71. Blocquiaux S, Gorski T, van Roie E, Ramaekers M, van Thienen R, Nielens H, et al. The effect of resistance training, detraining and retraining on muscle strength and power, myofibre size, satellite cells and myonuclei in older men. Exp Gerontol 2020;110860. [DOI] [PubMed] [Google Scholar]
- 72. Snijders T, Aussieker T, Holwerda A, Parise G, van Loon L, Verdijk LB. The concept of skeletal muscle memory: evidence from animal and human studies. Acta Physiolo 2020;229:e13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Turner DC, Seaborne RA, Sharples AP. Comparative transcriptome and methylome analysis in human skeletal muscle anabolism, hypertrophy and epigenetic memory. Sci Rep 2019;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. von Walden F, Rea M, Mobley CB, Fondufe‐Mittendorf Y, McCarthy JJ, Peterson CA, et al. The myonuclear DNA methylome in response to an acute hypertrophic stimulus. Epigenetics 2020;1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sharples AP, Polydorou I, Hughes DC, Owens DJ, Hughes TM, Stewart CE. Skeletal muscle cells possess a ‘memory’ of acute early life TNF‐α exposure: role of epigenetic adaptation. Biogerontology 2016;17:603–617. [DOI] [PubMed] [Google Scholar]
- 76. Murach KA, Vechetti IJ Jr, van Pelt DW, Crow SE, Dungan CM, Figueiredo VC, et al. Fusion‐independent satellite cell communication to muscle fibers during load‐induced hypertrophy. Function 2020;1(1):zqaa009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cui D, Drake JC, Wilson RJ, Shute RJ, Lewellen B, Zhang M, et al. A novel voluntary weightlifting model in mice promotes muscle adaptation and insulin sensitivity with simultaneous enhancement of autophagy and mTOR pathway. FASEB J 2020;34:7330–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Graber TG, Fandrey KR, Thompson LV. Novel individualized power training protocol preserves physical function in adult and older mice. GeroScience 2019;41:165–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hardee JP, Mangum JE, Gao S, Sato S, Hetzler KL, Puppa MJ, et al. Eccentric contraction‐induced myofiber growth in tumor‐bearing mice. J Appl Physiol 2016;120:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. von Haehling S, Morley JE, Coats AJ, Anker SD. Ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: update 2019. J Cachexia Sarcopenia Muscle 2019;10:1143–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Muscle mass, fiber type, size, and myonuclear number after 3 months of detraining in the soleus. (A) Soleus muscle mass in PoWeR trained (6M PoW) versus 3 month detrained (9M PoW+DT). (B&C) Repesentative IHC images from 6M PoW and 9M PoW+DT showing dystrophin (white), Type 1 (pink), Type 2a (green), unstained Type 2x and/or 2b‐containing fibers (black), and DAPI (blue). (D) Fiber type (FT) distribution. (E) Fiber cross sectional area (CSA). (F&G) Representative images of longitudinal single muscle fibers from (F) 6M PoW and (G) 9M PoW+DT. (H) Single fiber analysis of myonuclear number.
Data S1 Raw data for quantitative single molecule direct digital detection (NanoString) analysis of miRNAs in the gastrocnemius muscle


