Abstract
Most mammalian cytoplasmic tRNAs contain ribothymidine (T) and pseudouridine (Ψ) at positions 54 and 55, respectively. However, some tRNAs contain Ψ at both positions. Several Ψ54-containing tRNAs function as primers in retroviral DNA synthesis. The Ψ54 of these tRNAs is produced by PUS10, which can also synthesize Ψ55. Two other enzymes, TRUB1 and TRUB2, can also produce Ψ55. By nearest-neighbor analyses of tRNAs treated with recombinant proteins and subcellular extracts of wild-type and specific Ψ55 synthase knockdown cells, we determined that while TRUB1, PUS10, and TRUB2 all have tRNA Ψ55 synthase activities, they have different tRNA structural requirements. Moreover, these activities are primarily present in the nucleus, cytoplasm, and mitochondria, respectively, suggesting a compartmentalization of Ψ55 synthase activity. TRUB1 produces the Ψ55 of most elongator tRNAs, but cytoplasmic PUS10 produces both Ψs of the tRNAs with Ψ54Ψ55. The nuclear isoform of PUS10 is catalytically inactive and specifically binds the unmodified U54U55 versions of Ψ54Ψ55-containing tRNAs, as well as the A54U55-containing tRNAiMet. This binding inhibits TRUB1-mediated U55 to Ψ55 conversion in the nucleus. Consequently, the U54U55 of Ψ54Ψ55-containing tRNAs are modified by the cytoplasmic PUS10. Nuclear PUS10 does not bind the U55 versions of T54Ψ55- and A54Ψ55-containing elongator tRNAs. Therefore, TRUB1 is able to produce Ψ55 in these tRNAs. In summary, the tRNA Ψ55 synthase activities of TRUB1 and PUS10 are not redundant but rather are compartmentalized and act on different sets of tRNAs. The significance of this compartmentalization needs further study.
Keywords: tRNA modification, pseudouridine synthase, PUS10 isoforms, subcellular localization, Pus4, retroviral primer
INTRODUCTION
The “TΨC” or “common” arm of a typical tRNA is a 17-base stem–loop structure at position 49–65, with a consensus “GUUCRANUC” sequence at position 53–61, the “UUCRANU” of which forms a loop capping a five base pair stem (Gupta 1985; Juhling et al. 2009). The eukaryotic initiator tRNAs and some elongator tRNAs differ from this consensus. TΨC refers to T (ribothymidine or 5-methyluridine) and Ψ (pseudouridine) modifications of U54 and U55, respectively, followed by C56. The Ψ55 is present in nearly all tRNAs, again with the exception of eukaryotic initiators and a few others. T or a further modification of T is observed at position 54 in most bacterial and eukaryotic tRNAs, but rarely in archaeal tRNAs. Most archaeal tRNAs, in contrast, especially those of Euryarchaeota, contain Ψ54 or a modified Ψ54 instead of T54 (Gupta 1984, 1985, 1986; Juhling et al. 2009; Blaby et al. 2011; Chatterjee et al. 2012).
The tRNA methyltransferase TrmA/Trm2/TRMT2 and tRNA Ψ synthase TruB/Pus4 produce T54 and Ψ55, respectively, in most bacterial and eukaryotic tRNAs (Ny and Bjork 1980; Nurse et al. 1995; Becker et al. 1997a; Nordlund et al. 2000). In Archaea, Pus10 can produce both Ψ54 and Ψ55 (Gurha and Gupta 2008; Joardar et al. 2013). Pus10 is distinct from the TruB family of Ψ synthases (Watanabe and Gray 2000; Mueller and Ferre-D'Amare 2009; Rintala-Dempsey and Kothe 2017). Those Archaea that do have T54-containing tRNAs utilize an ortholog of bacterial RumA, an rRNA methyltransferase, instead of the bacterial TrmA ortholog (Urbonavicius et al. 2008). Pus10 produces only Ψ55 in these T54-containing archaeal tRNAs (Roovers et al. 2006; Gurha and Gupta 2008; Joardar et al. 2013). Humans have two paralogs of TruB, TRUB1, and TRUB2, that can produce Ψ55 (Zucchini et al. 2003; Rintala-Dempsey and Kothe 2017). TRUB1 is present in both the nucleus and cytoplasm, but its activity is predominantly in the nucleus (Safra et al. 2017). TRUB2 is present in the mitochondria and is suggested to produce Ψ55 in all four human mitochondrial tRNAs (out of 22 total) with this modification (Suzuki and Suzuki 2014; Antonicka et al. 2017). Humans also have two paralogs of TrmA, TRMT2A, and TRMT2B, located in the nucleus and mitochondria, respectively, that can produce T54 (Chang et al. 2019; de Crecy-Lagard et al. 2019).
Pus10 is present in most eukaryotes, but not in bacteria and dikaryon fungi, including yeast (Watanabe and Gray 2000; Gurha and Gupta 2008; Fitzek et al. 2018). This coincides with the presence of Ψ54 instead of T54 in some tRNAs of animals, such as tRNATrp and tRNAGln, and the absence of Ψ54 in the tRNAs of bacteria and yeast (Juhling et al. 2009). Several Ψ54-containing tRNAs function as primers in retroviral DNA synthesis (Mak and Kleiman 1997; Deogharia et al. 2019). Previously, we showed that PUS10 produces Ψ54 in these tRNAs (Deogharia et al. 2019). Maximum Ψ54 activity was observed when the consensus sequence of the TΨC arm was GUUCAm1AAUC (m1A is 1-methyladenosine and the underlined U is converted to Ψ54). Although human PUS10 is present in both the nucleus and cytoplasm, it is the cytoplasmic PUS10 that produces Ψ54. The nuclear PUS10 is involved in apoptosis and translocates to the mitochondria in TRAIL-treated cells (Jana et al. 2017). Nuclear PUS10 is also involved in miRNA processing and this activity is independent of tRNA pseudouridylation by cytoplasmic PUS10 (Song et al. 2020). Crystal structure of human PUS10 shows that this protein has two domains: an amino-terminal (Met1-His285) THUMP-containing domain and a carboxy-terminal (Gly286–Asp528) Ψ synthase domain (McCleverty et al. 2007). The carboxy-terminal domain has the full set of conserved pseudouridine (Ψ) synthase active site residues. Structural alignment of human PUS10 and Pus10 of Methanocaldococcus jannaschii, an archaeon, showed nearly superimposable catalytic domains (Joardar et al. 2013). However, the amino-terminal domain of eukaryotic Pus10 is much larger than that of archaeal Pus10 (McCleverty et al. 2007; Fitzek et al. 2018).
The presence of both TRUB and PUS10 in humans raises the question of whether, in vivo, their Ψ55-synthase activities are redundant. We show here that all three human recombinant proteins, TRUB1, PUS10, and TRUB2, have Ψ55 synthase activities, but differ slightly in their tRNA structural requirements. These differences are specifically reflected in the Ψ55 synthase activities of the nuclear, cytoplasmic, and mitochondrial extracts, respectively. Furthermore, individual knockdown of TRUB1, PUS10, and TRUB2 reduces the Ψ55 synthase activities of the nuclear, cytoplasmic, and mitochondrial extracts, respectively, suggesting compartmentalization of the activities of the three proteins. The Ψ55 of tRNAs that contain T54Ψ55 are produced by TRUB1 in the nucleus, but the two consecutive Ψs of the tRNAs that contain Ψ54Ψ55 are both produced by cytoplasmic PUS10. We also show that catalytically inactive nuclear PUS10 specifically binds the unmodified U54U55 versions of these Ψ54Ψ55-containing tRNAs and inhibits TRUB1-mediated Ψ55 production in these tRNAs in the nucleus. PUS10 binding can also explain why U55 remains unmodified in initiator tRNA but Ψ55 appears in certain tRNAAla, although both tRNAs contain A54 and U55, and in vitro TRUB1 can modify U55 in both cases.
RESULTS
Recombinant human TRUB1, TRUB2, and PUS10 have tRNA Ψ55 synthase activities that depend on specific structural requirements in tRNA
We showed previously that recombinant human PUS10 prepared from E. coli did not have any tRNA Ψ synthase activity, but that recombinant human PUS10 from SF9 insect cells (called i-PUS10 here) did show both Ψ54 and Ψ55 synthase activities (Deogharia et al. 2019). In vitro, i-PUS10 produced Ψ55 even in those tRNAs that are known to have T54. Here, we used i-PUS10 and E. coli–derived recombinant TRUB1 and TRUB2 to compare their Ψ55 synthase activities. These activities were tested on [α-32P]CTP-labeled transcripts of human tRNATrp and tRNAAla (sequences of tRNAs are shown in Supplemental Fig. S1), which served as representatives of the tRNAs that normally contain Ψ54 and T54, respectively. Both these tRNAs have Ψ55. RNase T2 digests of the products showed radioactive Ψp in all cases (Fig. 1A), suggesting that all three recombinant proteins can produce Ψ in both tRNA transcripts. RNase T2 digests RNAs to ribonucleoside-3′-monophosphates (Np). As a result, in this nearest-neighbor analysis, the labeled 5′-phosphate of the [α-32P]-labeled NTP used to produce the transcript is transferred to the 3′ side of the preceding residue. Therefore, labeled U (or modified U) spots in the RNase T2 digests of [α-32P]CTP-labeled transcripts would only be derived from the U of UC sequences. Although UC is present in several places in the transcripts, we believe that the Ψp observed in Figure 1A is derived from U55, because TRUB1 and TRUB2 are known to produce only Ψ55 and PUS10 only produces Ψ54 and Ψ55 (Zucchini et al. 2003; Rintala-Dempsey and Kothe 2017; Deogharia et al. 2019).
FIGURE 1.
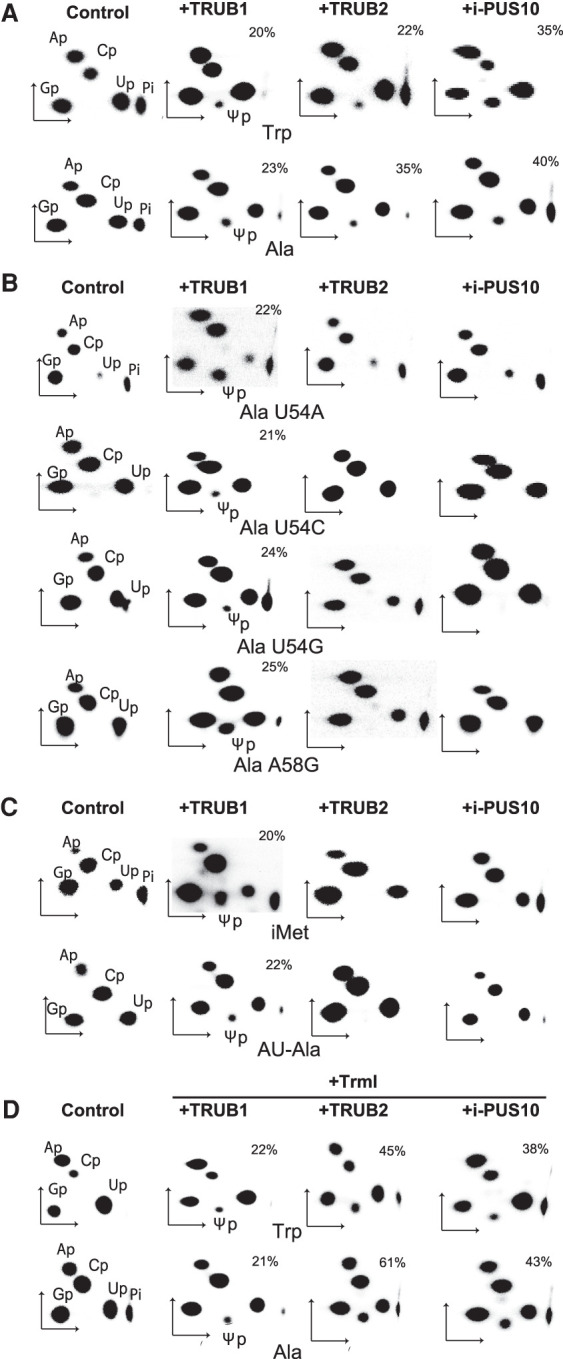
The tRNA Ψ55 synthase activities of recombinant proteins. Different [α-32P]CTP-labeled tRNA transcripts (indicated below panels) were incubated with recombinant TRUB1, TRUB2, and i-PUS10 (indicated above panels). Controls were untreated tRNAs. Purified RNA products were digested with RNase T2, and the digests were separated by 2D-TLC and phosphorimaged. As discussed in the text, Ψ in all cases is derived from position 55 of the tRNAs. The mole Ψ/mole tRNA ratio is indicated as a percentage in the panels where Ψ is observed. Representative autoradiograms of repeat experiments are shown. The labels indicate the 3′-monophosphates of corresponding ribonucleosides. Inorganic phosphate is labeled as Pi, which is cropped off in some panels. (A) Treatment of tRNATrp and tRNAAla. (B) Treatment of U54 and A58 mutants of tRNAAla. (C) Treatment of tRNAiMet and tRNAAU-Ala. (D) The tRNATrp and tRNAAla were first treated with TrmI to produce m1A58 and then treated with the three recombinant proteins.
Bacterial TruB and its yeast ortholog Pus4 require the presence of U54 and A58, which form a reverse Hoogsteen pair for their Ψ55 synthase activities (Becker et al. 1997b; Gu et al. 1998). Therefore, we tested the requirements of these residues for all three human proteins by mutating U54 of tRNAAla to A, C, or G, and A58 to G (Fig. 1B). Ψ55 was produced by TRUB1 in all cases. TRUB2 and i-PUS10 did not produce any Ψ55 in these mutants. These results suggest that the Ψ55 activities of TRUB2 and i-PUS10 depend on the presence of U54·A58 reverse Hoogsteen pair, but that of TRUB1 does not.
Eukaryotic initiator tRNAs (tRNAiMet) and several eukaryotic AGC anticodon-containing tRNAAla (called tRNAAU-Ala here) contain A54 instead of U54 (see tRNAAla and tRNAAU-Ala in Supplemental Fig. S1; Juhling et al. 2009; Westhof et al. 2020). Although the U55 of human tRNAiMet remains unmodified, that of tRNAAU-Ala is modified to Ψ in the cell (Bunn and Mathews 1987; Juhling et al. 2009). Recombinant TRUB1 could produce Ψ55 in both of these tRNAs (Fig. 1C), as was observed for the U54A mutant of tRNAAla (Fig. 1B). This confirms that the Ψ55 activity of TRUB1 does not specifically require U54. TRUB2 and i-PUS10 did not produce Ψ55 in either of these two tRNAs (Fig. 1C), confirming that both proteins require U54 to produce Ψ55.
Previously, we showed that Ψ54 synthesis by i-PUS10 was substantially enhanced when the transcript contained m1A58 (Deogharia et al. 2019). To determine whether Ψ55 synthesis by i-PUS10, TRUB1, and TRUB2 was also affected by the presence of m1A58, we treated tRNATrp and tRNAAla transcripts with TrmI to produce m1A58 as before (Deogharia et al. 2019) and then treated them independently with the three proteins (Fig. 1D). Only TRUB2 showed increased Ψ55 production in the presence of m1A58 (compare Fig. 1A with 1D). Unlike the Ψ54 activity of i-PUS10, its Ψ55 activity was not affected by m1A58.
Overall these results suggest that unlike TRUB1, the Ψ55 activity of both i-PUS10 and TRUB2 requires the presence of both U54 and A58. In addition, the m1A58 modification enhances this activity, but only for TRUB2.
The tRNA Ψ55 synthase activities of nuclear, cytoplasmic, and mitochondrial extracts are primarily due to TRUB1, PUS10, and TRUB2, respectively
Nuclear, cytoplasmic, and mitochondrial extracts prepared from human PC3 cells produced Ψ in both tRNATrp and tRNAAla (Fig. 2A). As shown later by knockdown of the three proteins (Fig. 2C), most, if not all, of the Ψ produced by the extracts is Ψ55. However, only the nuclear extract produced Ψ55 with the U54A and A58G mutants of tRNAAla (Fig. 2B). This was also the case with the recombinant TRUB1 (Fig 1B), which suggested that the Ψ55 synthase activity of TRUB1 is present in the nucleus.
FIGURE 2.
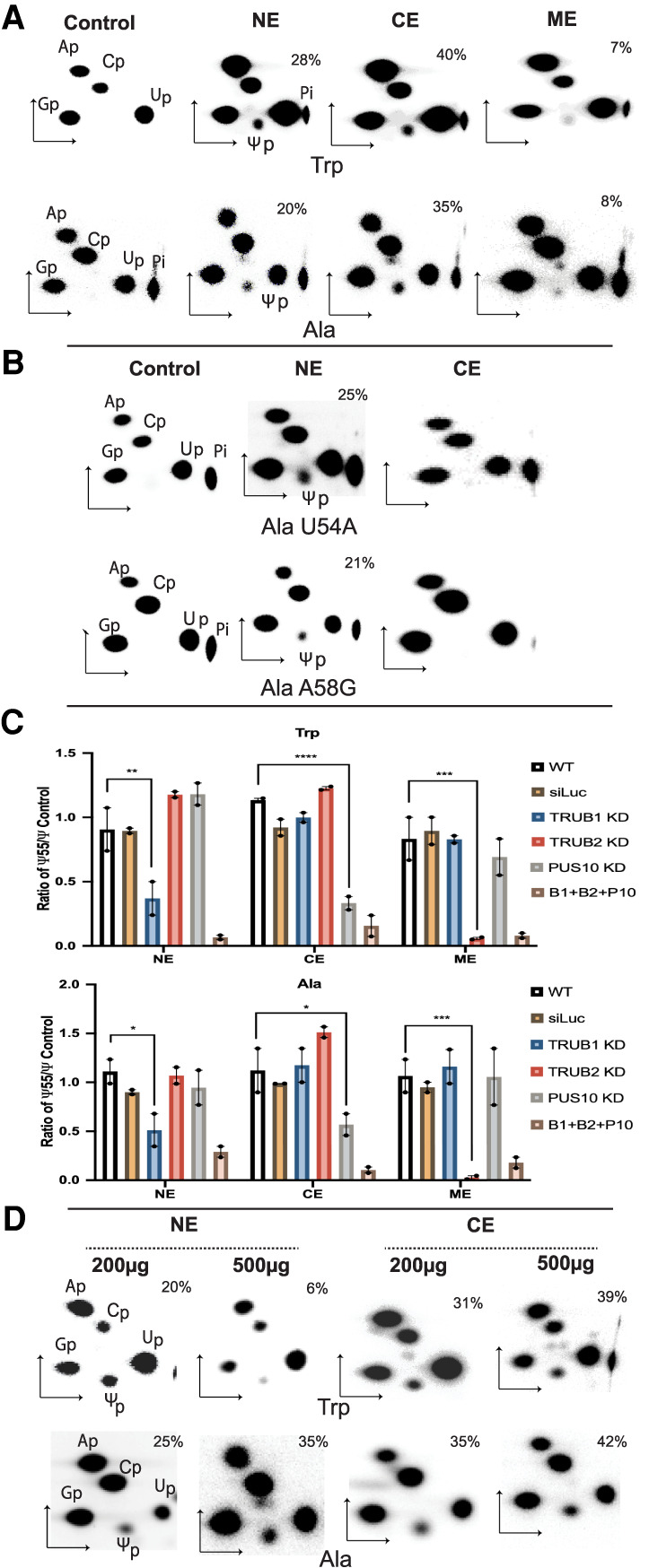
The tRNA Ψ55 synthase activities of TRUB1, PUS10, and TRUB2 are mainly in the nucleus, cytoplasm, and mitochondria, respectively. (A) 2D-TLC analyses, as in Figure 1, after treating [α-32P]CTP-labeled tRNATrp and tRNAAla with nuclear (NE), cytoplasmic (CE), and mitochondrial (ME) extracts of PC3 cells. (B) 2D-TLC analyses after treatment of U54A and A58G mutants of tRNAAla with NE and CE. (C) Ratio of Ψ55 synthesis in tRNATrp and tRNAAla versus control Ψ synthesis by NE, ME, and CE of PC3 strains individually knocked down (KD) for TRUB1, TRUB2, and PUS10 and all three together (B1 + B2 + P10). Untreated (WT) and luciferase siRNA-treated (siLuc) PC3 cells were used as controls. Control Ψ synthesis by the NE and ME were PUS1- and PUS2-mediated Ψ27 synthesis in tRNAPhe and by the CE was tRNA Ψ31 synthase-mediated Ψ31 synthesis in a mutant tRNAMet, as described in the Supplemental Methods and Supplemental Figure S2. Values are represented as mean ± SE. n = 2 replicates using independently prepared extracts. Each replicate is presented as a scattered dot (filled circle). ANOVA with Sidak's Multiple comparison test with respect to the WT was performed and significant P values for single KDs are presented as (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, and (****) P < 0.0001. P values of all comparisons are shown in Supplemental Table S1. (D) 2D-TLC analyses, as in A, after treating [α-32P]CTP-labeled tRNATrp and tRNAAla with 200 and 500 µg of NE and CE.
To confirm the localization of the Ψ55 synthase activities of the three proteins, we treated tRNATrp and tRNAAla with the extracts of PC3 cells that had individual knockdowns for each of the three protein genes, as well as the combined knockdown of all three genes. The Ψ55 synthase activities of the extracts were measured relative to Ψ27 synthesis in tRNAPhe mediated by PUS1 and PUS2 using the same nuclear and mitochondrial extracts and to Ψ31 synthesis in a mutant tRNAMet in the same cytoplasmic extracts. PUS1 and PUS2 are located in the nucleus and mitochondria, respectively (Rintala-Dempsey and Kothe 2017). Pus6 is known to produce Ψ31 in tRNAs of both the cytoplasm and mitochondria in yeast (Ansmant et al. 2001) and PUSD1 is suggested to do the same in human cells (Spenkuch et al. 2014). The activities of PUS1, PUS2, and tRNA Ψ31 synthase are not affected by knockdown of TRUB1, TRUB2, and PUS10 (see Supplemental Methods and Supplemental Fig. S2). Knockdown of TRUB1, TRUB2, and PUS10 reduced Ψ55 synthesis by the nuclear, mitochondrial, and cytoplasmic extracts, respectively, in both tRNATrp and tRNAAla (Fig. 2C). Combined knockdown of all three genes reduced Ψ55 synthesis in all three extracts. These results suggest that Ψ55 synthase activities of TRUB1, TRUB2, and PUS10 are located in the nucleus, mitochondria, and cytoplasm, respectively, and these three proteins account for synthesis of tRNA Ψ55 in vivo.
The tRNA Ψ55 synthase activity of the nuclear extract is not similar toward tRNATrp and tRNAAla
It is noteworthy that knockdown of both PUS10 and TRUB2 increased Ψ55 synthesis by the nuclear extract in tRNATrp, but not in tRNAAla (Fig. 2C). Previously, we showed that PUS10 produces Ψ54 in tRNATrp, but not in tRNAAla (Deogharia et al. 2019). Furthermore, we also showed that PUS10, although present mainly in the nucleus, has its catalytic activity present in the cytoplasm (Jana et al. 2017; Deogharia et al. 2019). Therefore, we tested whether there would be a difference in Ψ55 production in tRNATrp and tRNAAla by changing the amount of extracts relative to the amount of tRNAs. As expected, more cytoplasmic extract increased PUS10-mediated Ψ55 synthesis in both tRNATrp and tRNAAla (Fig. 2D). The same was observed for the activity of nuclear extract toward tRNAAla. However, increased amount of nuclear extract decreased Ψ55 synthesis in tRNATrp. This suggested that there is some factor(s) in the nuclear extract that suppressed TRUB1-mediated Ψ55 synthesis in tRNATrp but not in tRNAAla. This factor(s) increased when the amount of nuclear extract increased relative to the tRNA in the experiments shown in Figure 2D. As shown later, nuclear PUS10 is the factor that inhibits TRUB1-mediated Ψ55 formation in tRNATrp.
U54 and U55 of certain mammalian tRNAs are not modified in the nucleus
The presence of Ψ55 synthase activities both in the nucleus and cytoplasm initially suggested that the two activities are redundant and cytoplasmic PUS10 produces Ψ55 in those tRNAs that for some reason escape TRUB1-mediated modification in the nucleus. An alternative explanation could be that Ψ55 is produced in one set of tRNAs by TRUB1 in the nucleus and in another set by PUS10 in the cytoplasm. Mammalian cells contain multiple isoacceptor and isodecoder tRNAs for each amino acid. Therefore, we tested whether a significant proportion of nuclear tRNAs for certain amino acids in mouse liver cells contain unmodified U55. We used primer extension after 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluenesulfonate (CMCT) treatment of tRNA-enriched total RNA isolated from nuclear and total cytoplasmic fractions of mouse liver to determine the presence or absence of Ψ (Fig. 3). CMCT forms adducts with Ψ, U, and G. Alkali removes all CMCT groups except those attached to N3 of Ψ. Primer extension stops at one residue before the CMCT-modified Ψ. The tRNAAla and tRNAPhe, which normally contain T54 and Ψ55, showed Ψ at position 55 for both nuclear and cytoplasmic tRNAs. The tRNAAU-ALA, which normally contains A54 and Ψ55, also showed Ψ at position 55 in the nucleus. These results suggest that Ψ55 is produced in the nucleus in these tRNAs, before their transport to the cytoplasm.
FIGURE 3.
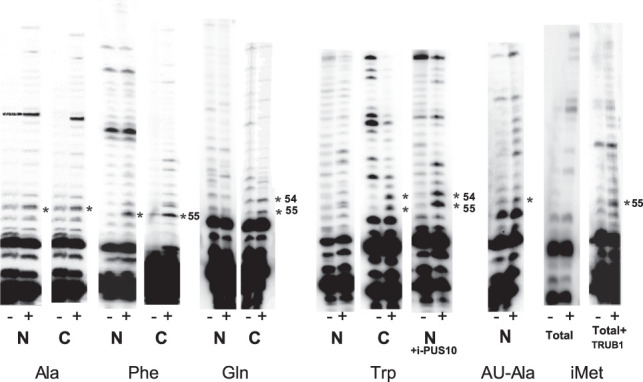
CMCT-primer extension analyses to determine the presence of Ψ55 in certain tRNAs in the nucleus and cytoplasm. The tRNA-enriched total RNAs prepared from the nuclear (N) and total cytoplasmic (C) extracts of mouse liver were first treated with AlkB to demethylate m1A58, which improves primer extension. These RNAs were either not treated (−) or treated with CMCT (+) for 20 min, followed by alkali treatment before primer extension reactions. The sequence of each primer for the tRNA of a particular amino acid corresponds to the 15 bases at position 62–76 of one isoacceptor for that amino acid (indicated below the panels). A dark band in the CMCT lane but not in the untreated lane indicates a Ψ. Dark bands in both lanes at the same position are caused by inhibition of primer extension by the presence of a modified residue at that position or due to a strong secondary and/or tertiary structure in that region. Unmodified U (and the T54 of tRNA) does not produce a dark band in either lane. The position of the band in the gel that corresponds to Ψ is determined by its distance from the end of the primer and correlation with the known sequence of the RNA. The asterisk next to a band indicates the presence of Ψ at that position. Positions 54 and 55 are indicated on the side. Reactions for tRNAAU-Ala were done only with the nuclear RNA. Reactions for tRNAiMet were done with the RNA isolated from total cell extract. The i-PUS10 treatment of nuclear RNA before the CMCT reaction converted both U54 and U55 of its tRNATrp to Ψ54 and Ψ55. Similarly, U55 of tRNAiMet, when treated with TRUB1 before the CMCT reaction was converted to Ψ55.
The tRNAGln and tRNATrp, which normally contain Ψ at both position 54 and 55, showed Ψ at these positions in the cytoplasmic fraction of tRNAs, but not in the nuclear fraction. This suggested that both U54 and U55 in these tRNAs remained unmodified in the nucleus and were converted to Ψ after transport to the cytoplasm. Treatment of the nuclear RNA fraction with recombinant i-PUS10 produced Ψ at both positions in tRNATrp, confirming that nuclear tRNATrp indeed contains unmodified U54 and U55. Had there been a T at position 54, it could not have been converted to Ψ. Since tRNAiMet normally contains unmodified U55, we used total cellular RNA. As expected, no Ψ55 was observed in this case. Recombinant TRUB1 treatment of total RNA produced Ψ55 in tRNAiMet, confirming that the U55 of this tRNA normally remains unmodified in the cell.
Cytoplasmic PUS10 can produce Ψ, but nuclear PUS10 cannot
Previously using an anti-PUS10 antibody, we showed that PUS10 is mainly present in the nucleus (Jana et al. 2017; Deogharia et al. 2019). However, PUS10-mediated Ψ54 formation in select tRNAs (tRNATrp and tRNAGln but not in some others, for example, tRNAAla and tRNAPhe) was observed only in the cytoplasmic extracts, not in the nuclear extracts (Deogharia et al. 2019). Unlike Ψ54 synthesis, PUS10 could produce Ψ55 in both tRNATrp and tRNAAla, and this activity too is cytoplasmic (Fig. 2). Although distinct, the nuclear and cytoplasmic versions of PUS10 are products of a single gene, not paralogous genes (Fitzek et al. 2018). This was confirmed when anti-His antibody could detect His-tagged PUS10 overexpressed from a plasmid-borne copy of PUS10, both in the nucleus and cytoplasm of HEK293T cells (Deogharia et al. 2019).
Therefore, we ascertained whether recombinant human PUS10 isolated from HEK293T cells also had Ψ synthase activity, and whether there was any difference between the recombinant PUS10 from the nuclear and cytoplasmic extracts. Here we refer to recombinant PUS10 proteins prepared from total cell, nuclear, and cytoplasmic extracts of HEK293T cells as h-PUS10, nh-PUS10, and ch-PUS10, respectively. Ψ55 synthesis was observed with ch-PUS10 in both tRNATrp and tRNAAla (Fig. 4A), although not as robust as with i-PUS10 (Fig. 1A). No Ψ55 was observed with either h-PUS10 or nh-PUS10 (Fig. 4A). We also confirmed that ch-PUS10 has Ψ54 synthase activity similar to that of i-PUS10, that is, it produces Ψ54 in tRNATrp, but not in tRNAAla, but again the activity was much less than that of i-PUS10 (Fig. 4B). Ψ54 synthesis was determined by nearest-neighbor analysis using [α-32P]UTP-labeled m1A58-modified tRNAs, as was done before (Deogharia et al. 2019). These results suggest that although recombinant cytoplasmic PUS10 is weakly catalytic, PUS10 isolated from total cell extract does not show any activity, because catalytically inactive nuclear PUS10 is its major component.
FIGURE 4.
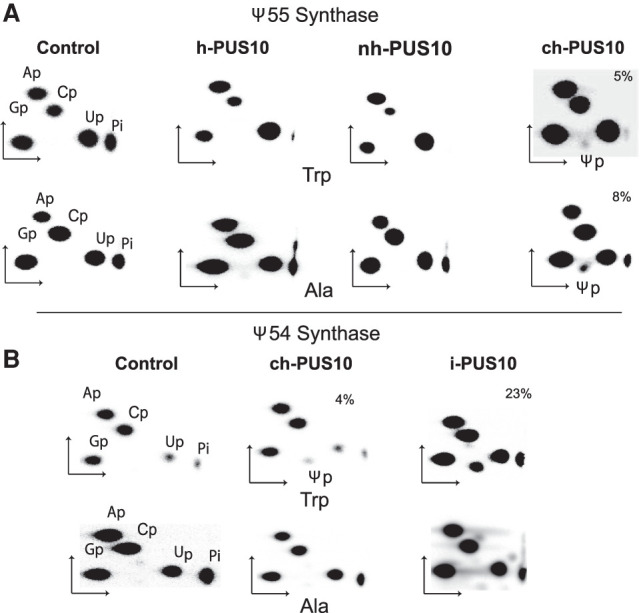
Cytoplasmic PUS10 has Ψ synthase activity. (A) Ψ55 synthase activities of h-PUS10, nh-PUS10, and ch-PUS10 were determined by 2D-TLC analyses as in Figure 1, after using these proteins to treat [α-32P]CTP-labeled tRNATrp and tRNAAla. (B) Ψ54 synthase activities of ch-PUS10 and i-PUS10 were determined by 2D-TLC analyses as in Figure 1, after using these proteins to treat [α-32P]UTP-labeled tRNATrp and tRNAAla. The tRNAs were pretreated with TrmI to produce m1A58, which enhances Ψ54 synthesis by PUS10.
Nuclear PUS10 binds some but not all tRNAs
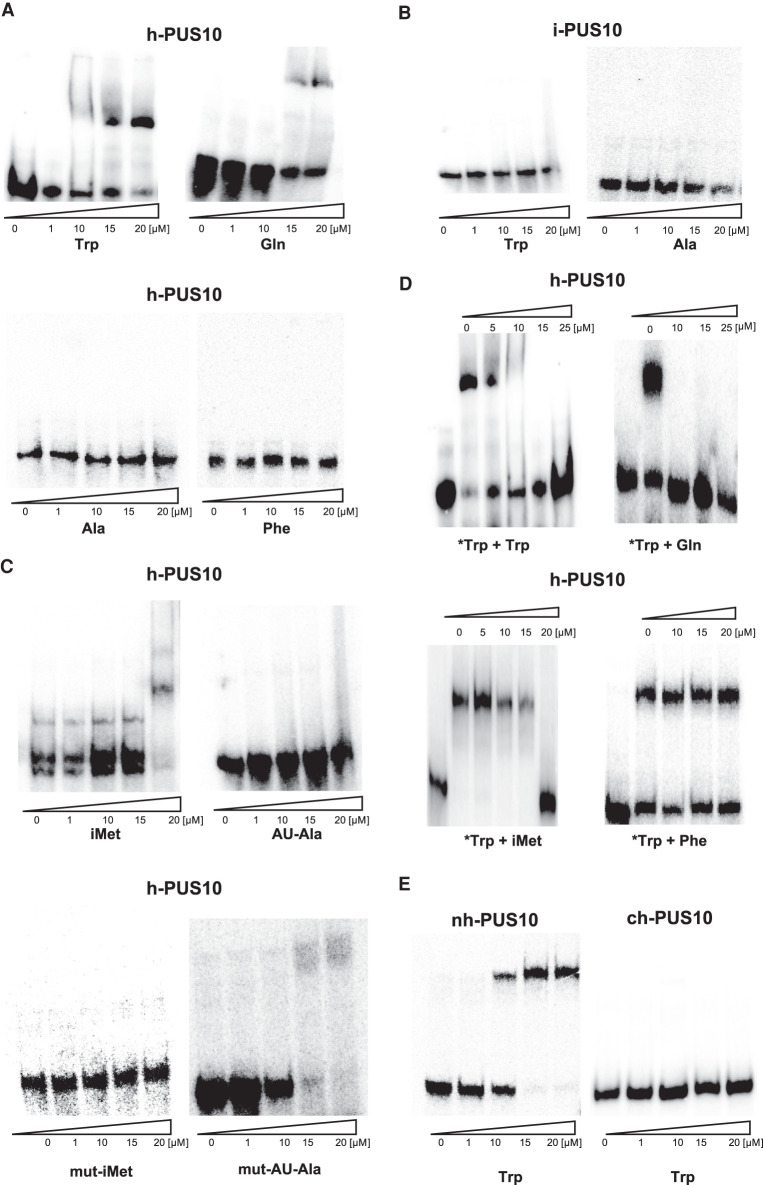
Previously, we showed that the AAAU sequence at positions 57–60, which is present in tRNATrp and tRNAGln, but not in tRNAAla and tRNAPhe, is needed for the Ψ54 synthase activity of i-PUS10 (Deogharia et al. 2019). Therefore, we tested whether h-PUS10 or i-Pus10 can bind to any tRNA. The h-PUS10 binds to tRNATrp and tRNAGln, but not to tRNAAla and tRNAPhe under our EMSA conditions (Fig. 5A). However, the i-PUS10 does not bind either to tRNATrp or to tRNAAla under the same conditions (Fig. 5B). The h-PUS10 also binds to tRNAiMet, although at higher concentrations than to tRNATrp (Fig. 5C), which may be due to the presence of GAAA in tRNAiMet, instead of AAAU at position 57–60. The h-PUS10 does not bind to tRNAAU-Ala (Fig. 5C), which has GAUG at position 57–60.
FIGURE 5.
Catalytically inactive PUS10 selectively binds only certain other tRNAs. (A) Radiolabeled tRNAs for Trp, Gln, Ala, and Phe (indicated below the panels) were incubated with increasing concentrations of h-PUS10, resolved by native PAGE, and visualized by phosphorimaging. (B) Reactions similar to those in A using i-PUS10 with tRNATrp and tRNAAla. (C) Reactions similar to those in A using h-PUS10 with tRNAiMet and tRNAAU-Ala and their mutants (mut-iMet and mut-AU-Ala) where sequences at positions 59–60 were interchanged as described in the text. (D) Separately radiolabeled tRNATrp (*Trp) along with increasing concentrations of unlabeled tRNAs (indicated above the panels) for Trp, Gln, initiator Met, and Phe were incubated with h-PUS (20 µM), resolved by native PAGE, and visualized by phosphorimaging. The first lane in each panel contains only the radiolabeled tRNATrp, with no unlabeled tRNA or h-PUS10. (E) Reactions similar to A after separate incubations of tRNATrp with nh-PUS10 and ch-PUS10.
We mutated AA at positions 59–60 of tRNAiMet to UG to make the sequence GAUG at position 57–60, as in tRNAAU-Ala, and did the reverse mutation in tRNAAU-Ala to make it like tRNAiMet. Now h-PUS10 binds to mutated tRNAAU-Ala but not to mutated tRNAiMet (Fig. 5C). The binding of h-PUS10 to tRNATrp is specific, because it could be competed out by addition of excess unlabeled tRNATrp or tRNAGln, and even by tRNAiMet at a higher concentration, but not by tRNAPhe (Fig. 5D). We also observed that it is only the nuclear fraction (nh-PUS10) that binds to these tRNAs, not the cytoplasmic fraction (ch-PUS10) (Fig. 5E).
Overall, these results suggest that catalytically inactive nuclear PUS10, which is the major fraction of total PUS10, binds to tRNAs that contain AAAU (or GAAA) at position 57–60, whereas catalytically active cytoplasmic PUS10 (and i-PUS10) does not bind under the EMSA conditions we tested. Probably the latter binds transiently to the tRNA during catalysis and dissociates after that.
Stably bound PUS10 blocks Ψ55 synthesis
Since nuclear h-PUS10 can stably bind certain tRNAs, we wanted to ascertain whether this binding blocks Ψ55 synthesis by TRUB1. We first bound recombinant h-PUS10 to different tRNAs and then compared recombinant TRUB1-mediated Ψ55 synthesis in these tRNAs with corresponding unbound tRNAs. As expected, Ψ55 synthesis was decreased in the tRNAs (tRNATrp and tRNAiMet) to which h-PUS10 binds, but not in the tRNAs (tRNAAla and tRNAAU-Ala) to which it does not bind (Fig. 6A).
FIGURE 6.
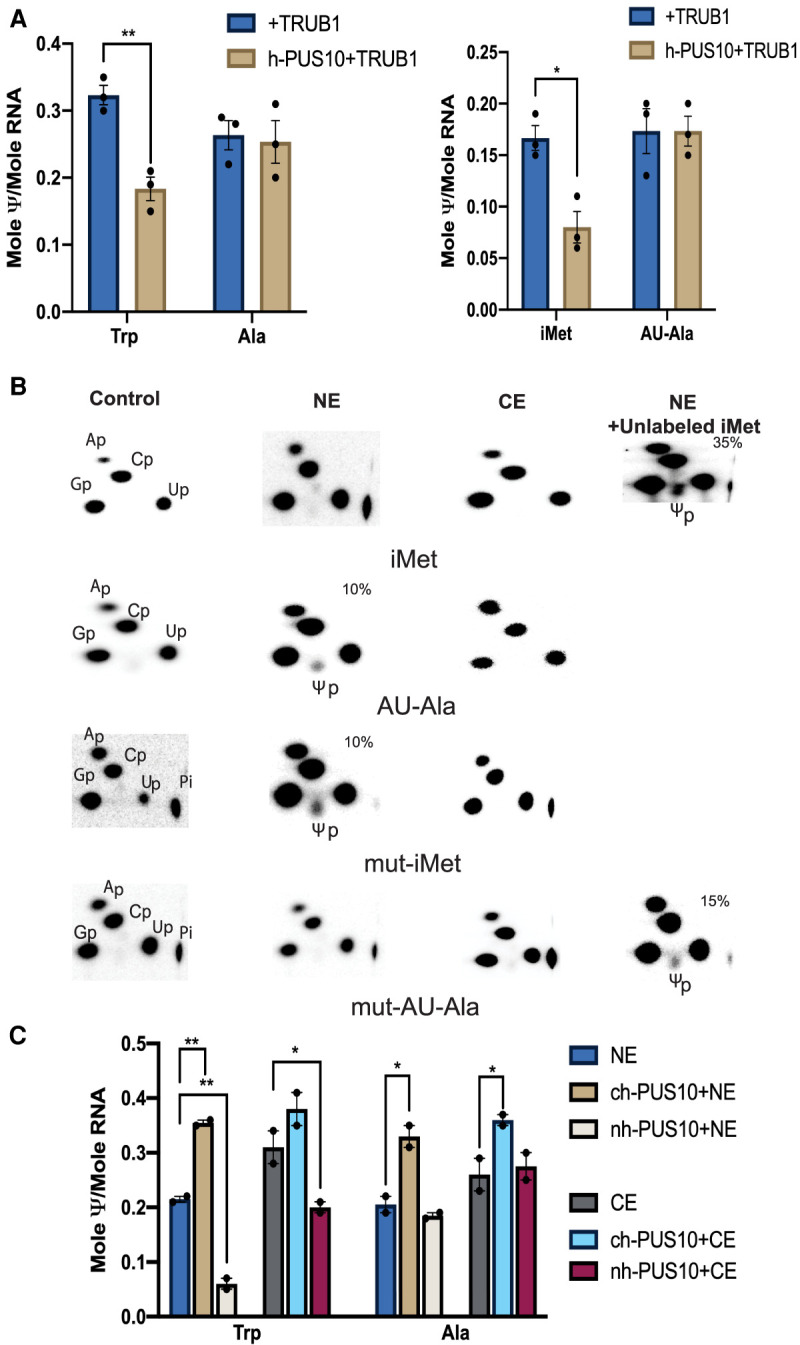
Bound PUS10 blocks Ψ55 synthesis. (A) Activity of recombinant TRUB1 was determined as in Figure 1A, using different [α-32P]CTP-labeled tRNAs (indicated below the panels), with or without pretreating the tRNAs with h-PUS10. Values are represented as mean ± SE. n = 3 replicates. Each replicate is presented as a scattered dot. ANOVA with Sidak's Multiple comparison test with respect to the “+TRUB1” was performed and significant P values are presented as (*) P < 0.05 and (**) P < 0.01. P values of all comparisons are shown in Supplemental Table S1. (B) Activity of nuclear (NE) and cytoplasmic (CE) extracts were determined as in Figure 2A using [α-32P]CTP-labeled tRNAiMet and tRNAAU-Ala and their interchanged mutants (same as in Fig. 5C). In separate NE reactions with tRNAiMet and mut-AU-Ala, extra unlabeled tRNAiMet (extreme right panels) was added to saturate the PUS10 present in the extract. (C) Activities of NE and CE were determined as in A, using [α-32P]CTP-labeled tRNATrp and tRNAAla, with or without pretreating the tRNAs with ch-PUS10 or nh-PUS10. Values are represented as mean ± SE. n = 2 replicates using independently prepared extracts. Each replicate is presented as a scattered dot. ANOVA with Sidak's Multiple comparison test with respect to the NE or CE, as appropriate, was performed and significant P values are presented as (*) P < 0.05 and (**) P < 0.01. P values of all comparisons are shown in Supplemental Table S1.
Although recombinant TRUB1 can produce Ψ55 both in tRNAiMet and tRNAAU-Ala (Figs. 1C and 6A), the former normally contains unmodified U55, whereas the latter has Ψ55 in the cell. Therefore, we tested the activities of nuclear and cytoplasmic extracts on these tRNAs, as done for tRNATrp and tRNAAla in Figure 2A. (Mitochondrial extract was not used with these tRNAs, because no human mitochondrial tRNA has AU at positions 54–55.) Under our normal reaction conditions (as used in Fig. 2), tRNAAU-Ala showed Ψ55 synthesis only with the nuclear extract, but neither extract produced Ψ55 in tRNAiMet (Fig. 6B). As expected, an AA to GU mutant of tRNAiMet (same mutant as used in Fig. 5C) recovered the nuclear Ψ55 activity, and the reverse change in tRNAAU-Ala lost it (Fig. 6B).
The absence of Ψ55 synthesis in tRNAiMet by the nuclear extract was not expected, because recombinant TRUB1 could produce Ψ55 in this tRNA (Fig. 1C). We hypothesized that the PUS10 present in the nuclear extract bound to tRNAiMet and did not allow TRUB1 to act. Therefore, to saturate the PUS10, we added excess unlabeled tRNAiMet to the nuclear extract reactions of tRNAiMet and mutant tRNAAU-Ala, that did not show Ψ55 synthesis. In both cases, Ψ55 formation was recovered (Fig. 6B).
To confirm that it is the nuclear PUS10, and not the cytoplasmic PUS10 that inhibits Ψ55 synthase activities of the extracts, we first independently treated tRNATrp and tRNAAla with recombinant nh-PUS10 and ch-PUS10. We then determined Ψ55 synthesis in these tRNAs, separately using nuclear and cytoplasmic extracts (Fig. 6C). As expected, in the case of tRNATrp, stable nh-PUS10 binding significantly reduced the activities of both nuclear and cytoplasmic extracts. Since nh-PUS10 does not bind to tRNAAla, there was hardly any effect on its Ψ55 synthesis by either extract. On the other hand, the addition of ch-PUS10 increased Ψ55 synthesis in both tRNAs by both extracts, because ch-PUS10 provides additional Ψ55 synthesizing capability.
Overall, these results suggest that nuclear PUS10 inhibits the activity of TRUB1 in the nucleus by binding to those tRNAs, for example, tRNATrp and tRNAGln, that contain the sequence 57-AAAU-60 (or its variant GAAA in tRNAiMet). Although stable binding of nuclear PUS10 to tRNATrp is also capable of inhibiting cytoplasmic PUS10, this does not occur normally in the cell. These results can also explain why knockdown of PUS10 increases TRUB1-mediated nuclear Ψ55 synthesis in tRNATrp (Fig. 2C), where the amount of PUS10 is decreased in nuclear extract. Similarly, increased amount of nuclear extract relative to tRNA decreases TRUB1-mediated Ψ55 synthesis in tRNATrp (Fig. 2D), because the amount of PUS10 relative to tRNA is increased.
DISCUSSION
We show here that although both mammalian TRUB1 and PUS10 can produce Ψ55 in U54U55-containing tRNAs in vitro, their activities inside the cell are compartmentalized and they modify different sets of tRNAs. TRUB1 primarily produces Ψ55 in the nucleus in those tRNAs that normally contain T54 and PUS10 produces Ψ55 in the cytoplasm in those tRNAs that normally contain Ψ54. Previously we showed that there are two versions of PUS10, nuclear and cytoplasmic (Jana et al. 2017; Deogharia et al. 2019). The nuclear version is noncatalytic and is involved in TRAIL-induced apoptosis, while the catalytic cytoplasmic version produces Ψ54. Here we show that nuclear PUS10 stably binds to unmodified U54U55 version of Ψ54-containing tRNAs and inhibits TRUB1-mediated Ψ55 production in these tRNAs. Both Ψ54 and Ψ55 are produced by the cytoplasmic PUS10 in these tRNAs. Specific binding of nuclear PUS10 to select tRNAs can also explain the absence of Ψ55 in tRNAiMet and its presence in tRNAAU-Ala.
Nuclear extracts do produce Ψ55 in both tRNATrp and tRNAAla (Fig. 2A), and TRUB1 knockdown reduces Ψ55 in both these tRNAs (Fig. 2C). These results suggest that TRUB1 activity is not affected by nuclear PUS10. However, increasing nuclear extract to tRNA ratio increases Ψ55 synthesis in tRNAAla, but decreases it in tRNATrp (Fig 2D), suggesting that nuclear PUS10 does decrease TRUB1 activity toward tRNATrp. We believe that the reason for these contradictory results is that only a limited amount of free PUS10 is available in the nuclear extract, which can bind only a small amount of added labeled tRNATrp. TRUB1 of the extract can then act on the remaining unbound tRNATrp as observed in Figure 2A. When the amount of the extract is increased, more tRNATrp becomes bound to PUS10 and less tRNATrp is available for TRUB1 to act on, as observed in Figure 2D. In case of knockdown of TRUB1, less TRUB1 would be available to act on both tRNAAla and unbound tRNATrp, decreasing Ψ55 synthesis in both tRNAs as seen in Figure 2C.
It is intriguing that TRUB2 knockdown increases Ψ55 synthesis by PUS10 of the cytoplasmic extract in both tRNATrp and tRNAAla, but only in tRNATrp by TRUB1 of the nuclear extract (Fig. 2C). It seems that somehow TRUB2 knockdown decreases the amount of PUS10 in the nucleus and increases it in the cytoplasm. A decrease of nuclear PUS10 would increase the amount of unbound tRNATrp, thus increase Ψ55 in this tRNA, and an increase of cytoplasmic PUS10 would increase Ψ55 in both tRNAs. TRUB2 may have an effect on the nuclear-cytoplasmic distribution of PUS10.
The dichotomy of activities between nuclear and cytoplasmic PUS10 has also been reported recently by others (Song et al. 2020). They showed that nuclear PUS10 is involved in miRNA processing, and, as here, cytoplasmic PUS10 catalyzes tRNA pseudouridylation. Their results agree with ours in some respects and differ in others. They observed that recombinant human PUS10 isolated from E. coli can both bind to several classes of RNAs and produce Ψ in tRNAs. As reported before, our E. coli–derived human PUS10 did not show any catalytic activity (Deogharia et al. 2019). Unlike their protein, our catalytic proteins (i-PUS10 and ch-PUS10) did not show binding to any tRNA under our EMSA conditions. Probably these proteins transiently bind during catalysis.
They used demethylase-pseudouridine sequencing to determine Ψ54 and Ψ55 production by PUS10 in natural tRNAs by comparing the Ψ's at these sites in the wild-type and PUS10 knockout (KO) cells. They concluded that PUS10 produces Ψ55 in an AGC anticodon-containing tRNAAla that has A54 instead of U54 (its TΨC loop is identical to our tRNAAU-Ala). However, as shown in Figures 1C and 6B, Ψ55 in tRNAAU-Ala can be produced by both recombinant TRUB1 and nuclear extract, but neither by recombinant PUS10 nor by cytoplasmic extract, because PUS10 cannot produce Ψ55 in the presence of A54. Furthermore, Ψ55 is observed in this tRNA in the nucleus (Fig. 3), again suggesting it is synthesized by TRUB1 inside the cell. They observed the absence of Ψ54 in select tRNAs (e.g., tRNATrp, tRNAGln, etc.) and the presence of Ψ55 in these tRNAs in the KO cells. Therefore, they assigned the production of Ψ54 but not of Ψ55 to PUS10 in these tRNAs. We believe that even Ψ55 in these tRNAs is normally produced by PUS10 in the cytoplasm, because we showed that stable binding of noncatalytic nuclear PUS10 to these tRNAs normally inhibits TRUB1-mediated Ψ55 in these tRNAs. We interpret their data differently. Both Ψ54 and Ψ55 in these tRNAs are produced by the cytoplasmic PUS10 under normal conditions. However, the absence of PUS10 in their KO cells allows nuclear TRUB1 to access these tRNAs and produce Ψ55.
Although PUS10 can produce both Ψ54 and Ψ55, the two activities show some differences. It can produce Ψ54 in select tRNAs that have a consensus AAAU sequence at position 57–60 and m1A58 modification enhances this activity (Deogharia et al. 2019). Its Ψ55 synthesis shows no such preference, but does require U at position 54. The catalytic activity of ch-PUS10 is much weaker than that of i-PUS10. There may be some structural or posttranslational differences between the two recombinant proteins. Also, there may be some associated factor(s) that are required to enhance the activity of PUS10 in the cytoplasm.
Both mammalian and yeast tRNAiMet have unmodified U55 and A54. An ortholog of TruB, Pus4 is present in yeast and is responsible for Ψ55 synthesis in both cytoplasmic and mitochondrial tRNAs (Becker et al. 1997a). Pus4 requires the presence of U54 in the tRNA for its activity and thus cannot produce Ψ55 in yeast tRNAiMet (Becker et al. 1997b). The reason for the absence of Ψ55 in mammalian tRNAiMet is different. Although mammalian TRUB1, unlike PUS10, does not require U54 in the tRNA for Ψ55 synthesis and can produce Ψ55 in tRNAiMet in vitro, it is blocked because the tRNA is bound by PUS10 in the nucleus. TRUB1 is reported to be present in both nucleus and cytoplasm, but its activity is predominantly in the nucleus (Safra et al. 2017). This may be the reason that cytoplasmic TRUB1 may not be able to produce Ψ55 in mammalian tRNAiMet even after its transport to the cytoplasm.
It is possible that nuclear PUS10 binding to select tRNAs, in addition to inhibiting TRUB1, also inhibits TRMT2A-mediated T54 synthesis in the nucleus in these tRNAs. TRMT2A is mainly observed in the nucleus in HeLa cells (Chang et al. 2019), and in yeast T54 is synthesized after Ψ55 (Barraud et al. 2019). Therefore, neither U54 nor U55 of the tRNAs to which PUS10 binds, is modified in the nucleus. We believe that the tRNATrp and tRNAGln in the KO cells of Song and colleagues (Song et al. 2020) would have T54Ψ55 instead of normal Ψ54Ψ55 due to the absence of PUS10, as is the case for yeast cells, which do not have PUS10.
We isolated two versions of recombinant human PUS10 from HEK293T cells: a nuclear and a cytoplasmic version. This suggested that there is a nuclear and a cytoplasmic isoform of PUS10 in human cells. The nuclear isoform is catalytically inactive and can stably bind to tRNATrp. Recombinant protein isolated from total extract of HEK293T cells behaves like nuclear isoform, suggesting nuclear isoform is the major component of the total protein. Nuclear isoform is also shown to be involved in TRAIL-induced apoptosis and miRNA processing (Jana et al. 2017; Song et al. 2020). On the other hand, cytoplasmic isoform is catalytically active and does not stably bind tRNATrp. Although both isoforms are of the same size (Supplemental Fig. S3B) and are products of the same single gene (Fitzek et al. 2018), the two probably differ in their structure or in posttranslational modifications. The two PUS10 isoforms are distinct because the addition of nuclear extract to cytoplasmic extract does not alter PUS10-mediated Ψ54 synthase activity of the latter (Deogharia et al. 2019). In principle, the two isoforms could be splice variants of the same gene product. However, based on the available information (https://www.proteinatlas.org/ENSG00000162927-PUS10/cell#human and http://useast.ensembl.org/Homo_sapiens/Transcript/ProteinSummary?db=core;g=ENSG00000162927;r=2:60940222-61018259;t=ENST00000421319), this is less likely. Human PUS10 is suggested to have six splice variants. Of these, only four are protein coding, two of which give rise to annotated full-length PUS10 (529 aa), and the other two are expected to produce proteins of 62 aa and 120 aa. The latter two shorter versions do not contain the catalytic domain (McCleverty et al. 2007) of the protein. Human Pus10 splice variants that code for the 529 aa protein contain 18 exons. Conversion of the same PUS10 gene product into two isoforms appears to occur in mammalian cells but not in insect cells, because recombinant human PUS10 isolated from SF9 cells behaves only like human cytoplasmic isoform (i.e., it has catalytic activity and does not stably bind tRNATrp). Although the insect Pus10 gene has a nuclear localization signal, so far there is no report of any catalytic or noncatalytic function assigned to insect nuclear or cytoplasmic Pus10 and roles of PUS10 and TRUB in tRNA Ψ55 synthesis.
There appears to be some association between Ψ54 modification of tRNA and retroviral DNA synthesis. Several Ψ54-containing tRNAs function as primers for the retroviral reverse transcriptases (Mak and Kleiman 1997; Deogharia et al. 2019). Interaction of avian and mammalian tRNATrp with avian myeloblastosis virus reverse transcriptase either requires or is stabilized by Ψ at positions 54 and 55 (Hu and Dahlberg 1983). Although tRNALys3, the primer for HIV reverse transcriptase, is reported to contain 2′-O-methylribothymidine (Tm) at position 54 (Juhling et al. 2009), we showed previously that human PUS10 can produce Ψ54 in this tRNA (Deogharia et al. 2019). Therefore, it may be worth investigating whether PUS10 knockout cells, where Ψ54Ψ55 in the tRNAs would be replaced by T54Ψ55 are resistant to infection by retroviruses and to transduction by lentiviral vectors.
Based on this work, compartmentalization of Ψ55 synthesis between the nucleus and cytoplasm is diagrammatically presented in Figure 7. TRUB1 produces Ψ55 in most U54U55-containing tRNAs, for example, tRNAAla and tRNAPhe, and A54U55-containing elongator tRNAs, for example, tRNAAU-Ala in the nucleus, and then these tRNAs are exported to the cytoplasm. Nuclear PUS10 (nh-PUS10) inhibits nuclear TRUB1 activity by binding to a specific set of U54U55-containing tRNAs, for example, tRNATrp, tRNAGln, that have the consensus sequence AAAU at position 57–60. These tRNAs are exported to the cytoplasm with both U55 and U54 unmodified. Cytoplasmic PUS10 (ch-PUS10) produces both Ψ54 and Ψ55 in these tRNAs. Nuclear PUS10 also binds to tRNAiMet, which has A54U55 and GAAA at position 57–60 and inhibits its U55 to Ψ55 conversion by nuclear TRUB1. However, U55 of tRNAiMet remains unmodified even in the cytoplasm because PUS10 cannot produce Ψ55 in those tRNAs that contain A54.
FIGURE 7.
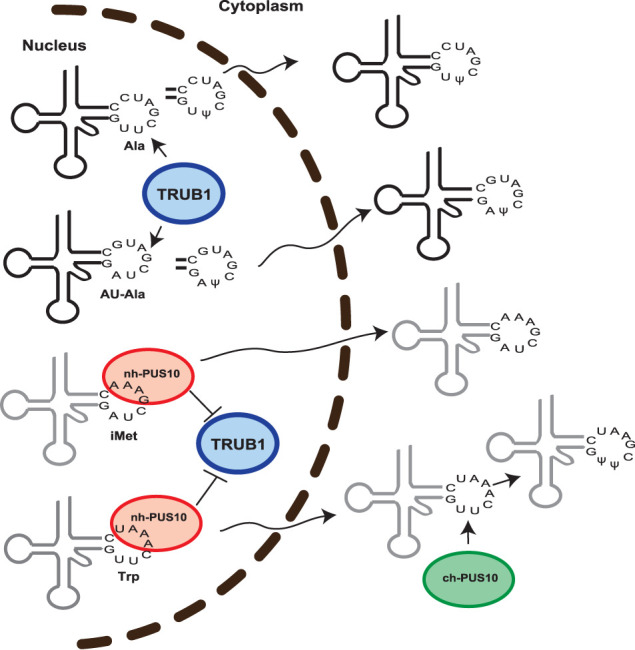
Overview of Ψ55 synthesis in different tRNAs. TRUB1 produces Ψ55 in U54U55-containing tRNAAla and A54U55-containing tRNAAU-Ala in the nucleus. Modified tRNAs are then exported to the cytoplasm. Conversion of U54 to T54 in tRNAAla is not shown here. Nuclear PUS10 (nh-PUS10) inhibits nuclear TRUB1 activity by binding to U54U55-containing tRNATrp that has AAAU at position 57–60, and to A54U55-containing tRNAiMet, that has GAAA at position 57–60. These tRNAs are consequently exported to the cytoplasm with an unmodified U55. Cytoplasmic PUS10 (ch-PUS10) produces both Ψ54 and Ψ55 in tRNATrp, but not in tRNAiMet.
This work raises a question. How is the catalytically inactive Pus10 replaced by the active Pus10 in the cytoplasm? A possibility is that it dissociates during nucleus to cytoplasm transport of tRNAs. Nuclear PUS10 remains in the nucleus and free cytoplasmic PUS10 produces Ψ54 and Ψ55 in the tRNAs. An alternative possibility is that the noncatalytic nuclear PUS10-bound tRNAs are exported to the cytoplasm, where some structural change in the bound PUS10 converts it into catalytic PUS10. This change can occur either while crossing the nuclear membrane or after reaching the cytoplasm. The catalytic PUS10 after producing Ψ54 and Ψ55 dissociates from the tRNA. Since we observed that unless overexpressed, normal PUS10 is detected only in the nucleus and not in the cytoplasm, it is possible that after release from the tRNA most of the cytoplasmic PUS10 is either degraded or transported back to the nucleus.
Unanswered basic questions still remain: Why are U54U55 of most but not all mammalian tRNAs converted to T54Ψ55 by two different enzymes (TRUB1 and TRMT2a) in the nucleus, and why are the same Us of certain other tRNAs protected by another protein (PUS10) from these two nuclear enzymes, transported to the cytoplasm, and there converted to Ψ54Ψ55 by the catalytic version of the same protein? Further studies are needed to answer these questions.
MATERIALS AND METHODS
Standard molecular biology procedures (Green and Sambrook 2012) were used unless specifically described. Oligonucleotides used in this work are listed in Supplemental Table S2.
Cell cultures
PC3 cells were cultured in RPMI 1640 (ATCC), and HEK293T cells were grown in DMEM media (HyClone, GE Healthcare), both with 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin (15140122, Gibco, Thermo Fisher) under standard culture conditions (37°C and 5% CO2).
Preparation of recombinant proteins and assays
Recombinant human PUS10 from SF9 insect cells (i-PUS10) was prepared and used for reactions as described before (Deogharia et al. 2019). Recombinant human PUS10 proteins from HEK293T cells were prepared by using PUS10 gene in a clone in pEF6/V5-His-TOPO (Invitrogen) described before (Deogharia et al. 2019), to transiently transfect the cells using TurboFect (Thermo Fisher). The cells were harvested after 48 h incubation. Total cell extracts from these cells were prepared as described before (Deogharia et al. 2019). Recombinant protein (h-PUS10) from the total cell extract was purified using an Ni-NTA column and used for the reactions as described before for i-PUS10 (Deogharia et al. 2019). Although PUS10 is not normally detected in the immunoblots of WT cytoplasmic extracts, overexpressed His-tagged PUS10 protein is detectable in these extracts (Deogharia et al. 2019). Therefore, recombinant nuclear (nh-PUS10) and cytoplasmic (ch-PUS10) PUS10 proteins were also prepared from the nuclear and total cytoplasmic fractions of total cell extract (Supplemental Fig. S3B). The fractions were prepared as described before (Jana et al. 2017). The purity of these and other subcellular fractions described later was determined by immunoblotting using antibodies against appropriate markers. These are shown in Supplemental Figure S3.
To clone human TRUB1 and TRUB2 genes, cDNA was prepared first from the total RNA of HEK293T cells using the Superscript III First Strand Synthesis System for RT-PCR kit with oligo dT primers (Invitrogen). TRUB1 and TRUB2 were amplified from the cDNA using primers corresponding to their coding regions. The resulting PCR product for TRUB1 was cloned between the NcoI and XhoI sites of the pET28a vector (Novagen, EMD Millipore). TRUB2 was cloned between the XbaI and XhoI sites of the same vector. E. coli BL21 (DE3) pLysS cells (Novagen) were transformed with these clones and grown to an O.D600 of 0.5 and induced with 1 mM IPTG for 2 h at 37°C. After harvesting, the cells were used for protein purification using an Ni-NTA column as previously mentioned.
Concentrations of recombinant proteins were determined by the Coomassie Protein Assay reagent (Thermo Scientific 1856209). Reaction conditions were similar to those used for the i-PUS10 protein (Deogharia et al. 2019). Reactions contained 4 pmol tRNA and 50 µg protein (∼600 pmol of PUS10 proteins or approximately 1 nmol of TRUB proteins) in a 50 µL volume. Some reactions with TRUB1 were done after pretreating 4 pmol of a tRNA with 50 µg h-PUS10.
TrmI and AlkB were purified and used as described before (Deogharia et al. 2019).
Preparation of tRNA transcripts
Cloned genes of tRNATrp, tRNAAla, tRNAPhe, and tRNAGln were used to prepare transcripts as described before (Deogharia et al. 2019). Transcripts for tRNAiMet, tRNAAU-Ala, and tRNAMet were prepared from the PCR-amplified DNAs using two primers that overlapped at their 3′-end. Forward primers for all PCR contained T7 RNA polymerase promoter sequences. PCR primers containing the desired mutations were used to prepare mutant tRNAs. In vitro transcriptions to prepare 32P-labeled and unlabeled transcripts were carried out as described before (Gurha et al. 2007; Gurha and Gupta 2008).
Pseudouridylation detection by thin layer chromatography
Reaction products of 32P-labeled tRNAs treated with recombinant proteins or cellular fractions were phenol–chloroform extracted, ethanol precipitated, and digested with RNase T2. The digests were resolved by two-dimensional TLC on cellulose plates (EMD Millipore). The solvents used were Solvent I (isobutyric acid/0.5 N NH4OH, 5:3, v/v) for the first dimension and Solvent II (isopropanol/HCl/H2O, 70:15:15, v/v/v) for the second dimension (Gupta 1984; Gurha et al. 2007; Gurha and Gupta 2008). Radioactivity in the plates was revealed and quantified by phosphorimaging using Optiquant software. All assays were repeated at least twice. The mole Ψ/mole RNA was determined by the formula (radioactivity in Ψp spot × the number of labeled nucleotides per transcript)/(sum of the radioactivity in all Np spots). This number was multiplied by 100 to determine percent Ψ produced. We could detect as low as 1% pseudouridylation in our TLC assays. We prepared RNase T2 according to a published procedure (Lichtler et al. 1992). These preparations contained some phosphatase activity. Since this activity nonspecifically removed phosphate (Pi) from all nucleotides, we did not include radioactivity in the Pi spot in calculating mole Ψ/mole RNA.
Determination of pseudouridylation activity of subcellular fractions
Nuclear, cytoplasmic, and mitochondrial extracts of PC3 cells were prepared according to a published procedure (Dewe et al. 2017). Briefly, cells from 100 cm tissue culture plates were harvested after trypsinization, washed twice with PBS (137 mM NaCl, 27 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4), and resuspended in 3–5 mL homogenization buffer (10 mM Tris-HCl, pH 6.7, 10 mM KCl, 0.15 mM MgCl2, 1 mM PMSF, and 1 mM DTT) per plate. The suspension was lysed with 40 strokes of a Dounce homogenizer and then transferred to a 15-mL conical tube. An appropriate amount of concentrated sucrose solution in homogenization buffer was added to bring the final sucrose concentration to 0.25 M. The suspension was centrifuged at 1000g for 5 min in a swinging-bucket rotor at 4°C to pellet nuclei. The supernatant was transferred to a fresh tube and centrifuged at 5000g at 4°C for 10 min in a fixed-angle rotor to pellet mitochondria. The pellet containing mitochondria was washed with sucrose-containing buffer (10 mM Tris-HCl, pH 6.7, 0.15 mM MgCl2, 0.25 M sucrose, 1 mM PMSF, 1 mM DTT). The supernatant was saved as the cytoplasmic extract. The mitochondrial and nuclear pellets were resuspended in PBS containing 0.1% NP40 and protease inhibitor cocktail (Pierce) and sonicated three times for 3–5 s at 3 watts each, to prepare the nuclear and mitochondrial extracts, respectively. Protein concentrations of the extracts were determined by the Coomassie Protein Assay reagent. Activities of the extracts were determined as described before (Deogharia et al. 2019), except that 100 µg (protein) of mitochondrial extract was used in each reaction instead of 200 µg, as used for the nuclear and cytoplasmic extracts. Some reactions with nuclear and cytoplasmic extracts were done after pretreating 4 pmol of a tRNA with 50 µg nh-PUS10 or ch-PUS10. Each reaction was done twice using two independently prepared extracts.
Preparation of knockdown cells
siRNAs were used for transient knockdown of TRUB1 and TRUB2 in PC3 cells. The siRNA for each gene was designed using the server (http://sirna.wi.mit.edu/). These are listed in Supplemental Table S2. Sense and antisense strands were hybridized at 95°C for 5 min followed by slow cooling to 25°C to create a duplex and then placed on ice for 10 min. The transfections using jetPRIME transfection reagent (Polypus) were performed as recommended by the manufacturer. The cells were harvested 72 h posttransfection and assayed for the gene of interest using immunoblotting and qPCR as described before (Jana et al. 2017; Deogharia et al. 2019). The data are shown in Supplemental Figure S4. Image J was used for densitometric analysis of the immunoblots and the values obtained relative to the WT PC3 cells are represented as fold change.
PUS10 knockdown strain KD1 of PC3 cells prepared before (Jana et al. 2017) was used here to study the effect of reduction of PUS10. We could not produce an shRNA-mediated PC3 strain that had a more efficient knockdown than the one used here. Triple knockdown cells were prepared by simultaneous transfection of this PUS10 knockdown strain by siRNAs for TRUB1 and TRUB2.
Determination of the presence of Ψ at specific positions in the tRNAs
Total RNA from C57BL/6 mouse livers was prepared using Tri Reagent (Molecular Research Center) according to the manufacturer's protocol. To prepare total cellular RNA, a homogenous suspension of the liver tissue in PBS containing 0.1% NP40 and protease inhibitor (88665, Pierce Thermo Fisher) was prepared by a few strokes of Dounce homogenizer. Nuclear and total cytoplasmic extracts were prepared from this suspension as for HEK293T cells. RNAs were isolated separately from the nuclear and total cytoplasmic extracts, as well as from the total cellular extracts. tRNAs were separated from other RNAs by PAGE on 6% denaturing gel and eluted. The presence of Ψ in the tRNA was determined by primer extension following CMCT treatment, as described previously (Deogharia et al. 2019). Briefly, 20 µg tRNA-enriched total RNA (6 µg tRNA and 14 µg total RNA) was treated with CMCT for 20 min at 37°C. Controls were simply untreated and incubated samples. CMCT-treated RNAs after precipitation were treated with 50 mM Na2CO3, pH 10.4, for 3 h at 37°C. The RNAs were precipitated and used for primer extension with [5′-32P]-labeled primers using M-MLV reverse transcriptase (Promega) according to the manufacturer's instructions. Reaction products were separated by 12% sequencing gels. The tRNA-enriched total RNAs from the nuclei and cytoplasm of mouse liver were used for the CMCT reactions because the yield of RNAs were higher than those from human cell lines.
Electrophoretic mobility shift assays (EMSA)
Complexes were assembled by incubating ∼1 pmol of 3′-end-labeled tRNAs with varying concentrations of recombinant proteins in 20 µL reactions in a buffer (20 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.75 mM DTT, 1.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol) for 15 min at 37°C. Complexes were resolved on native 6% polyacrylamide gel in 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA) run at 4°C. The bands were visualized by using a phosphorimager.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Clark and Mike Bosmeny for critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) grant number GM055945 to R.G.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.076810.120.
REFERENCES
- Ansmant I, Motorin Y, Massenet S, Grosjean H, Branlant C. 2001. Identification and characterization of the tRNA:psi 31-synthase (Pus6p) of Saccharomyces cerevisiae. J Biol Chem 276: 34934–34940. 10.1074/jbc.M103131200 [DOI] [PubMed] [Google Scholar]
- Antonicka H, Choquet K, Lin ZY, Gingras AC, Kleinman CL, Shoubridge EA. 2017. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep 18: 28–38. 10.15252/embr.201643391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, Gato A, Heiss M, Catala M, Kellner S, Tisne C. 2019. Time-resolved NMR monitoring of tRNA maturation. Nat Commun 10: 3373 10.1038/s41467-019-11356-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HF, Motorin Y, Planta RJ, Grosjean H. 1997a. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res 25: 4493–4499. 10.1093/nar/25.22.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HF, Motorin Y, Sissler M, Florentz C, Grosjean H. 1997b. Major identity determinants for enzymatic formation of ribothymidine and pseudouridine in the T psi-loop of yeast tRNAs. J Mol Biol 274: 505–518. 10.1006/jmbi.1997.1417 [DOI] [PubMed] [Google Scholar]
- Blaby IK, Majumder M, Chatterjee K, Jana S, Grosjean H, de Crecy-Lagard V, Gupta R. 2011. Pseudouridine formation in archaeal RNAs: the case of Haloferax volcanii. RNA 17: 1367–1380. 10.1261/rna.2712811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn CC, Mathews MB. 1987. Two human tRNA(Ala) families are recognized by autoantibodies in polymyositis sera. Mol Biol Med 4: 21–36. [PubMed] [Google Scholar]
- Chang YH, Nishimura S, Oishi H, Kelly VP, Kuno A, Takahashi S. 2019. TRMT2A is a novel cell cycle regulator that suppresses cell proliferation. Biochem Biophys Res Commun 508: 410–415. 10.1016/j.bbrc.2018.11.104 [DOI] [PubMed] [Google Scholar]
- Chatterjee K, Blaby IK, Thiaville PC, Majumder M, Grosjean H, Yuan YA, Gupta R, de Crecy-Lagard V. 2012. The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA. RNA 18: 421–433. 10.1261/rna.030841.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crecy-Lagard V, Boccaletto P, Mangleburg CG, Sharma P, Lowe TM, Leidel SA, Bujnicki JM. 2019. Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic Acids Res 47: 2143–2159. 10.1093/nar/gkz011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deogharia M, Mukhopadhyay S, Joardar A, Gupta R. 2019. The human ortholog of archaeal Pus10 produces pseudouridine 54 in select tRNAs where its recognition sequence contains a modified residue. RNA 25: 336–351. 10.1261/rna.068114.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewe JM, Fuller BL, Lentini JM, Kellner SM, Fu D. 2017. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol Cell Biol 37: e00214–e00217. 10.1128/MCB.00214-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzek E, Joardar A, Gupta R, Geisler M. 2018. Evolution of eukaryal and archaeal pseudouridine synthase Pus10. J Mol Evol 86: 77–89. 10.1007/s00239-018-9827-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Gu X, Yu M, Ivanetich KM, Santi DV. 1998. Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry 37: 339–343. 10.1021/bi971590p [DOI] [PubMed] [Google Scholar]
- Gupta R. 1984. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem 259: 9461–9471. [PubMed] [Google Scholar]
- Gupta R. 1985. Transfer ribonucleic acids of archaebacteria. In The archaebacteria; the bacteria: a treatise on structure and function (ed. Woese CR, Wolfe RS), Vol. 8, pp. 311–343. Academic, New York. [Google Scholar]
- Gupta R. 1986. Transfer RNAs of Halobacterium volcanii: sequences of five leucine and three serine tRNAs. System Appl Microbiol 7: 102–105. 10.1016/S0723-2020(86)80131-X [DOI] [Google Scholar]
- Gurha P, Gupta R. 2008. Archaeal Pus10 proteins can produce both pseudouridine 54 and 55 in tRNA. RNA 14: 2521–2527. 10.1261/rna.1276508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurha P, Joardar A, Chaurasia P, Gupta R. 2007. Differential roles of archaeal box H/ACA proteins in guide RNA-dependent and independent pseudouridine formation. RNA Biol 4: 101–109. 10.4161/rna.4.2.5177 [DOI] [PubMed] [Google Scholar]
- Hu JC, Dahlberg JE. 1983. Structural features required for the binding of tRNATrp to avian myeloblastosis virus reverse transcriptase. Nucleic Acids Res 11: 4823–4833. 10.1093/nar/11.14.4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S, Hsieh AC, Gupta R. 2017. Reciprocal amplification of caspase-3 activity by nuclear export of a putative human RNA-modifying protein, PUS10 during TRAIL-induced apoptosis. Cell Death Dis 8: e3093 10.1038/cddis.2017.476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joardar A, Jana S, Fitzek E, Gurha P, Majumder M, Chatterjee K, Geisler M, Gupta R. 2013. Role of forefinger and thumb loops in production of Psi54 and Psi55 in tRNAs by archaeal Pus10. RNA 19: 1279–1294. 10.1261/rna.039230.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. 2009. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162. 10.1093/nar/gkn772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtler A, Barrett NL, Carmichael GG. 1992. Simple, inexpensive preparation of T1/T2 ribonuclease suitable for use in RNase protection experiments. BioTechniques 12: 231–232. [PubMed] [Google Scholar]
- Mak J, Kleiman L. 1997. Primer tRNAs for reverse transcription. J Virol 71: 8087–8095. 10.1128/JVI.71.11.8087-8095.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. 2007. Crystal structure of human Pus10, a novel pseudouridine synthase. J Mol Biol 373: 1243–1254. 10.1016/j.jmb.2007.08.053 [DOI] [PubMed] [Google Scholar]
- Mueller EG, Ferre-D'Amare AR. 2009. Pseudouridine formation, the most common transglycosylation in RNA. In DNA and RNA modification enzymes: structure, mechanism, function and evolution (ed. Grosjean H), pp. 363–376. Landes Bioscience, Austin, TX. [Google Scholar]
- Nordlund ME, Johansson JO, von Pawel-Rammingen U, Bystrom AS. 2000. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA 6: 844–860. 10.1017/S1355838200992422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse K, Wrzesinski J, Bakin A, Lane BG, Ofengand J. 1995. Purification, cloning, and properties of the tRNA psi 55 synthase from Escherichia coli. RNA 1: 102–112. [PMC free article] [PubMed] [Google Scholar]
- Ny T, Bjork GR. 1980. Cloning and restriction mapping of the trmA gene coding for transfer ribonucleic acid (5-methyluridine)-methyltransferase in Escherichia coli K-12. J Bacteriol 142: 371–379. 10.1128/JB.142.2.371-379.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintala-Dempsey AC, Kothe U. 2017. Eukaryotic stand-alone pseudouridine synthases—RNA modifying enzymes and emerging regulators of gene expression? RNA Biol 14: 1185–1196. 10.1080/15476286.2016.1276150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. 2006. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res 34: 4293–4301. 10.1093/nar/gkl530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Nir R, Farouq D, Slutzkin IV, Schwartz S. 2017. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res 27: 393–406. 10.1101/gr.207613.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhuang Y, Zhu C, Meng H, Lu B, Xie B, Peng J, Li M, Yi C. 2020. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat Chem Biol 16: 160–169. 10.1038/s41589-019-0420-5 [DOI] [PubMed] [Google Scholar]
- Spenkuch F, Motorin Y, Helm M. 2014. Pseudouridine: still mysterious, but never a fake (uridine)!. RNA Biol 11: 1540–1554. 10.4161/15476286.2014.992278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Suzuki T. 2014. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 42: 7346–7357. 10.1093/nar/gku390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Auxilien S, Walbott H, Trachana K, Golinelli-Pimpaneau B, Brochier-Armanet C, Grosjean H. 2008. Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol Microbiol 67: 323–335. 10.1111/j.1365-2958.2007.06047.x [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Gray MW. 2000. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res 28: 2342–2352. 10.1093/nar/28.12.2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Liang S, Tong X, Ding X, Zheng L, Dai F. 2020. Unusual tertiary pairs in eukaryotic tRNA-Ala. RNA 26: 1519–1529. 10.1261/rna.076299.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchini C, Strippoli P, Biolchi A, Solmi R, Lenzi L, D'Addabbo P, Carinci P, Valvassori L. 2003. The human TruB family of pseudouridine synthase genes, including the Dyskeratosis Congenita 1 gene and the novel member TRUB1. Int J Mol Med 11: 697–704. 10.3892/ijmm.11.6.697 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.