Abstract
The coronavirus disease 2019 (COVID-19) is caused by a novel coronavirus known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is associated with several fatal cases worldwide. The rapid spread of this pathogen and the increasing number of cases highlight the urgent development of vaccines. Among the technologies available for vaccine development, DNA vaccination is a promising alternative to conventional vaccines. Since its discovery in the 1990s, it has been of great interest because of its ability to elicit both humoral and cellular immune responses while showing relevant advantages regarding producibility, stability, and storage. This review aimed to summarize the current knowledge and advancements on DNA vaccines against COVID-19, particularly those in clinical trials.
Keywords: Immunization, DNA vaccine, Nucleic acid-based vaccines, Coronavirus, COVID-19
Graphical abstract
1. Introduction
In the second week of December 2019, patients with an atypical form of pneumonia were diagnosed in Wuhan, Hubei Province, China [1]. At this time, it was detected as a new coronavirus, initially named nCoV-2019. In mid-January, the genome of this pathogen was made public, with its precise definition as “severe acute respiratory syndrome coronavirus 2” (SARS-CoV-2), thus named “coronavirus disease 2019” (COVID-19). This virus has spread rapidly across mainland China and worldwide. On March 11, the World Health Organization declared a pandemic state to the whole world [[2], [3], [4], [5]].
Coronaviruses are a large family of viruses known to cause illnesses in different animals, ranging from the common cold to more severe diseases. As of this year, seven different coronaviruses from alpha and beta genera are known to infect and cause disease in humans. These include the betacoronaviruses that cause severe acute respiratory syndrome (SARS-CoV), the Middle East respiratory syndrome (MERS-CoV) [3], and SARS-CoV-2, all of which were responsible for a relatively high number of cases with high mortality rates. As of mid-August 2020, COVID-19 has already been responsible for more than 20 million infections and 750 thousand deaths [2]. As transmission rates are shown to be higher than other coronaviruses, SARS-CoV-2 is considered a concern for public health, mainly because of the possibility of overloading intensive care units, thus causing health systems to collapse.
Considering this upcoming threat, the majority of countries adopted measures to reduce the transmission rate, which reduced the burden of COVID-19, though causing a significant economic loss globally. Therefore, the search for a vaccine against SARS-CoV-2 was the main topic of research in the world to help restabilize the normal pace [6]. However, no coronavirus vaccine to prevent respiratory infections in humans has been licensed yet [7].
In this context, various groups have made remarkable advances in developing new vaccines in a very short time [8]. Traditional vaccine development methods, although extremely effective in combating highly contagious diseases such as measles, require a large number of active viruses during production and can even take a longer time for the development in case attenuated pathogens are needed. Thus, more advanced vaccine technologies, such as DNA, RNA, subunit, and virus-like particles, have been extensively tested [9]. The aim of this study was to elucidate the current knowledge and discuss the use of DNA vaccines against COVID-19.
2. Vaccine development during the SARS-CoV-2 pandemic
For human vaccine development, regulatory agencies of different countries have historically requested an increasing number of clinical studies with a large number of patients and complexity. These measures assure that the final vaccine product will be safe and effective for different population subgroups before licensure for commercial use. Typically, 10–15 years are required from conducting preclinical studies to obtaining vaccine licensure. The fastest approval, though, was the Ebola vaccine, which took five years [10].
In brief, clinical tests in humans can be divided into four phases: phase 1, wherein the vaccine is given to the healthy volunteers, and its safety and dosing are determined; phase 2, wherein initial immune stimulation is evaluated, and safety is further explored in small numbers of healthy people; phase 3 wherein the vaccine efficacy in preventing the disease is determined after it is given to a large cohort; and phase 4 which is conducted after vaccine approval to guarantee its safety and study long-term effects [11]. The necessity of these phases is unanimous in the vaccinology field. However, the time required for each of these steps needs to be better planned to fasten the process and obtain the approval of a vaccine during a pandemic situation.
The vast majority of viral vaccines currently licensed for humans can be categorized as virus or protein based. The virus-based group consists of an inactivated or live-attenuated virus, and even though these vaccines are more immunogenic, several limitations are associated with these approaches. In the case of SARS-CoV-2, large quantities of viruses need to be produced under biosafety level 3 conditions for an inactivated vaccine, and extensive safety testing is required to ensure they do not revert to infective [12]. In contrast, subunit vaccines such as those with purified proteins present higher safety and scalability than whole-pathogen vaccines. Nevertheless, they may show less immunogenicity and require multiple immunizations [13,14].
The current life-threatening infectious diseases, such as influenza (H5N1 subtype), Zika, Ebola, and MERS-CoV, are driving the demand for new, faster, and effective vaccine platforms [15,16]. Considering recent experiences from the SARS-CoV-2 pandemic, next-generation platforms must allow having a vaccine formulation useful for clinical trials in less than 16 weeks from genome sequencing to application in humans. Furthermore, these technologies elicit consistent immune responses across different pathogens and are suitable for large-scale manufacturing even as a pathogen-agnostic platform. Multiple approaches are still under development; however, the DNA- and RNA-based approaches are among those with the greatest potential for speeding up the production of effective vaccines when urgently needed [12,17].
On August 11, 2020, Russia was the first country to approve a vaccine against COVID-19 named “Sputnik V,” formerly known as the “Gam-COVID-Vac,” which is based on a viral vector technology. However, the world scientific community has raised doubts about its clinical trials to prove its safety and efficacy [11]. Although there is a consensus that developing a safe and effective vaccine against SARS-CoV-2 is crucial to end the COVID-19 pandemic, cautious measures are necessary to ensure that new vaccines are safe for the entire population [18]. In addition, efficacy and safety are not the only crucial criteria, as a vaccine candidate must also have scalable production. Thus, it is necessary to select an adequate vaccine platform for accelerating vaccine development.
3. DNA vaccine
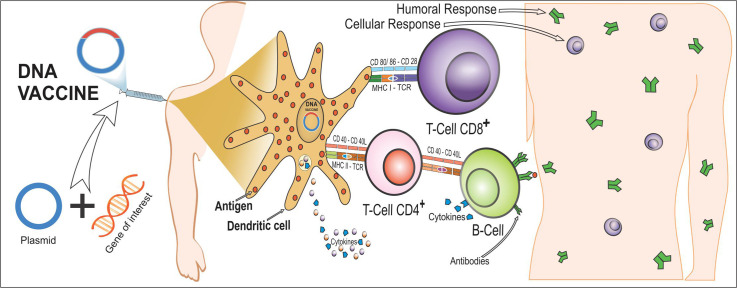
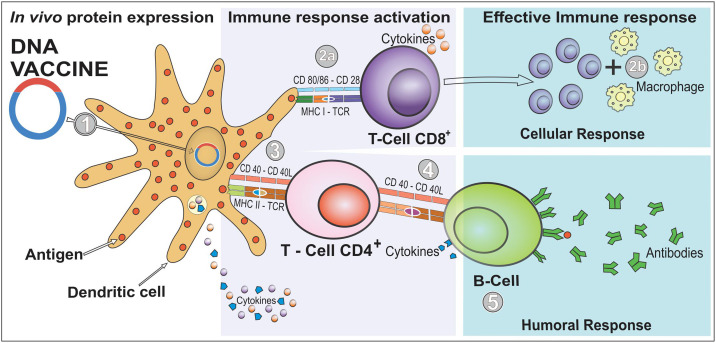
Immunization with nucleic acids has received considerable attention in the field of new generation vaccines. The first proof of concept of a DNA vaccine was made in 1990 and involved the injection of RNA or DNA molecules, expressing chloramphenicol acetyltransferase, luciferase, and beta-galactosidase into mouse skeletal muscle, and the expression of reporter genes in vivo, which can be detected for up to two months after infection [19,20]. In brief, DNA vaccine consists of delivering genes or fragments of it, encoding immunogenic antigens to the host's cells by using DNA plasmids as a vector. This approach induces both humoral and cell-mediated immune responses efficiently [21,22]. The vaccine formulation is made such that the genetic material is translocated to the host's cell nucleus (Fig. 1 ). Once it reaches there, the mammalian promoter present in the vector structure is activated, triggering the transcription of the gene used for the vaccine through the host's cellular machinery. The antigen-presenting cells (APCs) are the major target cells to receive the genetic material. In addition, myocytes have been reported to play a crucial role [19]. After the translation of the translocated gene into a protein or protein fragment, it is further processed into peptides that bind to major histocompatibility complex (MHC) class I or II. Cells other than APC, such as the myocytes, use MHC-I for the antigen presentation, and APC, such as dendritic cells (DCs), can use MHC-II, resulting in cross-priming and presentation of antigens to both CD4+ and CD8+ T cells [[23], [24], [25], [26]]. Regarding COVID-19 immune regulation, a recent study showed that at-risk patients with pericardial effusion with a worse prognosis show elevated CD3+CD8+ T cells together with reduced Tregs and CD14+HLA-DR+ monocytes [26]. These findings show that the main course of the disease occurs because of a misbalance of the immune response, often leading to misregulation and worsening of the infection [27]. Thus, the vaccines in development against COVID-19 aim to build a proper and effective immune response without causing such misbalance. Of note, not only this cellular immune response can be activated, but also humoral responses can be triggered if the produced immunogen is released from the cells and recognized by B cell receptors [25].
Fig. 1.
Induction of cellular and humoral immunity after immunization with DNA vaccines. A DNA vaccine consists of a plasmid produced in bacteria that encodes the protein of interest (an antigen) in the presence of a mammalian promoter. It is placed in a way that it reaches the cell nucleus, enabling the transcription and translation in the transfected human cells (step 1). After the plasmid uptake in vivo, the encoded protein is expressed in the host's cells, and the vaccine antigen can be then presented to antigen-presenting cells (APCs), such as dendritic cells (DCs), through the major histocompatibility complex (MHC) pathways and be presented to activate naïve T cells. CD8+ T cell immunity is predominantly activated by endogenously expressed antigens presented on MHC class I molecules (step 2a). The active CD8+ T cell stimulates the release of cytokines (e.g., interferon-gamma [IFN-γ] and tumor necrosis factor-alpha [TNF-α]) that inhibit viral replication and increase the expression of MHC I molecules. Therefore, macrophages are also activated to support cell-mediated immune responses (step 2b). However, CD4+ T helper cell activation is triggered through MHC class II from APC (step 3). In case the vaccine proteins are secreted, these targets are recognized by B cell receptors in naïve B cells, which also use MHC-II to get activated (step 4). In this immune pathway, activated B cells will produce different classes of antibodies (mainly IgG) to protect against the disease (step 5). Furthermore, immunization with DNA vaccine expresses proinflammatory cytokines and chemokines. DCs are responsible for producing IL-10, IL-12, and TNF-α that induce the cellular response by activating CD8+ T and IL-4 is involved in activating CD4+ T.
In addition, intrinsic elements of plasmid DNA, such as CpG unmethylated sequences, can activate innate immune responses, thereby enhancing adaptive immune responses against the expressed antigens. Although clinical trials using DNA vaccines in humans induced both cellular and humoral responses, these responses are often not sufficient to elicit significant clinical benefits. Therefore, DNA vaccines have only been licensed for use in veterinary medicine [21,24,25]. Because of this limitation, several research lines focus on DNA vaccine optimization and delivery, including promoter design, codon optimization, adjuvants, use of electroporation, prime/boost immunization, or “omics” approaches for refined vaccine design [28].
Compared with traditional live or attenuated vaccines, DNA vaccines have several advantages, such as induction of broad immune responses without any risk being associated with replicating microorganisms; stimulation of both cellular and humoral immunity; construction of a vector encoding different antigens in a single vaccine; efficient large-scale, low-cost, production; and high storage stability [21,29]. In the vaccinology field, storage is a crucial factor, as preserving the high quality of the vaccine contents and, thus, the protective potential is necessary. Hence, cold storage is essential to ensure the survival of live vaccines and preserve their content. On the other hand, DNA vaccines are highly stable and have less need for refrigeration, which may be highly practical for use in endemic areas [23].
4. SARS-CoV-2
The SARS-CoV-2 has a single-stranded, positive-sense RNA genome with approximately 26–32 kilobases in size. This virus belongs to the family Coronaviridae (order Nidovirales). The Coronaviridae family contains four genera to include alphacoronavirus (alphaCoV), betacoronavirus (betaCoV), deltacoronavirus (deltaCoV), and gammacoronavirus (gammaCoV). Although bats and rodents are believed to be the reservoir for alphaCoV and betaCoV, it is less clear, which animals can be the reservoir for deltaCoV and gammaCoV [3,30].
Among the coronaviruses that infect humans, two betaCoV (HCoV-229E and HCoV-HKU1) and two alphaCoV (HCoV-NL63 and HCoV-OC43) circulate among the population during the past few decades and were identified as causative agents of approximately one-third of the common colds [31]. It has been reported that betaCoV used 9-O-acetylsialic acids as a receptor and alphaCoV used host proteins, including polypyrimidine tract binding and hnRNP-A1 as receptors [31].
In addition to these common cold viruses, the three CoVs responsible for high mortality rates in humans, i.e., MERS-CoV, SARS-CoV, and SARS-CoV-2, are from betaCoV genus. The genome sequencing of SARS-CoV-2 has shown an overall similarity of 98% with the bat CoV RaTG13 [32]. Moreover, SARS-CoV-2 can enter the host's cells through different mechanisms, including an endosomal and nonendosomal entry through the action of proteases. Currently, the main receptor for SARS-CoV-2 entry in humans is angiotensin-converting enzyme 2 (ACE2), which is also used by SARS-CoV [[32], [33], [34], [35]].
Structurally, SARS-CoV-2 contains four structural proteins, including spike (S), envelope (E), membrane (M), and nucleocapsid (N). Moreover, these proteins share high sequence similarity to those from SARS-CoV and MERS-CoV [32]. Thus, the knowledge from SARS-CoV and MERS-CoV vaccines provides some insights and lessons concerning the development of SARS-CoV-2 vaccine design and thus helps in accelerating the development of new vaccines for COVID-19 [18]. Some successful DNA vaccines expressing S, M, and N proteins have been developed against SARS-CoV. The obtained results confirmed the strong protective humoral and cellular immune responses in mice, macaques, and camels [[36], [37], [38], [39], [40]]. Furthermore, the first DNA vaccine candidate against MERS-CoV to enter clinical trials, named GLS-5300, was well tolerated with no vaccine-associated serious adverse events. Immune responses were dose-independent, detected in more than 85% of participants after two immunization regimens, and durable through one year of follow-up [41,42].
5. The immune response and COVID-19
The immune system can be categorized into innate immunity (rapid and nonspecific response) and adaptive immunity (slow and specific response). The adaptive immunity can be further divided into cellular responses mainly characterized by T cell maturation and humoral responses characterized by B cell maturation. Considering this, vaccines are prepared to induce both arms of the adaptive immune system and stimulate a sufficient number of memory T and B cells. Furthermore, immunity can be divided into two types: active and passive. Active immunity refers to the process of exposing a patient to an antigen to generate an adaptive immune response, and passive immunity refers to the transfer of antibodies from one individual to another [43].
Moreover, passive immunity can occur naturally when maternal antibodies are transferred to the fetus through the placenta or from breast milk to the gut of the infant [43,44]. Regarding immunity against COVID-19, it is still not completely clear, which responses occur in natural COVID-19, and if people who recover from COVID-19 infection are protected from a second infection [45,46].
SARS-CoV-2 is associated with a robust adaptive immune response of both T and B cells. In addition, both immunoglobulin (Ig)M and IgG antibodies are produced mainly against N and S proteins. The antibodies appear around the 10th day of infection, and most patients seroconvert within three weeks. If reinfections occur, it would indicate that immune response against SARS-CoV-2 is not protective and, therefore, the use of vaccines could not be associated with protection [47].
On the other hand, SARS-CoV-2 antibodies are found to be protective, although it remains uncertain if a high level of circulating antibodies lasts for enough duration to avoid further infections. This question still cannot be properly answered, as SARS-CoV-2 has been in the community only for the past few months. However, results obtained with other closely related coronaviruses, mainly SARS-CoV and MERS-CoV, showed robust, long-lasting T and B cell immune responses [[48], [49], [50]].
6. DNA vaccine in clinical trials
As of January 2020, the complete genome of SARS-CoV-2 was published. Understanding the genome structure of the virus is a critical step for the development of new vaccines [33,51]. During infection, antibodies are raised mainly against both N and S proteins. The N protein covers the viral genome and is also involved in the release of virus particles from cells. Whereas the S protein plays a significant role in pathogenesis by binding to the host cell through its receptor-binding domain and thus initiating the infection to the host cell [52]. The S protein has 1273 amino acid residues and can be divided into three subunits, S1, S2, and S2', each having a different role during the adherence to the host cell [52].
The S1 subunit is involved in the attachment of virions to the host cell membrane by interacting with human ACE2, which initiates the infection process. Furthermore, during this process, the S protein undergoes conformational changes induced by its entry into the endosomes of the host's cell. The S2 subunit acts as a fusion protein that helps in the union of the viral and host cell membranes [53,54]. During the fusion process, the S2 protein appears in three consecutive conformational states: 1) prefusion (native state), 2) prehairpin (intermediate state), and 3) ensuing postfusion (hairpin state). Subsequently, surface proteases cleave S2. SARS-CoV-2 S protein has a furin site as an additional cleavage spot, which may be associated with the broader infectivity in comparison with SARS-CoV [55]. Understanding these dynamic conformation states that are associated with the mechanism of viral entry into the host cell membrane could lead to the development of effective therapeutics [32,56].
The S protein has been used as the antigen of all DNA vaccines currently being tested in clinical trials (Table 1 ). One of them, named AG0301-COVID19 (ClinicalTrials.gov number, NCT04463472), uses a two-immunization scheme, the first with a low dose (1.0 mg) and the second with a high dose (2.0 mg). Both injections are administered intramuscularly within a two-week interval. At present, healthy individuals aged between 20 and 65 years are being recruited to evaluate the immunogenicity of this vaccine.
Table 1.
Overview of the ongoing clinical trials of nucleic acid vaccines against COVID-19 (assess at ClinicalTrials.gov as of November 02, 2020).
| Study start date/study identifier | Technology | Study phase/ECD | Project title | Immunogen | Via | Subjects | Number of subjects | Study location |
|---|---|---|---|---|---|---|---|---|
| June 17, 2020/NCT04445389 | DNA vaccine | Phase 1–2 June 2022 |
Safety and Immunogenicity Study of GX-19, a COVID-19 Preventive DNA Vaccine in Healthy Adults | S protein | IM | Adults (18–50 years old) | 210 | Korea |
| July 29, 2020/NCT04463472 | DNA vaccine | Phase 1–2 July 2021 |
Study of COVID-19 DNA Vaccine (AG0301-COVID19) | S protein | IM | Adults (20–65 years old) | 30 | Japan |
| August 31, 2020/NCT04527081 | DNA vaccine | Phase 1–2 September 2021 |
Study of COVID-19 DNA Vaccine (AG0302-COVID19) | S protein | IM | Adults (20–65 years old) | 30 | Japan |
| April 3, 2020/NCT04336410 | DNA vaccine | Phase 1 July 2021 |
Safety, Tolerability and Immunogenicity of INO-4800 for COVID-19 in Healthy Volunteers | S protein | ID (EP) | Adults (18 years and older) | 120 | United States |
| July 2020/NCT04334980 | DNA vaccinea | Phase 1/ February 2022 |
Evaluating the Safety, Tolerability and Immunogenicity of bacTRL-Spike Vaccine for Prevention of COVID-19 | S protein | oral | Adults (18 years and older) | 12 | Australia |
| November 2020/NCT04591184 | DNA vaccine | Phase 1 June 2021 |
A Clinical Trial of a Plasmid DNA Vaccine for COVID-19 [Covigenix VAX-001] in Adults | S protein | IM | Adults (18 to 84 years old) | 72 | Canada |
| March 16, 2020/NCT04470427 | mRNA vaccineb | Phase 3 October 2022 |
Safety and Immunogenicity Study of 2019-nCoV Vaccine (mRNA-1273) for Prophylaxis of SARS-CoV-2 Infection (COVID-19) | S protein | IM | Adults (18 years to 99 Years old) | 30,000 | United States |
| June 18, 2020/NCT04515147 | mRNA vaccine | Phase 2 November 2021 |
A Study to Evaluate the Safety, Reactogenicity and Immunogenicity of Vaccine CVnCoV in Healthy Adults | S protein | IM | Adults (18–60 years old) | 691 | Germany |
| April 23, 2020/NCT04368728 | mRNA Vaccineb | Phase 2–3 December 2022 |
A Trial Investigating the Safety and Effects of Four BNT162 Vaccines Against COVID-2019 in Healthy Adults | S protein | IM | Adults (18–85 years old) | 29,481 | Germany |
| January 2021/NCT04566276 | mRNA vaccineb | Phase 1 June 2021 |
ChulaCov19 mRNA Vaccine in Healthy Adults | S protein | IM | Adults (18–75 years old) | 96 | Thailand |
ECD, Estimated Study Completion Date; IM, intramuscular; EP, electroporation; ID, intradermal.
Bifidiobacterium longum.
Lipid nanoparticle-encapsulated mRNA.
The biotechnology company (Inovio Pharmaceuticals, Plymouth Meeting, PA, USA) has previously developed experimental vaccines against MERS-CoV (INO-4700) and is currently evaluating a DNA vaccine against COVID-19. A plasmid pGX9501 designed to encode the SARS-CoV-2 S protein has been evaluated as an antigen. The INO-4800 vaccine induced both cellular and humoral immune responses that were observed within days following a single immunization in mice and guinea pigs during preclinical testing [57].
Therefore, the company started a phase 1, open-label study to evaluate the safety, tolerability, and immunogenicity of INO-4800. This vaccine is administered intradermally through electroporation [41,57]. Moreover, a preliminary study showed that INO-4800 induced neutralizing antibodies that blocked the binding of SARS-CoV-2 S protein to the host receptor ACE2 [57].
Electroporation uses short electrical pulses at the vaccine application site, resulting in an increased cell membrane permeability; improved absorption of the antigen; and, consequently, a more effective immune response [58]. Moreover, electroporation was associated with an enhancement of immune response by recruiting inflammatory cells and APCs to the application site [59].
Vaccine vectors have several advantages: low cost, noninvasive administration, and high safety levels [60]. The bacTRL-Spike vaccine uses live, recombinant Bifidobacterium longum that contains synthetic plasmid DNA encoding the S protein of SARS-CoV-2. Bifidobacterium is a nonpathogenic anaerobic bacterium that is part of the human microbiota. In addition, it has been proposed that this bacterium improves the host's endurance by increasing the immune response against viral infection [61].
Some strains of Bifidobacterium were tested as carriers of antigens for its use as a recombinant vaccine against hepatitis C virus, enterovirus, and cancer [60,62,63]. A clinical trial was designed to evaluate the safety and tolerability of orally immunized bacTRL-Spike vaccine in healthy adults. However, this study has not yet started recruiting, and the estimated completion date is December 31, 2021 (ClinicalTrials.gov number, NCT04334980). Moreover, the safety and immunogenicity of GX-19, another DNA vaccine against COVID-19 that uses intramuscular immunizations are already in clinical trials.
7. mRNA vaccines: a fast and consistent strategy to control COVID-19
Over the past decade, major technological innovation and research investment have enabled mRNA to become a promising therapeutic tool in the fields of vaccine development. At present, multiple mRNA vaccine platforms against infectious diseases and several types of cancer have demonstrated encouraging results in both animal models and humans. Moreover, the area of mRNA vaccine is very rapidly developing. The mRNA vaccine does not need to reach the cell nucleus like the DNA ones, which is one of the potential practical advantages [64]. Thus, the mode of application and effectiveness of mRNA vaccines may be increased [65]. In fact, several preclinical studies and human clinical trials are using mRNA technology, particularly during the SARS-CoV-2 pandemic [66,67].
The mRNA-based vaccines comprise mRNA that encodes a protein antigen. Although RNA is known to be a relatively unstable molecule, novel vaccine designs were developed to improve its stability and protein translation efficiency, which enhanced immune response. The mRNA-1273 (Moderna Inc. Cambridge, MA, USA) was the first mRNA vaccine to be designed against COVID-19 and has achieved a time record of 63 days from vaccine design to human trials. Phase 1 clinical trials in the USA were already published, whereas phases 2 and 3 are either analyzing results or ongoing (ClinicalTrials.gov number, NCT04470427). This vaccine uses lipid nanoparticle (LNP)-encapsulated mRNA that encodes for a full-length, prefusion stabilized S protein of SARS-CoV-2 [[68], [69], [70]].
Previous studies with similar formulations have demonstrated that the delivery of the mRNA vaccine has been optimized by using LNPs for intramuscular or intradermal administration [71,72]. Data showed that mRNA-1273 induced both potent neutralizing antibody and CD8 T cell responses. In addition, it protected against SARS-CoV-2 infection in the lungs and nose of mice without evidence of immunopathology [73,74].
The mRNA-1273 vaccine is currently in phase 3 clinical trial. A preliminary report published after phase 1 showed that the vaccine-induced anti–SARS-CoV-2 immune responses in 45 participants. Antibody responses were increased with a higher dose (250 μg) after the first vaccination. Moreover, after the second vaccination, neutralizing antibodies were detected in all evaluated participants, with generally similar values to those in the upper half of the distribution of a panel of control convalescent serum specimens. Adverse events such as fatigue, chills, headache, myalgia, and pain at the injection site occurred in more than half of the participants. Moreover, systemic adverse events were more common after the second vaccination, particularly with the highest dose [73,75]. Another mRNA vaccine, called CVnCoV (ClinicalTrials.gov number, NCT04515147), encoding the full-length S protein and formulated with LNPs is recruiting volunteers (as of August 2020).
Another mRNA vaccine, the BNT162 (BioNTech, Mainz, Germany) is in advanced clinical trials and has four variants, namely, a1, b1, b2, and c2. It is another LNP mRNA vaccine that recently made the results of the phase 1/2 trial public but not yet peer reviewed [76]. In this study, only the BNT162b1 variant was tested in a prime-boost regimen within 21 days in three different doses [77]. Similar to mRNA-1273, the adverse events were dose-dependent and only mild to moderate, while the immune response showed a high level of neutralizing antibodies. Overall, these data suggest the potential to develop DNA and mRNA vaccines that are easier to design and can quickly proceed into clinical trials, which will be helpful for pandemic states such as the one caused by COVID-19 [77].
8. Nucleic acid-based vaccines and SARS-CoV-2
Immunoinformatics is a branch of bioinformatics used to design vaccines against several infectious diseases [[78], [79], [80]]. This approach involves computational analysis of immunological data by predicting appropriate antigens, epitopes, carriers, and adjuvants for vaccine development. Therefore, immunoinformatics can reduce the time and cost of vaccines [81,82]. Considering this, the design of novel multiepitope mRNA vaccines consisting of cytotoxic T lymphocyte, helper T lymphocyte, and linear B lymphocyte epitopes derived from SARS-CoV-2 S protein, as well as adjuvant highly immunogenic, have been analyzed [53,81,[83], [84], [85]].
Furthermore, different DNA vaccine candidates expressing different forms of the SARS-CoV-2 S protein have been evaluated in 35 rhesus macaques. Vaccinated animals developed humoral and cellular immune responses, including neutralizing antibody titers that are comparable to those found in convalescent humans [86]. From a general perspective, although mRNA vaccines quickly advanced to clinical trials in humans, the majority of candidate DNA vaccines are currently in the preclinical stage.
9. Considerations and challenges
COVID-19 is an unusual global health threat wherein the vaccine is needed immediately. Because of the high risk of collapsing healthcare systems in several countries, governments had to implement lockdown measures, including no international travel and other public containment measures in attempting to reduce the virus morbidity and mortality worldwide [[87], [88], [89]]. Although effective, these actions led to vast economic devastation.
Until date, there is no effective drug to reduce the infection and pandemic burden [90,91]. However, exceptional efforts have been made in attempting to develop an effective and safe vaccine at the earliest, which should be available to all countries affected by the pandemic at an affordable price [92,93]. Previous advancements made since the SARS and MERS outbreaks have accelerated our understanding of the epidemiology, pathogenesis, and diagnosis of SARS-CoV-2, as well as the development of therapies to treat viral infection and possible vaccines [68].
In this pandemic situation, it is crucial to ensure that rigorous and adequate clinical trials are performed to evaluate drugs with antiviral effects for avoiding the usage of ineffective and unsafe drugs [68]. Current clinical trials of candidate vaccines are undergoing phase 1 or parallel 1/2 studies to initiate phase 3 at the earliest and decrease the time required for the development of the new vaccine [74].
Although advances in genetic sequencing and other technological developments have sped up the establishment of various vaccine platforms, several uncertainties still remain. Questions have arisen regarding the mutation rates of SARS-CoV-2, which could lead to immune evasion. Understanding mutations in the coding and non-coding regions, genetic diversity, pathogenicity, and host-pathogen interactions is essential. Mutations in the S protein seem to induce conformational changes, which may alter antigenicity and, thus, may affect the vaccine design [32,94,95]. Moreover, previous studies suggested that various mutations in the target proteins of the coronaviruses can be associated with drug resistance and changes in the protein structures of target proteins that may lead to vaccine inefficacy [32].
It is crucial to highlight that the uncertainty over long-lasting protection against COVID-19 still remains. Patients with reinfection have been reported, and it is necessary to determine how long a protective immune response can be maintained in an individual [96,97]. Considering the immunogenicity of the SARS-CoV-2 mRNA-1273 vaccine in older adults, it has been reported that after the second immunization, serum neutralizing activity was detected in all the participants, and binding and neutralizing antibody responses appeared to be similar to those described among vaccine recipients between the ages of 18 and 55 years and were above the median of a panel of control individuals who had donated convalescent serum [98]. Ideally, vaccination would induce long-lived immunity, but annual vaccination would be feasible based on experiences with the annual influenza vaccine [12].
In the current pandemic, several specialists have suggested that immune responses against SARS-CoV-2 could lead to antibody-dependent enhancement (ADE) [99]. Although this phenomenon has major importance for Flavivirus and some feline coronaviruses, SARS-CoV and SARS-CoV-2 have shown not to cause this effect in living animals or humans [100]. However, if ADE becomes crucial during the current pandemic, both DNA- and RNA-based platforms could quickly provide alternatives to circumvent such issues, as regions or motifs of the vaccine antigen responsible for causing ADE could be easily engineered or removed [101].
The great advantage of the current advancements in nucleic acid-based technologies for vaccine development is the short time required from the design to clinical trials. Therefore, it may be soon possible to test together, in the same vaccine, different variants of antigens that cover circulating mutations. This would represent a major step forward in vaccine development against rapidly emerging threats such as the current SARS-CoV-2 pandemic. Therefore, developing vaccines against COVID-19 require further studies on gene mutations and how to avoid vaccine failure because of them.
Declaration of competing interest
The authors have declared no conflict of interest.
References
- 1.Ghaebi M, Osali A, Valizadeh H, Roshangar L, Ahmadi M. Vaccine development and therapeutic design for 2019-nCoV/SARS-CoV-2: challenges and chances. J. Cell. Physiol.. 2020. [DOI] [PMC free article] [PubMed]
- 2.Bhagavathula AS, Aldhaleei WA, Rovetta A, Rahmani J. Vaccines and drug therapeutics to lock down novel coronavirus disease 2019 (COVID-19): a systematic review of clinical trials. Cureus. 2020;12:e8342. [DOI] [PMC free article] [PubMed]
- 3.Abd El-Aziz TM, Stockand JD. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2) - an update on the status. Infection, genEtics and Evolution : Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases. 2020;83:104327. [DOI] [PMC free article] [PubMed]
- 4.Alsuliman T., Alasadi L., Alkharat B., Srour M., Alrstom A. A review of potential treatments to date in COVID-19 patients according to the stage of the disease. Current Research in Translational Medicine. 2020;68(3):93–104. doi: 10.1016/j.retram.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO . 11 March 2020. Director-General’s Opening Remarks at the Media Briefing on COVID-19. [Google Scholar]
- 6.Covian C, Retamal-Diaz A, Bueno SM, Kalergis AM. Could BCG vaccination induce protective trained immunity for SARS-CoV-2? Front. Immunol.. 2020;11:970. [DOI] [PMC free article] [PubMed]
- 7.Ong E, Wong MU, Huffman A, He Y. COVID-19 Coronavirus Vaccine Design Using Reverse Vaccinology and Machine Learning. bioRxiv : The Preprint Server for Biology. 2020. [DOI] [PMC free article] [PubMed]
- 8.de Alwis R., Chen S., Gan E.S., Ooi E.E. Impact of immune enhancement on Covid-19 polyclonal hyperimmune globulin therapy and vaccine development. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calina D, Docea AO, Petrakis D, Egorov AM, Ishmukhametov AA, Gabibov AG, et al. Toward effective COVID19 vaccines: updates, perspectives and challenges (review). Int. J. Mol. Med.. 2020;46:3–16. [DOI] [PMC free article] [PubMed]
- 10.Steve Blacka DEB, David C. Kaslow, Simone Pecetta, Rino Rappuolide. Transforming vaccine development. Semin. Immunol.. 2020. [DOI] [PMC free article] [PubMed]
- 11.Rab S, Afjal, Javaid M, Haleem A, Vaishya R. An update on the global vaccine development for coronavirus. Diabetes & metabolic syndrome. 2020;14:2053–5. [DOI] [PMC free article] [PubMed]
- 12.van Riel D., de Wit E. Next-generation vaccine platforms for COVID-19. Nat. Mater. 2020;19:810–812. doi: 10.1038/s41563-020-0746-0. [DOI] [PubMed] [Google Scholar]
- 13.Brisse M., Vrba S.M., Kirk N., Liang Y., Ly H. Emerging concepts and technologies in vaccine development. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.583077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mubarak A, Alturaiki W, Hemida MG. Middle East respiratory syndrome coronavirus (MERS-CoV): infection, immunological response, and vaccine development. J Immunol Res. 2019;2019:6491738. [DOI] [PMC free article] [PubMed]
- 15.Cockrell A.S., Baric R.S. An effective DNA vaccine platform for Middle East respiratory syndrome coronavirus. Annals of Translational Medicine. 2016;4:499. doi: 10.21037/atm.2016.11.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flemming A. New vaccine platform? Nat. Rev. Drug Discov. 2010;9:191. doi: 10.1038/nrd3233. [DOI] [PubMed] [Google Scholar]
- 17.Lurie N, Saville M, Hatchett R, Halton J. Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 2020;382:1969–73. [DOI] [PubMed]
- 18.Linares-Fernandez S., Raguindin P.F. Vaccine development in the SARS-CoV-2 pandemic: a balancing act on accuracy and speed. International Journal of Public Health. 2020;65:1433–1434. doi: 10.1007/s00038-020-01511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Petrovsky N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Review of Vaccines. 2016;15:313–329. doi: 10.1586/14760584.2016.1124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 21.Silveira M.M., Oliveira T.L., Schuch R.A., McBride A.J.A., Dellagostin O.A., Hartwig D.D. DNA vaccines against leptospirosis: A literature review. Vaccine. 2017;35:5559–5567. doi: 10.1016/j.vaccine.2017.08.067. [DOI] [PubMed] [Google Scholar]
- 22.Lee LYY, Izzard L, Hurt AC. A review of DNA vaccines against influenza. Front. Immunol.. 2018;9:1568. [DOI] [PMC free article] [PubMed]
- 23.Ingolotti M., Kawalekar O., Shedlock D.J., Muthumani K., Weiner D.B. DNA vaccines for targeting bacterial infections. Expert Review of Vaccines. 2010;9:747–763. doi: 10.1586/erv.10.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coban C., Kobiyama K., Jounai N., Tozuka M., Ishii K.J. DNA vaccines: a simple DNA sensing matter? Hum Vaccin Immunother. 2013;9:2216–2221. doi: 10.4161/hv.25893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobernik D, Bros M. DNA vaccines-how far from clinical use? Int. J. Mol. Sci.. 2018;19. [DOI] [PMC free article] [PubMed]
- 26.Duerr G.D., Heine A., Hamiko M., Zimmer S., Luetkens J.A., Nattermann J., et al. Parameters predicting COVID-19-induced myocardial injury and mortality. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L., Saade F., Petrovsky N. The future of human DNA vaccines. J. Biotechnol. 2012;162:171–182. doi: 10.1016/j.jbiotec.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y., Yuen P.W., Lam J.K. Intranasal DNA vaccine for protection against respiratory infectious diseases: the delivery perspectives. Pharmaceutics. 2014;6:378–415. doi: 10.3390/pharmaceutics6030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randhawa GS, Soltysiak MPM, El Roz H, de Souza CPE, Hill KA, Kari L. Machine learning using intrinsic genomic signatures for rapid classification of novel pathogens: COVID-19 case study. PLoS One 2020;15:e0232391. [DOI] [PMC free article] [PubMed]
- 31.Hazafa A, Ur-Rahman K, Haq IU, Jahan N, Mumtaz M, Farman M, et al. The broad-spectrum antiviral recommendations for drug discovery against COVID-19. Drug Metab. Rev.. 2020:1–17. [DOI] [PMC free article] [PubMed]
- 32.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochimica et Biophysica Acta Molecular Basis of Disease. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amawi H, Abu Deiab GI, AA AA, Dua K, Tambuwala MM. COVID-19 pandemic: an overview of epidemiology, pathogenesis, diagnostics and potential vaccines and therapeutics. Ther. Deliv.. 2020;11:245–68. [DOI] [PMC free article] [PubMed]
- 34.Bourgonje AR, Abdulle AE, Timens W, Hillebrands JL, Navis GJ, Gordijn SJ, et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J. Pathol.. 2020. [DOI] [PMC free article] [PubMed]
- 35.Albini A, Di Guardo G, Noonan DM, Lombardo M. The SARS-CoV-2 receptor, ACE-2, is expressed on many different cell types: implications for ACE-inhibitor- and angiotensin II receptor blocker-based cardiovascular therapies. Intern. Emerg. Med.. 2020. [DOI] [PMC free article] [PubMed]
- 36.Al-Amri S.S., Abbas A.T., Siddiq L.A., Alghamdi A., Sanki M.A., Al-Muhanna M.K., et al. Immunogenicity of candidate MERS-CoV DNA vaccines based on the spike protein. Sci. Rep. 2017;7 doi: 10.1038/srep44875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roper R.L., Rehm K.E. SARS vaccines: where are we? Expert Review of Vaccines. 2009;8:887–898. doi: 10.1586/erv.09.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheung Y.K., Cheng S.C., Sin F.W., Chan K.T., Xie Y. Induction of T-cell response by a DNA vaccine encoding a novel HLA-A*0201 severe acute respiratory syndrome coronavirus epitope. Vaccine. 2007;25:6070–6077. doi: 10.1016/j.vaccine.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muthumani K., Falzarano D., Reuschel E.L., Tingey C., Flingai S., Villarreal D.O., et al. A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aac7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modjarrad K. MERS-CoV vaccine candidates in development: the current landscape. Vaccine. 2016;34:2982–2987. doi: 10.1016/j.vaccine.2016.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modjarrad K., Roberts C.C., Mills K.T., Castellano A.R., Paolino K., Muthumani K., et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019;19:1013–1022. doi: 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon I.K., Kim J.H. First clinical trial of a MERS coronavirus DNA vaccine. Lancet Infect. Dis. 2019;19:924–925. doi: 10.1016/S1473-3099(19)30397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcotte H., Hammarström L. Passive immunization toward magic bullets. Mucosal Immunol. 2015;2:1403–1434. [Google Scholar]
- 44.Homberger F.R. Maternally-derived passive immunity to enterotropic mouse hepatitis virus. Arch. Virol. 1992;122:133–141. doi: 10.1007/BF01321123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang H, Wang Y, Tong Z, Liu X. Retest positive for SARS-CoV-2 RNA of “recovered” patients with COVID-19: persistence, sampling issues, or reinfection? J. Med. Virol.. 2020. [DOI] [PMC free article] [PubMed]
- 46.Ota M. Will we see protection or reinfection in COVID-19? Nat. Rev. Immunol. 2020;20:351. doi: 10.1038/s41577-020-0316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khuroo MS, Khuroo M, Khuroo MS, Sofi, AA, Khuroo NS. COVID-19 vaccines: a race against time in the middle of death and devastation! Journal of clinical and experimental hepatology. 2020. [DOI] [PMC free article] [PubMed]
- 48.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yong CY, Ong HK, Yeap SK, Ho KL, Tan WS. Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front. Microbiol.. 2019;10:1781. [DOI] [PMC free article] [PubMed]
- 50.Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang ML, et al. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with high attack rate. J. Clin. Microbiol.. 2020. [DOI] [PMC free article] [PubMed]
- 51.Xiong X, Tortorici MA, Snijder J, Yoshioka C, Walls AC, Li W, et al. Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections. J. Virol.. 2018;92. [DOI] [PMC free article] [PubMed]
- 52.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 53.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., et al. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12 doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–92 e6. [DOI] [PMC free article] [PubMed]
- 56.Qing E., Gallagher T. SARS coronavirus Redux. Trends Immunol. 2020;41:271–273. doi: 10.1016/j.it.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villarreal D.O., Talbott K.T., Choo D.K., Shedlock D.J., Weiner D.B. Synthetic DNA vaccine strategies against persistent viral infections. Expert Review of Vaccines. 2013;12:537–554. doi: 10.1586/erv.13.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J., Kjeken R., Mathiesen I., Barouch D.H. Recruitment of antigen-presenting cells to the site of inoculation and augmentation of human immunodeficiency virus type 1 DNA vaccine immunogenicity by in vivo electroporation. J. Virol. 2008;82:5643–5649. doi: 10.1128/JVI.02564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Z., Huang Z., Sao C., Huang Y., Zhang F., Ma G., et al. Oral immunization of mice using Bifidobacterium longum expressing VP1 protein from enterovirus 71. Arch. Virol. 2013;158:1071–1077. doi: 10.1007/s00705-012-1589-z. [DOI] [PubMed] [Google Scholar]
- 61.Saavedra J.M., Bauman N.A., Oung I., Perman J.A., Yolken R.H. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet. 1994;344:1046–1049. doi: 10.1016/s0140-6736(94)91708-6. [DOI] [PubMed] [Google Scholar]
- 62.Takei S., Omoto C., Kitagawa K., Morishita N., Katayama T., Shigemura K., et al. Oral administration of genetically modified Bifidobacterium displaying HCV-NS3 multi-epitope fusion protein could induce an HCV-NS3-specific systemic immune response in mice. Vaccine. 2014;32:3066–3074. doi: 10.1016/j.vaccine.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 63.Shirakawa T., Kitagawa K. Antitumor effect of oral cancer vaccine with Bifidobacterium delivering WT1 protein to gut immune system is superior to WT1 peptide vaccine. Hum Vaccin Immunother. 2018;14:159–162. doi: 10.1080/21645515.2017.1382787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verbeke R, Lentacker I, De Smedt S, Dewitte H. Three decades of messenger RNA vaccine development. Nano Today 2019:100766.
- 65.Jackson N., Kester K., Casimiro D., Gurunathan S., DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. npj Vaccines. 2020;5 doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rauch S, Jasny E, Schmidt KE, Petsch B. New vaccine technologies to combat outbreak situations. Front. Immunol.. 2018;9:1963. [DOI] [PMC free article] [PubMed]
- 68.Shih H.I., Wu C.J., Tu Y.F., Chi C.Y. Fighting COVID-19: a quick review of diagnoses, therapies, and vaccines. Biom. J. 2020;43(4):341–354. doi: 10.1016/j.bj.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang C, Maruggi G, Shan H, Li J. Advances in mRNA vaccines for infectious diseases. Front. Immunol.. 2019;10:594. [DOI] [PMC free article] [PubMed]
- 70.Chakraborty R., Parvez S. COVID-19: an overview of the current pharmacological interventions, vaccines, and clinical trials. Biochem. Pharmacol. 2020;180 doi: 10.1016/j.bcp.2020.114184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Englezou P.C., Sapet C., Demoulins T., Milona P., Ebensen T., Schulze K., et al. Self-amplifying replicon RNA delivery to dendritic cells by cationic lipids. Molecular therapy Nucleic Acids. 2018;12:118–134. doi: 10.1016/j.omtn.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. U. S. A. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Corbett KS, Edwards D, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, et al. SARS-CoV-2 mRNA Vaccine Development Enabled by Prototype Pathogen Preparedness. bioRxiv : The Preprint Server for Biology. 2020. [DOI] [PMC free article] [PubMed]
- 74.Koirala A., Joo Y.J., Khatami A., Chiu C., Britton P.N. Vaccines for COVID-19: the current state of play. Paediatr. Respir. Rev. 2020;35:43–49. doi: 10.1016/j.prrv.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med.. 2020. [DOI] [PMC free article] [PubMed]
- 76.Mulligan M., Lyke K., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. 2020. Phase 1/2 Study to Describe the Safety and Immunogenicity of a COVID-19 RNA Vaccine Candidate (BNT162b1) in Adults 18 to 55 Years of Age: Interim Report. [Google Scholar]
- 77.Pandey S.C., Pande V., Sati D., Upreti S., Samant M. Vaccination strategies to combat novel corona virus SARS-CoV-2. Life Sci. 2020;256 doi: 10.1016/j.lfs.2020.117956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moise L., McMurry J.A., Buus S., Frey S., Martin W.D., De Groot A.S. In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine. 2009;27:6471–6479. doi: 10.1016/j.vaccine.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eickhoff C.S., Van Aartsen D., Terry F.E., Meymandi S.K., Traina M.M., Hernandez S., et al. An immunoinformatic approach for identification of Trypanosoma cruzi HLA-A2-restricted CD8(+) T cell epitopes. Hum Vaccin Immunother. 2015;11:2322–2328. doi: 10.1080/21645515.2015.1061160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bounds C.E., Terry F.E., Moise L., Hannaman D., Martin W.D., De Groot A.S., et al. An immunoinformatics-derived DNA vaccine encoding human class II T cell epitopes of Ebola virus, Sudan virus, and Venezuelan equine encephalitis virus is immunogenic in HLA transgenic mice. Hum Vaccin Immunother. 2017;13:2824–2836. doi: 10.1080/21645515.2017.1329788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ahammad I., Lira S.S. Designing a novel mRNA vaccine against SARS-CoV-2: an immunoinformatics approach. Int. J. Biol. Macromol. 2020;162:820–837. doi: 10.1016/j.ijbiomac.2020.06.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He L., Zhu J. Computational tools for epitope vaccine design and evaluation. Current Opinion in Virology. 2015;11:103–112. doi: 10.1016/j.coviro.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and Bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 2020;27:671–80 e2. [DOI] [PMC free article] [PubMed]
- 84.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerging Microbes & Infections. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Enayatkhani M, Hasaniazad M, Faezi S, Guklani H, Davoodian P, Ahmadi N, et al. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. J. Biomol. Struct. Dyn.. 2020:1–16. [DOI] [PMC free article] [PubMed]
- 86.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jamrozik E., Selgelid M.J. COVID-19 human challenge studies: ethical issues. Lancet Infect. Dis. 2020;35:43–49. doi: 10.1016/S1473-3099(20)30438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamin M. Counting the cost of COVID-19. International journal of information technology : an official journal of Bharati Vidyapeeth's Institute of Computer Applications and Management. 2020:1–7. [DOI] [PMC free article] [PubMed]
- 89.Ceylan RF, Ozkan B, Mulazimogullari E. Historical evidence for economic effects of COVID-19. The European Journal of Health Economics : HEPAC : Health Economics in Prevention and Care. 2020;21:817–23. [DOI] [PMC free article] [PubMed]
- 90.Alam A, Siddiqui MF, Imam N, Ali R, Mushtaque M, Ishrat R. Covid-19: current knowledge, disease potential, prevention and clinical advances. Turkish journal of biology = Turk biyoloji dergisi. 2020;44:121–31. [DOI] [PMC free article] [PubMed]
- 91.Mishra B.K., Keshri A.K., Rao Y.S., Mishra B.K., Mahato B., Ayesha S., et al. COVID-19 created chaos across the globe: three novel quarantine epidemic models. Chaos, Solitons, and Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dyer O. Covid-19: trump sought to buy vaccine developer exclusively for US, say German officials. Bmj. 2020;368:m1100. [DOI] [PubMed]
- 93.Afrough B., Dowall S., Hewson R. Emerging viruses and current strategies for vaccine intervention. Clin. Exp. Immunol. 2019;196:157–166. doi: 10.1111/cei.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang X.W., Li J.S., Jin M., Zhen B., Kong Q.X., Song N., et al. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deng X., StJohn S.E., Osswald H.L., O’Brien A., Banach B.S., Sleeman K., et al. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J. Virol. 2014;88:11886–11898. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bongiovanni M. COVID-19 reinfection in an healthcare worker. J. Med. Virol.. 2020. [DOI] [PMC free article] [PubMed]
- 97.To KK, Hung IF, Ip JD, Chu AW, Chan WM, Tam AR, et al. COVID-19 reinfection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clinical infectious Diseases : an Official Publication of the Infectious Diseases Society of America. 2020. [DOI] [PMC free article] [PubMed]
- 98.Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med.. 2020. [DOI] [PMC free article] [PubMed]
- 99.Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol.. 2020;94. [DOI] [PMC free article] [PubMed]
- 100.Quinlan B.D., Mou H., Zhang L., Guo Y., He W., Ojha A., et al. 2020. The SARS-CoV-2 Receptor-Binding Domain Elicits a Potent Neutralizing Response without Antibody-Dependent Enhancement. bioRxiv : The Preprint Server for Biology. (2020.04.10.036418) [Google Scholar]
- 101.Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]