Abstract
Coronaviruses are known to infect respiratory tract and intestine. These viruses possess highly conserved viral macro domain A1pp having adenosine diphosphate (ADP)-ribose binding and phosphatase activity sites. A1pp inhibits adenosine diphosphate (ADP)-ribosylation in the host and promotes viral infection and pathogenesis. We performed in silico screening of FDA approved drugs and nucleoside analogue library against the recently reported crystal structure of SARS-CoV-2 A1pp domain. Docking scores and interaction profile analyses exhibited strong binding affinity of eleven FDA approved drugs and five nucleoside analogues NA1 (−13.84), nadide (−13.65), citicholine (−13.54), NA2 (−12.42), and NA3 (−12.27). The lead compound NA1 exhibited significant hydrogen bonding and hydrophobic interaction at the natural substrate binding site. The root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), solvent accessible surface (SASA), hydrogen bond formation, principle component analysis, and free energy landscape calculations for NA1 bound protein displayed stable complex formation in 100 ns molecular dynamics simulation, compared to unbound macro domain and natural substrate adenosine-5-diphosphoribose bound macro domain that served as a positive control. The molecular mechanics Poisson–Boltzmann surface area analysis of NA1 demonstrated binding free energy of −175.978 ± 0.401 kJ/mol in comparison to natural substrate which had binding free energy of −133.403 ± 14.103 kJ/mol. In silico analysis by modelling tool ADMET and prediction of biological activity of these compounds further validated them as putative therapeutic molecules against SARS-CoV-2. Taken together, this study offers NA1 as a lead SARS-CoV-2 A1pp domain inhibitor for future testing and development as therapeutics against human coronavirus.
Keywords: SARS-CoV-2, A1pp domain, Macro domain, Nucleoside analogues, FDA approved Drugs, In silico
1. Introduction
In December 2019, SARS-CoV-2, a novel coronavirus emerged in China, thereafter infecting more than seventy million individuals worldwide. Discovery of novel targeted drug(s) is needed and has taken the center stage in combating coronavirus disease-19 (COVID-19) pandemic. The nonstructural protein 3 (Nsp3) is the largest protein encoded by coronavirus (CoV) genome following proteolytic cleavage of polyprotein 1a (pp1a), which plays a central role in viral genome transcription and its subsequent replication. Characteristic macro domain of Nsp3 binds to ADP-ribose-1′-phosphate and has ADP-ribose-1′-phosphatase (ADRP) activity [1]. This catalytically active domain is present in several positive sense SSRNA viruses including all CoVs that infect humans. Crystal structure functional analysis of three viral ADRP domains (HCoV-229E, SARS-CoV and IBV) demonstrates the presence of a highly conserved sequence and structural superposition with variations in substrate binding site. Reports suggest the role of viral ADRP domains in hydrolyzing ADP-ribose-1′-monophosphate to ADP-ribose with high specificity [2,3]. Genetically engineered mutants of SARS-CoV expressing ADRP-deficient macro domains display enhanced sensitivity to the antiviral effect of α-interferon compared with their wild-type counterparts [4,5]. Macro domain-associated ADRP activities may have significant role in viral escape from the innate immune responses of the host, contributing resistance to antiviral cytokines [6]. During viral infection, interferon (IFN) induces human poly ADP ribose polymerase (PARPs) which covalently attach ADP ribose (mono or poly) on cellular protein amino acids (known as ADP ribosylation) that serves as a recruitment platform. Thus, IFN-induced ADP-ribosylation of human proteins specify their role in antiviral defense system. ADP-ribosylation is a reversible process and viral macro domains are known to possess de-mono-ADP-ribosylation (de-MARylation) or ADP ribose hydrolase activity [7]. It has been reported that de-MARylation is an important function executed by coronavirus macro domains during infection [8]. Alhammad and Fehr (2020) suggested that viral macro domain plays imperative dominant role in virulence and pathogenesis of coronavirus by countering host innate immune response [9]. PARPs are known to induce pro-inflammatory cytokine production that results in maintaining the innate immunity. Studies have shown that viral macro domain inhibits IFN production during infection and impedes PARP-mediated antiviral host defense system [8]. Overall, it is evident that targeting macro domain of coronavirus is an important strategy for SARS-CoV-2 drug discovery.
Various synthetic, non-synthetic and natural compounds are well recognized for possessing anti-microbial activity [10]. Nucleoside analogues (NA) are one of the important class of small molecule based antiviral drugs that play important role as therapeutics against chronic viral infections. It has been reported that NAs show potent antiviral activity and favorable pharmacokinetics. Various NAs exhibited strong efficacy against several viruses including Coronaviridae family [[11], [12], [13]]. The present study was aimed to screen nucleoside analogue and FDA approved drug libraries to identify SARS-CoV-2 A1pp domain potential inhibitors applying molecular docking approach. In addition, the lead compound was subjected to molecular dynamics simulation to study the protein-ligand complex stability and favorable conformational changes.
2. Materials and methods
2.1. Compounds database
The structures of FDA approved drugs and nucleoside analogues were downloaded from selleckchem (https://www.selleckchem.com/) in. sdf format and converted to different formats using Open Babel software.
2.2. Molecular docking
For molecular docking studies, the receptor (PDB ID: 6W02) and the ligands were loaded onto Auto Dock Tools 1.5.6 (ADT) [14,15]. Gestgeiger partial charges were assigned after merging non-polar hydrogen bonds and torsions to the ligands. Molecular docking calculations were performed on the protein model. The Kollman charges, polar hydrogen atoms, and the solvation parameters were evaluated by the module of Auto Dock tools. The Lamarckian Genetic Algorithm (LGA) option provided in the Auto Dock 4.2 was used to explore the binding site on the protein. The grid box comprises the whole binding site of the target proteins and offers enough space for the ligand movement. A maximum number of 27,000 GA operations created on a solitary population of 150 individuals for each of the 30 independent runs. Operator weights for the rate of crossover (0.80), rate of gene mutation (0.02), and elitism (1) was defaulting parameters. UCSF Chimera and LigPlot+ were used for visualization of protein-ligand complex [[16], [17], [18]].
2.3. MM/GBSA energy calculation
The molecular mechanics with generalized born and surface area solvation (MM/GBSA) approaches were utilized to calculate the free energy. The free energy calculations for docking were performed using the Glide module of Maestro. The grid was prepared using default parameters from the receptor grid generation wizard. The MM/GBSA free energy was calculated subsequently after molecular docking [19].
2.4. Molecular dynamics simulation
Compound with ADRP inhibition activity was identified from molecular docking studies. Molecular dynamics simulation was performed using GROningen MAchine for Chemical Simulations (GROMACS) version 5.1.1 [20]. Protein parameters were generated using gromos43a1 force field. Ligand parameters for same force field were generated using PRODRG server [21]. Gmxeditconf tool was used to generate dodecahedron simulation box. Solvation was performed with SPC water model using gmx solvate tool. Gmxgenion tool was utilized to electro-neutralize the system. Following neutralization, energy minimization was performed to remove steric clashes to optimize the structure. After energy minimization, system was equilibrated in two steps. In first step of 100 ps of NVT equilibration, system was heated up to 300 K to stabilize the temperature of the system. In the second step of 100 ps of NPT ensemble, pressure and density of system was stabilized. Pressure was maintained using Parrinello-Rahman barostat [22]. Bonds length were kept conserved using linear constraint solver (LINCS) algorithm [23]. Long range interactions were handled using particle mesh ewald (PME) summation method [24]. This well equilibrated system with desired temperature and pressure was used to compute trajectory of 100 ns on a Linux machine with Intel Core i-7 processor (32 GB RAM).
2.5. Trajectory analysis
Trajectory analysis was performed using various GROMACS analysis tools. The root mean square deviation (RMSD) and root mean square fluctuations (RMSF) of proteins were calculated using gmx rms, and gmxrmsf tools, respectively. Solvent accessible surface area (SASA) and radius of gyration (Rg) were computed by gmxsasa and gmx gyrate tools. Various energy related parameters were estimated using gmx energy tool whereas secondary structure estimation was performed by gmxdodssp tool. Hydrogen bonds were analyzed using gmxhbond tool. VMD [25] and PyMol [26] were used for the visualization. The plots were prepared by using Grace Software.
2.6. Principal component analysis and free energy landscape
Principal component analysis (PCA) is a widely used analytical technique to illustrate the slow and functional motions for bio-molecules [27]. The principal components of the proteins were obtained by diagonalizing and solving the eigenvalue and eigenvectors for the covariance matrix. The eigenvectors are a representation of the direction of motion whereas eigenvalues represent the magnitude of motion along with the direction. The covariance matrix for illustration of PCA was calculated using GROMACS analysis tool gmx cover. gmx cover builds and also diagonalizes the covariance matrix. Another GROMACS analysis tool gmxanaeig was utilized to calculate the overlap between principal components and coordinates of the trajectory.
Free energy landscape (FEL) is a representation of possible conformation taken by a protein in molecular dynamics simulation along with the Gibbs free energy. FEL represents two variables that reflect specific properties of the system and measure conformational variability [19]. FEL was calculated using probability distribution from the essential plane composed of first two eigenvectors. gmx sham tool was used for construction of FEL.
2.7. g_mmpbsa analysis
The g_mmpbsa is an additional GROMACS utility applied to compute the binding free energy of protein-ligand complexes. g_mmpbsa uses Molecular Mechanic/Poisson-Boltzmann Surface Area (MM-PBSA) method to compute the binding free energy of protein-ligand complexes [28]. The binding free energy was computed for reference and lead compound in complex with target protein. Molecular dynamics simulation trajectories for the last 20 ns (80–100 ns) were used for the calculation. The binding free energy (ΔGbinding) of the protein-ligand complex in aqueous environment can represented as following,
| ΔGbinding = Gcomplex - (Gprotein + Gligand) |
Where, Gcomplex denotes energy of the protein-ligand complex, Gprotein and Gligand denote the energy of protein and ligand in aqueous environment, respectively. Estimation of binding free energy of protein-ligand complex provides an insight of the bio-molecular interaction.
2.8. ADMET parameter and bioactivity prediction
The free web tool SwissADME (http://www.swissadme.ch/) was used to evaluate the physicochemical properties and pharmacokinetics of the identified compounds [29]. Different bioactivity (G-protein-coupled receptors ligand; Ion channel modulator; Kinase inhibitor; Nuclear receptor ligand; Protease inhibitor; Enzyme inhibitor) score of lead FDA approved drug and lead nucleoside analogue were predicted using molinspiration cheminformatics online server (https://www.molinspiration.com/).
3. Results
3.1. Molecular docking
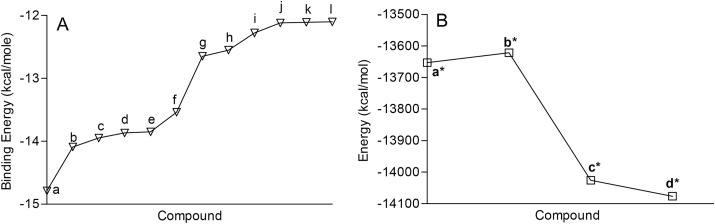
Screening of FDA approved drugs (n = 2682) and nucleotide analogues (n = 135) against SARS-CoV-2 Macro (ADP-ribose-1″-phosphatase) domain active site revealed compounds that exhibited significant binding potential in comparison to natural substrate. Eleven FDA approved drugs showed tight binding with the active site and binding energy was in the range of −12.01 to −14.09 kcal/mol (Fig. 1 A). Lead nucleoside analogue [5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyfuran-2-yl](sodiylium)methyl [({[5-(3-carbamoyl-1,4-dihydropyridin-1-yl)-3,4-dihydroxyfuran-2-yl]methoxy}(sodiooxo)phosphoryl)oxy]phosphonate (NA1); nadide; 1-[5-({[({[5-(6-amino-9H-purin-9-yl)-4-(hydrogen phosphonatooxy)-3-hydroxyfuran-2-yl](sodiylium)methyl phosphonato}oxy) (sodiooxo)phosphoryl]oxy}methyl)-3,4-dihydroxyfuran-2-yl]-3-carbamoyl-1λ⁵-pyridin-1-ylium (NA2); citicholine; and sodium 4-amino-1-[3,4-dihydroxy-5-({[hydroxy ({[hydroxy (sodiooxo)phosphoryl phosphonato]oxy})phosphoryl]oxy}methyl)furan-2-yl]-1,2-dihydropyrimidin-2-one (NA3), interacted with the protein active site with binding energy −13.84, −13.65, −13.54, −12.42, and −12.27 kcal/mol, respectively. Natural substrate of the macro domain showed −14.09 kcal/mol binding energy. MM-GBSA analysis showed that lead nucleotide analogue (NA1) ([5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyfuran-2-yl](sodiylium)methyl [({[5-(3-carbamoyl-1,4-dihydropyridin-1-yl)-3,4-dihydroxyfuran-2-yl]methoxy} (sodiooxo) phosphoryl) oxy] phosphonate) and FDA approved lactobionic acid make stable protein-ligand complex (Fig. 1 B). The position of lead FDA approved drugs bound to target protein are shown in supplementary figure 1.
Fig. 1.
Binding energy, and energy calculation of the lead FDA approved drugs at SARS-CoV-2 macro (ADP-ribose-1″-phosphatase) domain active site. (A) Binding energy of FDA approved lead drugs at ADP-ribose-1″-phosphatase active site. (B) MM-GBSA analysis of receptor (SARS-CoV-2 Macro (ADP-ribose-1″-phosphatase) domain), receptor-lactobionic acid complex, recptor-NA1 complex and receptor-adenosine-5-diphosphoribose complex a = adenosine-5-diphosphoribose; b = Lactobionic acid; c = Neohesperidin; d = Salvianolic acid B; e = Ribostamycin Sulfate; f = NADIDE; g = Lactitol; h = Folic acid; i = Melibiose; j = Naringin; k = Maltose; l = Rutin DAB10; a* = A1pp receptor; b* = A1pp-lactobionic acid complex; c* = A1pp-NA1 complex; d* = A1pp-adenosine-5-diphosphoribose complex.
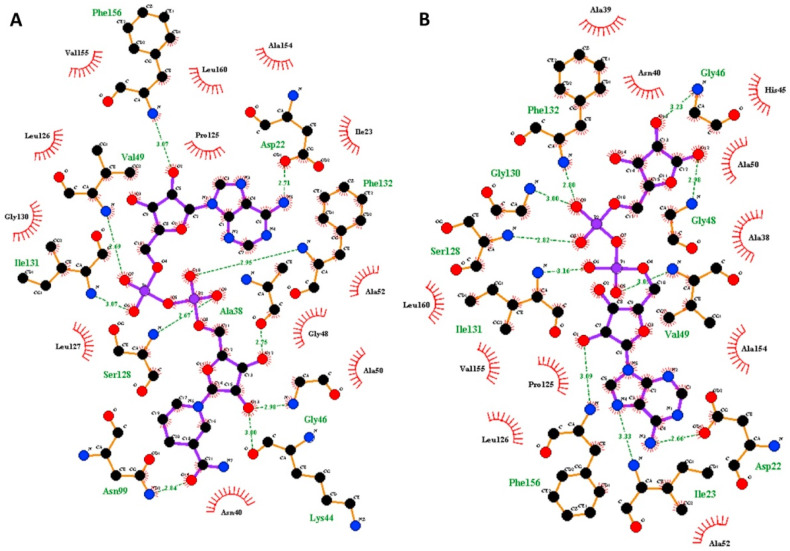
The names of amino acid residue involved in the protein-ligand complex formation and type of interaction such as hydrogen bonding and hydrophobic interaction is summarized in Table 1 and supplementary table 1. Structure of lead FDA approved drugs (FAD) and nucleoside analogues (NA) are shown in supplementary figure 2 . Surface structure of lead FDA/NA and natural substrate complexed with SARS-CoV-2 macro (ADP-ribose-1″-phosphatase) domain active site is visualized in Fig. 2 A, B and C and supplementary figure 3. Further, docking pose of natural substrate and lead nucleoside inhibitor (NA1) molecule at the macro domain active site is depicted in supplementary figure 4. Lead nucleoside analogue NA1 formed 10 hydrogen bond and interacted with 11 hydrophobic amino acid residues of the macro domain (Fig. 3 A and Table 1). FDA approved lead compound lactobionic acid, neohesperidin, salvianolic acid, ribostamycin sulfate, nadide, lactitol, folic acid, malibiose, naringin, maltose, and rutin formed 7, 7, 7, 9, 9, 7, 9, 6, 6, 5 and 6 hydrogen bonds with active site of the protein respectively. The lead FAD compounds showed hydrophobic interaction with plenty of amino acids (Table 1).
Table 1.
List of amino acid residue and type of interaction involved in SARS-CoV-2 Macro (ADP-ribose-1″-phosphatase) domain active site and lead compound and natural substrate.
| Sr. no. | Compound name | Hydrogen bond forming residues | Hydrophobic interaction residues |
|---|---|---|---|
| 1. | Lactobionic acid | Phe156, Ala154, Ile23, Asp22, Asp 157, Gly130, Leu126 | Val49, Val155, Pro125, Ala 29 |
| 2 | NA1 | Phe156, Val49, Asp22, Phe132, Gly46, Lys44, Asn99, Ser128, Ile131, Ala38 | Leu160, Ala154, Ile23, Pro125, Ala52, Gly48, Ala50, Asn40, Leu127, Leu126, Val155 |
| 3 | ADP | Gly46, Gly48, Val49, Asp22, Ile23, Phe156, Ile131, Ser128, Gly130, Phe132 | Ala 39, Asn40, His 45, Ala50, Ala38, Ala154, Ala52, Leu126, Pro125, Val155, Leu160 |
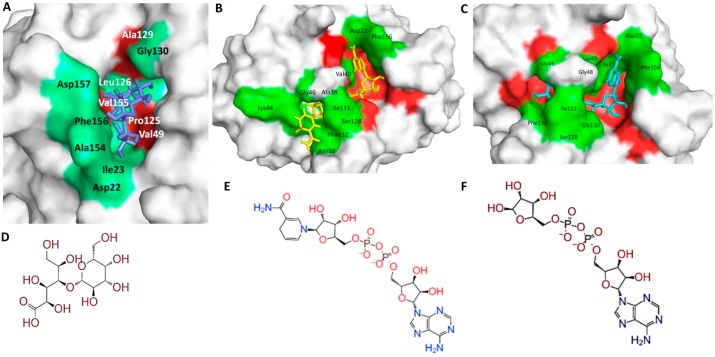
Fig. 2.
Surface structure of lead FDA approved drug, nucleoside inhibitor and natural substrate at SARS-CoV-2 A1pp domain active site. (A) Surface structure of lactobionic acid at ADP-ribose-1″-phosphatase active site. (B) Surface structure of NA1 at ADP-ribose-1″-phosphatase active site. (C) Surface structure of adenosine-5-diphosphoribose at ADP-ribose-1″-phosphatase active site. (D) 2D structure of lactobionic acid. (E) 2D structure of NA1. (F) 2D structure of adenosine-5-diphosphoribose.
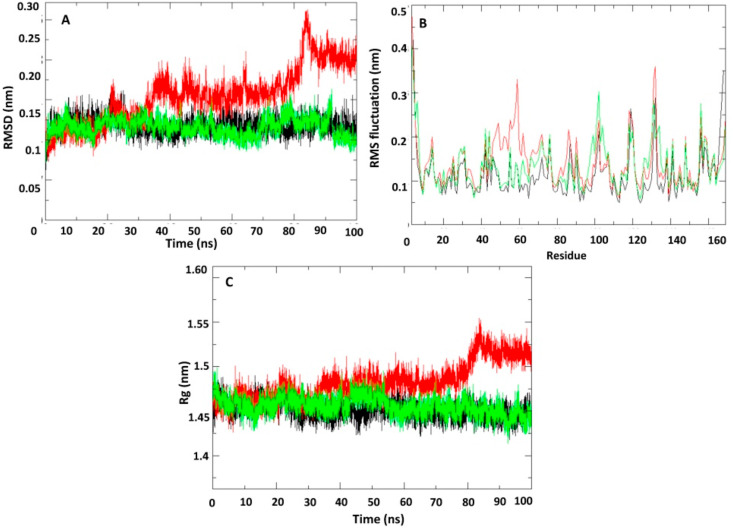
Fig. 4.
Plot of molecular dynamic simulation trajectories of SARS-CoV-2 macro (ADP-ribose-1″-phosphatase) domain and in unbound and ligand/natural substrate bound complex during 100 ns simulation. (A) The root mean square deviation (RMSD) of solvated macro domain backbone in unbound and complexed with lead nucleotide analogue/natural substrate during 100 ns molecular dynamics simulation. (B) The root mean square fluctuation (RMSF) values of solvated macro domain backbone in unbound and complexed with lead nucleotide analogue/natural substrate plotted against residue numbers. (C) Plot of radius of gyration (Rg) during 100 ns molecular dynamics simulation of macro domain backbone in unbound and complexed with lead nucleotide analogue/natural substrate during simulation period. Unbound protein parameters are depicted in black color while natural substrate and lead inhibitor bound protein complex parameters are shown in red and green color respectively.
Fig. 3.
Ligplot showing name of amino acid residue and type of interaction involved in the binding with SARS-CoV-2 Macro (ADP-ribose-1″-phosphatase) domain active site. (A) Type of interaction and target protein amino acid residues involved in interaction with NA1. (B) Type of interaction and target protein amino acid residues involved in interaction with substrate molecule (adenosine-5-diphosphoribose). Amino acid residue involved in hydrogen bond is shown in green color while red color depicts residues involve in hydrophobic interaction. Green dotted line shows hydrogen bond between amino acid residue and substrate/lead molecule atom. Numerical values given in-between the dotted lines show length of hydrogen bond in Å.
4. Molecular dynamics simulation
We utilized X-ray crystal structure of SARS-CoV-2 NSP3 ADP ribose phosphatase (PDB ID: 6W02) to dock the FDA approved drugs and nucleotide analogue compounds and the natural substrate. Among the screened compounds (FDA approved drugs and nucleotide analogues) lead nucleotide inhibitor NA1 showed more number of hydrogen bonds (10) and hydrophobic interaction (11) than the natural substrate. Therefore, we selected NA1 for molecular dynamic simulation. Natural substrate adenosine-5-diphosphoribose was also subjected to perform molecular dynamic simulation as a positive control. NA1, adenosine-5-diphosphoribose and unbound macro domain were subjected to 100ns MD simulations for comparative conformation dynamics. Density, temperature, and pressure of the unbound protein and in complex with adenosine-5-diphosphoribose and NA1 were stable for 100 ns simulation (Supplementary Fig. 5 A, B, and C). The average backbone RMSD for unbound protein and NA1 bound protein remained stable at ≈0.13 nm throughout the simulation period (Fig. 4 A). However, the natural substrate adenosine-5-diphosphoribose bound protein backbone showed flexible stability at ≈0.17–0.25 nm RMSD. Similarly, the average solvated macro domain whole protein RMSD for unbound protein and NA1 bound protein was found to stable at 0.22 nm and remained stable during the 100ns MD simulation period. The natural substrate adenosine-5-diphosphoribose bound solvated macro domain whole protein showed flexible stability at ≈0.25–0.35 nm RMSD. Next, we studied the effect of adenosine-5-diphosphoribose and NA1 binding on internal dynamics and compared with the unbound protein by calculating the root mean square fluctuation (RMSF) (Fig. 4 B). Average RMSF value for the free protein, adenosine-5-diphosphoribose and NA1 bound protein was mostly in the range of 0.1–0.3 nm. Moreover, the result showed that NA1 binding to macro domain active site comparatively reduced the fluctuations at 45–65 amino acid residues in comparison to unbound and natural substrate adenosine-5-diphosphoribose bound protein. The effect of adenosine-5-diphosphoribose and NA1 binding on the macro domain protein radius of gyration (Rg) was studied and compared with unbound protein (Fig. 4 C). Binding of NA1 continuously decreased the Rg value however, after 20ns of simulation period significant low gyration value was obtained in comparison to unbound and natural substrate bound protein. Rg value for unbound protein, adenosine-5-diphosphoribose and NA1 bound protein were in the range of 1.44–1.48, 1.45 to 1.55 and 1.44 to 1.47 respectively during 100 ns simulation.
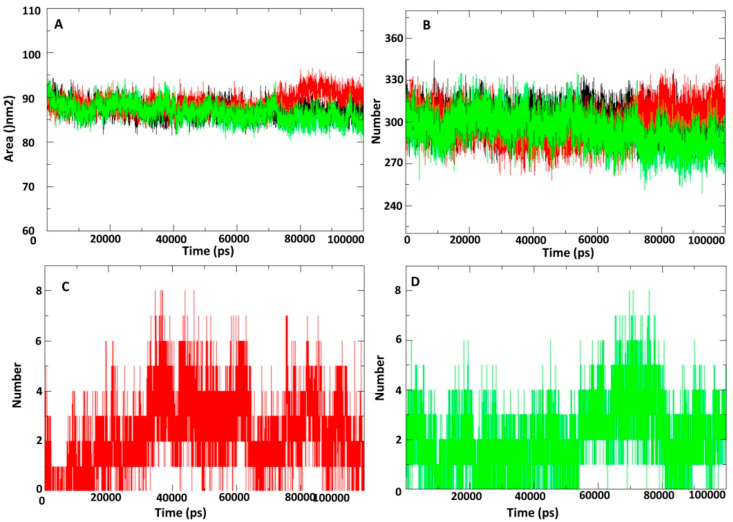
Next, changes in solvent accessibility surface area (SASA) for adenosine-5-diphosphoribose, and NA1 bound and free protein was analyzed and results are depicted in Fig. 5 A. Binding of NA1 significantly decreased the SASA value throughout the 100 ns simulation period in comparison to natural substrate adenosine-5-diphosphoribose. Average SASA value of both the NA1bound and free conformation of the protein during the 100 ns simulation was in the range of ≈82–92 nm2. However, the SASA value for natural substrate adenosine-5-diphosphoribose bound protein was 85–95 nm2. Pattern of hydrogen bond formation with surrounding water and unbound protein and NA1/natural substrate bound protein were studied (Fig. 5 B). About 300 intermolecular hydrogen bonds were formed between NA1 bound protein and water (Fig. 5 B). However, natural substrate adenosine-5-diphosphoribose bound protein formed about 320 intermolecular hydrogen bonds with water throughout the 100ns simulation period. Hydrogen bonds between macro domain and natural substrate adenosine-5-diphosphoribose and NA1 were about 8 and 8 at 100ns simulation time (Fig. 5 C and D).
Fig. 5.
Plot of solvent accessible surface (SASA) region and hydrogen bond formation during 100 ns MD simulation. (A) Plot of solvent accessible surface area (SASA) during 100 ns molecular dynamics simulation of Macro Domain in unbound and complexed and with natural substrate or lead nucleotide analogue. (B) Plot of number of hydrogen bond formation between water and macro domain in unbound and complexed with natural substrate or lead nucleotide analogue. (C) Plot of number of hydrogen bond formation between macro domain complexed with natural substrate. (D) Plot of number of hydrogen bond formation between macro domain complexed with lead nucleotide analogue. Unbound protein parameters are depicted in black color; Natural substrate and lead inhibitor bound protein complex parameters are shown in red and green color respectively.
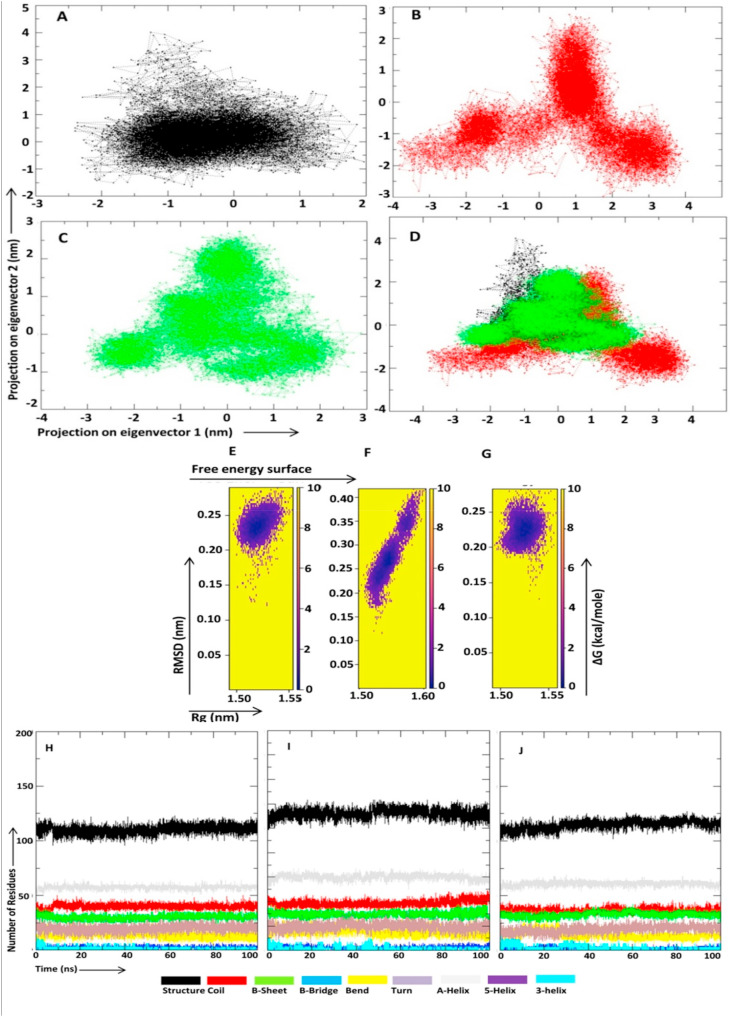
Principal component analysis (PCA) of molecular dynamics trajectories was conducted to study the collective motion of the macro domain bound adenosine-5-diphosphoribose/NA1 and free macro domain protein and results are shown in Fig. 6 A-D. PCA analysis showed that NA1 binding to macro domain protein decreased the collective motion of the protein in comparison to unbound protein structure and adenosine-5-diphosphoribose bound macro domain (Fig. 6 A, B, C and D). To visualize the energy minima landscape of, adenosine-5-diphosphoribose bound, NA1 bound and free target protein, the free energy landscape (FEL) against first two principal components PC1 (Rg) and PC2 (RMSD) were studied. The analysis revealed ΔG value in the range of 0–10 kJ/mol. Concise minimal energy area (blue color) was found for NA1 bound protein in comparison to natural substrate adenosine-5-diphosphoribose bound and unbound protein (Fig. 6 E, F and G).
Fig. 6.
Projection of SARS-CoV-2 macro (ADP-ribose-1″-phosphatase) domain atoms in phase space along the first two principal eigenvectors, free energy landscape and protein-ligand interaction energy. (A–C) Projection of the motion of the macro domain in phase space along the first two principal eigenvectors for unbound macro domain, adenosine-5-diphosphoribose bound macro domain, and NA1 bound macro domain. (D) Merged projection of the motion of the macro domain in phase space along the first two principal eigenvectors for unbound macro domain, adenosine-5-diphosphoribose bound macro domain, and NA1 bound macro domain. Unbound macro domain is shown in black, adenosine-5-diphosphoribose bound macro domain in red, and NA1 bound macro domain in green. (E–G) Free energy landscape of the SARS-CoV-2 macro domain unbound, complexed with adenosine-5-diphosphoribose and NA1 respectively. (H–J) Secondary structure changes during the course of 100ns MD simulation in unbound macro domain, adenosine-5-diphosphoribose bound macro domain, and NA1 bound macro domain respectively. Short range interaction analysis.
The propensity of secondary structural content remains an important component to study the structural behavior of the macro domain protein. We investigated changes in the secondary structure in unbound macro domain, adenosine-5-diphosphoribose bound macro domain and NA1 bound macro domain as shown in Fig. 6 H-J. The NA1 ligand does not induce any significant change in the secondary structure content (Fig. 6 J), while a little fluctuations were recorded in natural substrate adenosine-5-diphosphoribose bound macro domain (Fig. 6 I).
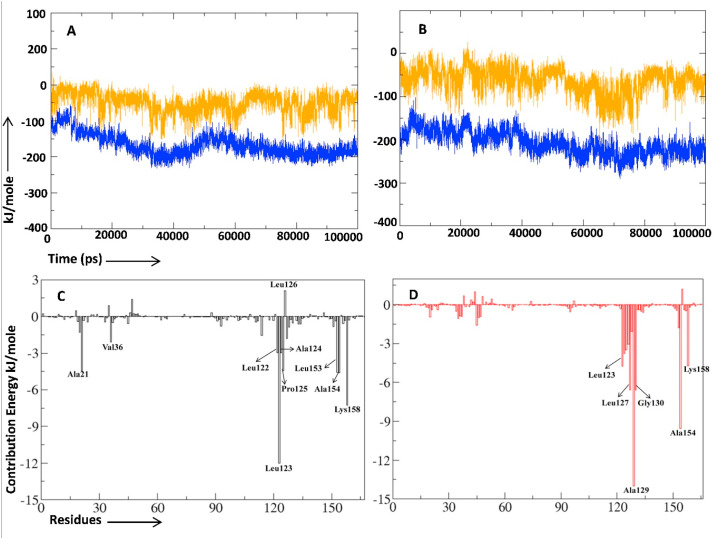
To acquire further insights of the binding strength of NA1-macro domain complex and adenosine-5-diphosphoribose macro domain complex, we evaluated the non-bonded interaction energy between protein and ligands. Short range non-bonded interaction energy is composed of two quantities, Coulombic short range interaction energy (CsrIE) and Lennard Jones short range interaction energy (LJsrIE) [19]. NA1-macro domain complex had lower values for both CsrIE (−62.8912 kJ/mol) and LJsrIE (−207.9721 kJ/mol) in comparison to the adenosine-5-diphosphoribose macro domain complex which had higher values for both CsrIE (−50.7681 kJ/mol) and LJsrIE (−168.2841 kJ/mol) (Fig. 7 A–B). From above results it evident that NA1-macro domain complex possesses strong non-bonded interaction between protein and ligand in comparison to the adenosine-5-diphosphoribose macro domain complex.
Fig. 7.
Interaction energies pattern for natural substrate adenosine-5-diphosphoribose and NA1 with macro domain and residue mediated binding energy calculations (A–B) Interaction energies pattern for natural substrate adenosine-5-diphosphoribose bound macro domain and NA1 bound macro domain respectively. Orange color representing the coulombic interaction energy while blue color representing the Lennard Jones interaction energy. Binding energy calculations for (C) Adenosine-5-diphosphoribose macro domain complex and (D) NA1 bound macro domain complex.
4.1. g_mmpbsa analysis
The last 20 ns (80–100 ns) of trajectory obtained during MD simulations were used to compute the binding free energy of the protein-ligand complexes by applying the g_mmpbsa tool of GROMACS. For each complex, binding free energy (ΔEbinding), van der Waals energy (Evdw), electrostatic energy (Eelec), polar solvation energy (ΔEpolar), and SASA were calculated (Table 2 , supplementary Fig. 6 A, B, C and D, and supplementary table 2). It is evident from Table 2 that NA1 has significantly lower value (−175.978 kJ/mol) of binding free energy in comparison to natural substrate adenosine-5-diphosphoribose (−133.403 Kj/mol) which suggest strong binding of NA1 in comparison to adenosine-5-diphosphoribose. Further contribution of each residue was evaluated by decomposing the total binding free energy into per residue contribution energy (Fig. 7C–D). In binding of NA1 to target protein amino acid residues Leu123, Leu127, Ala129, Gly130, Ala154, and Lys158 contributed significantly in binding. Next in binding of adenosine-5-diphosphoribose to the target protein amino acid residues Leu123, Ala 124, Pro125, Leu 153, Ala154, and Lys158 contributed significantly in binding (Supplementary table 1). In NA1-macro domain complex, Ala129 showed highest contribution (−13.9812 kJ/mol) to the binding free energy whereas in adenosine-5-diphosphoribose macro domain complex Leu123 showed highest contribution (−12.0236 kj/mol).
Table 2.
MM-PBSA calculations of binding free energy for lead molecule and natural substrate bound protein complex.
| Type of Binding energy | Binding energy values (Protein Lead Compound complex) | Binding energy values (Protein Natural Substrate complex) |
|---|---|---|
| ΔEbinding (kj/mol) | −175.978±0.401 | −133.403±14.103 |
| SASA (kj/mol) | −21.726±0.033 | −18.994±1.252 |
| ΔEpolar solvation (kj/mol) | 133.686±0.458 | 134.794±15.382 |
| ΔEElectrostatic (kj/mol) | −36.203±0.264 | −34.575±15.045 |
| ΔEVan der Waal (kj/mol) | −251.714±0.344 | −214.628±13.536 |
4.2. ADMET analysis
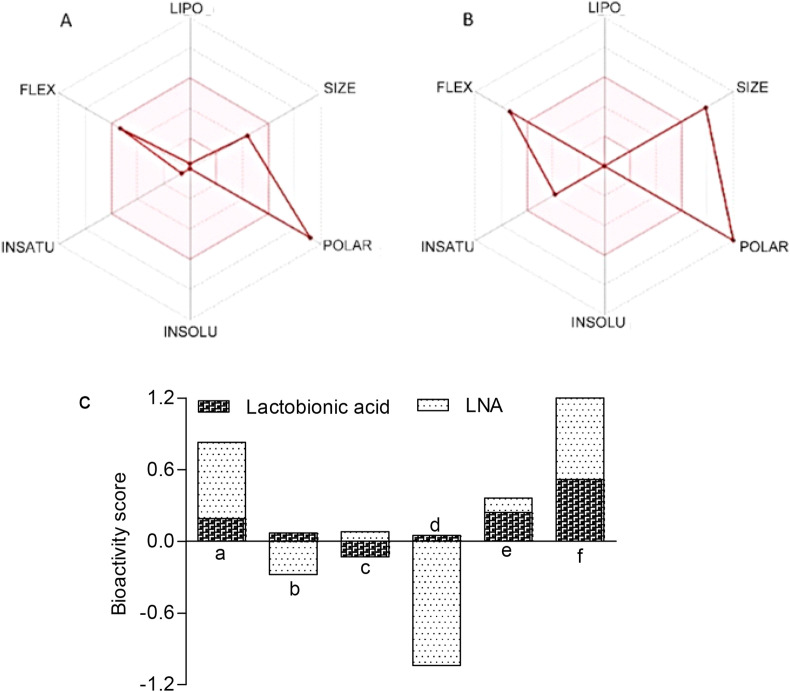
The oral absorption of our suggested bioactive molecule is represented in the bioavailability radar plots (Fig. 8 A and B). The plot shows a graphical representation of the drug-likeness parameters of the lead molecules. Bioavailability radar showed that FDA approved drug lactobionic acid has suitable flexibility and acceptable size for a drug (Fig. 8 A). Lead nucleotide analogue, NA1 has acceptable unsaturation physicochemical properties for their oral bioavailability (Fig. 8 B). Bioactivity score was checked for different target like GPCR, ion channel, kinase, nuclear receptor, protease and enzymes. Result indicate lead nucleotide analogue NA1 is superior ligand for enzyme inhibition as per in silico data prediction (Enzyme inhibition > GPCR ligand > Protease inhibitor > Kinase inhibitor > Ion channel modulation > Nuclear receptor) (Fig. 8C). FDA approved drug lactobionic acid also acts as ligand for enzyme inhibition (Enzyme inhibition > Protease inhibitor > GPCR ligand > Ion channel modulation > Nuclear receptor > Kinase inhibitor) (Fig. 8 C).
Fig. 8.
Bioavailability radar and bioactivity profile of FDA approved lactobionic acid and lead nucleotide analogue NA1 (A) and (B) represents the bioavailability radar status of FDA approved lactobionic acid and lead nucleotide analogue NA1. (C) Predicted bioactivity score of FDA approved lactobionic acid and lead nucleotide analogue NA1. a = G-protein-coupled receptors ligand; b = Ion channel modulator; c = Kinase inhibitor; d = Nuclear receptor ligand; e = Protease inhibitor; f = Enzyme inhibitor; LNA = Lead nucleotide analogue.
Different physicochemical properties and pharmacokinetics of lactobionic acid and lead nucleotide analogue NA1 ([5-(6-amino-9 H-purin-9-yl)-3, 4-dihydroxyfuran-2-yl] (sodiylium) methyl [({[5-(3-carbamoyl-1,4-dihydropyridin-1-yl)-3,4-dihydroxyfuran-2-yl]methoxy} (sodiooxo) phosphoryl) oxy] phosphonate) are provided in Table 3 .
Table 3.
Physicochemical Properties and pharmacokinetics of lactobionic acid and lead nucleotide analogue NA1.
| Properties | Parameter | Lead nucleotide analogue (NA1) | Lactobionic acid |
|---|---|---|---|
| Physicochemical Properties | Formula | C21H27N7O14P2 | C12H22O12 |
| Molecular weight (g/mol) | 663.43 | 358.3 | |
| Num. heavy atoms | 44 | 24 | |
| Num. arom. heavy atoms | 9 | 0 | |
| Fraction Csp3 | 0.52 | 0.92 | |
| Num. rotatable bonds | 11 | 8 | |
| Num. H-bond acceptors | 17 | 12 | |
| Num. H-bond donors | 6 | 9 | |
| Molar Refractivity | 142.18 | 70.92 | |
| TPSA (Å2) | 342.90 | 217.6 | |
| Pharmacokinetics | GI absorption | Low | Low |
| BBB permeant | No | No | |
| P-gp substrate | Yes | Yes |
Fraction Csp3 = Fraction of sp3 carbon; TPSA = The Polar Surface Area; GI = Gastrointestinal tract; BBB= Blood brain barrier; P-gp = P-glycoprotein.
5. Discussion
It has been reported that the SARS-CoV-2 ADP-ribose phosphatase enzyme activity plays an important role in viral pathogenesis [9,30]. In the present study, we identified a lead nucleoside inhibitor having potential to bind the substrate binding site of the SARS-CoV-2 ADRP enzyme. It has been reported that amino acid residue involved in active site formation of SARS-CoV ADP ribose phosphatase are critical for catalytic process and substrate binding [2]. Michalska et al. (2020) demonstrated that the crystal structure of SARS-CoV-2 ADRP enzyme possesses adenine, distal/proximal ribose and di-phosphate binding sites at the substrate binding pocket [31]. In our study we found that the natural substrate (ADP) binds to the similar amino acids corresponding to adenine, distal/proximal ribose and di-phosphate binding sites (Table 1, Fig. 2C). In another study Debanth et al. (2020) reported some inhibitors against SARS-CoV-2 ADRP using molecular docking approach [32]. In our study the lead inhibitor formed additional binding to Gly46, Lys44, Asn99, Leu160, Pro125, Ala52, Gly48, Leu127, Val155 amino acids (Table 1 and Fig. 2A). We found that nucleotide analogue (NA1) form hydrogen bond with the same seven amino acid residues (Asp22, Val49, Gly46, Ser128, Ile131, Phe132, Phe156) involved in binding with the natural substrate (Fig. 3 A and B). This data indicate that NA1 competes for the active site binding with the natural substrate. It has been reported that aspartate and asparagine residue are involved in catalysis and substrate binding at the active site of coronaviruses and other viral macro domain [2]. In the present study, we found that lead nucleotide analogue and natural substrate showed hydrogen bond with Asp22 (Fig. 3 A and B). Moreover, NA1 showed hydrogen binding with asparagine at the active site. Overall, the binding of similar and critical amino acids at the macro domain active site indicate the ADRP inhibitory potential of the identified NA1 compound. In the present study three nucleoside lead inhibitors (NA1, nadide, citicholine) showed ≈ -14.00 kJ/mol docking score. There is no report on the biological activity of NA1. Although the identified lead nucleoside inhibitors (except NA1) are experimental drugs till date, but the clinical trials have been reported on nadide, and citicholine. It should be noted that recently a clinical trial has been designed to study the effect of low dose of nadide (also known as nicotinamide adenine dinucleotide) in patients with post-COVID-19 syndrome [33]. In the present study five FDA approved lead inhibitors (Lactobionic acid, Neohesperidin, Salvionic acid B, Ribostamycin Sulfate and nadide) showed ≈ -14.00 kJ/mol docking score. Lactobionic acid is used in the intravenous delivery of erythromycin and act as an excipient for formulations in pharmaceutical industry [34, 35]. Neosperidin possess antidiabetic potential and used to improve and promote skin microcirculation [36]. Salvionic acid has been patented as antithrombotic agent and possesses other therapeutic value [37,38]. Ribostamycin sulfate is known to inhibit the protein synthesis in bacteria [39]. Overall the published reports provide information about the therapeutic potential of the identified lead compounds.
The average distance between the protein atoms was computed in terms of root-mean-square deviation (RMSD) value. The analysis provides insights into protein conformation, stability and equilibrium of the system during simulation [40]. In the present study, binding of NA1 to the active of target protein did not increase the RMSD of whole protein and its backbone atoms during simulation. The result indicates that protein-ligand complex was stable during 100 ns period (Fig. 4 A). Binding of ligands to the active site of protein stabilize the overall structure and decreases the fluctuation of amino acid residues. The study showed that NA1 formed 5 hydrogen bonds in the region of 45–65 residues (Figs. 3 A and 4 B). The decrease fluctuation in this region may be due to the hydrogen bond formation between the ligand and macro domain amino acids. Further, binding of NA1 to target protein did not increase the residue fluctuation during the simulation period which indicates the overall stable binding interaction among NA1 and macro domain active site. Radius of gyration (Rg) is used to assess the folding of regular 2D structures into 3D protein structure. It shows change in structure compactness and its overall dimension. Significant decrease in Rg value (Fig. 4 C) during the simulation suggested tight packing of the protein after ligand binding, thus, making a stable complex. Solvent accessibility surface area (SASA) is an important parameter of biomolecule which indicates its surface area able to make contact with surrounding solvent molecules. Thus, increase in SASA indicates the unfolding and native protein structure disruption. The present study indicates that binding of NAI binding to active site of the protein stabilize its native structure by maintaining the SASA during the simulation period (Fig. 5 C). Formation of hydrogen bond between ligand and target protein minimize the energy of the protein-ligand complex. The study showed that binding of NAI binding to active site of the protein modestly decreased the number of hydrogen-bond interaction between protein and surrounding water molecule (Fig. 5 B and C). Moreover, the pattern of hydrogen bond formation was precisely stable during the simulation period (Fig. 5 D). Overall, intra-molecular, surrounding water-protein, and ligand-protein hydrogen bonding indicate the energetically favorable and stable NAI binding to the targeted active site. PCA analysis of unbound and ligand bound protein is strenuous motion of Cα atom of the protein through magnitude (eigenvalues) and direction (eigenvectors) [41].
Examination of molecular dynamics trajectories of NAI bound and unbound protein indicated lesser comparative motion of the protein in ligand bound conformation (Fig. 6 A). Furthermore, lesser motion in ligand bound protein conformation indicates lesser flexibility, and increased stability of the complex. Compact and smaller energy area in free energy landscape analysis for ligand bound protein in comparison to unbound structure further indicates the stability of NA1-protein ligand complex formation (Fig. 6 B). g_mmpbsa analysis demonstrates the therapeutic potentials of NA1 against A1pp domain of COVID-19. Non-bonded interaction energy between unbound macro domain and NA1 and adenosine-5-diphosphoribose showed that the NA1 have excellent potential to target the A1pp domain to overcome the effect of SARS-CoV-2.
6. Conclusion
ADP-ribosylation is an important process involved in cellular signaling, genome stability, cellular proliferation and apoptosis. Viral macro domains are known to inhibit and/or reverse ADP-ribosylation thereby affecting the normal consequence of ribosylation in the host. The present study identifies FDA approved drug (lactobionic acid) and nucleotide analogue (NA1) as novel SARS-CoV-2 A1pp domain inhibitor. Our study concludes that nucleoside analogue (NA1) possess SARS-Cov-2 A1pp inhibitory potential and future testing is needed for its development as a SARS-CoV-2 therapeutic.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgement
PPK and MS acknowledge financial support from Indian Council of Medical Research (ICMR), India in the form of ICMR-Senior Research Fellowship. SK acknowledges Department of Science and Technology, India for providing financial support in the form of DST-SERB Grant [EEQ/2016/000350] and DST-FIST Departmental Grant. AKS and KSP acknowledge CSIR-India and DBT-India funding agencies respectively for providing financial assistance in the form of Senior and Junior Research Fellowships respectively. Efforts are supported by the Department of Defense Grants W81XWH-19-1-0720 and W81XWH18-1-0618 and VA Merit Review 1I01BX002494 to SG.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2020.104185.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Xu Y., Cong L., Chen C., Wei L., Zhao Q., Xu X., Ma Y., Bartlam M., Rao Z. Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain. J. Virol. 2009;83(2):1083–1092. doi: 10.1128/JVI.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., Ahola T., Canard B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80(17):8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Putics A., Gorbalenya A.E., Ziebuhr J. Identification of protease and ADP-ribose 1''-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 2006;87(Pt 3):651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- 4.Hosking M.P., Lane T.E. The role of chemokines during viral infection of the CNS. PLoS Pathog. 2010;6(7):1–4. doi: 10.1371/journal.ppat.1000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Randall R.E., Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 6.Kuri T., Eriksson K.K., Putics A., Züst R., Snijder E.J., Davidson A.D., Siddell S.G., Thiel V., Ziebuhr J., Weber F. The ADP-ribose-1''-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J. Gen. Virol. 2011;92(Pt 8):1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- 7.Fontana P., Bonfiglio J.J., Palazzo L., Bartlett E., Matic I., Ahel I. Serine ADP-ribosylation reversal by the hydrolase ARH3. Elife. 2017;6:1–20. doi: 10.7554/eLife.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7(6):1–12. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alhammad Y.M.O., Fehr A.R. The viral macrodomain counters host antiviral ADP-ribosylation. Viruses. 2020;12(4):1–12. doi: 10.3390/v12040384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mishra A., Kumar S., Bhargava A., Sharma B., Pandey A.K. Studies on in vitro antioxidant and antistaphylococcal activities of some important medicinal plants. Cell. Mol. Biol. (Noisy-Le-Grand) 2011;57(1):16–25. [PubMed] [Google Scholar]
- 11.Warren T.K., Wells J., Panchal R.G., Stuthman K.S., Garza N.L., Van Tongeren S.A., Dong L., Retterer C.J., Eaton B.P., Pegoraro G., Honnold S., Bantia S., Kotian P., Chen X., Taubenheim B.R., Welch L.S., Minning D.M., Babu Y.S., Sheridan W.P., Bavari S. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508(7496):402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters H.L., Jochmans D., de Wilde A.H., Posthuma C.C., Snijder E.J., Neyts J., Seley-Radtke K.L. Design, synthesis and evaluation of a series of acyclic fleximer nucleoside analogues with anti-coronavirus activity. Bioorg. Med. Chem. Lett. 2015;25(15):2923–2926. doi: 10.1016/j.bmcl.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruijssers A.J., Denison M.R. Nucleoside analogues for the treatment of coronavirus infections. Curr Opin Virol. 2019;35:57–62. doi: 10.1016/j.coviro.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senapati S., Kumar S., Singh A.K., Banerjee P., Bhagavatula S. Assessment of risk conferred by coding and regulatory variations of TMPRSS2 and CD26 in susceptibility to SARS-CoV-2 infection in human. J. Genet. 2020;99(1):1–5. doi: 10.1007/s12041-020-01217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 17.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 18.Pushpendra S., Kushwaha P.P., Shashank K. Novel potent inhibitors of Plasmodium vivax dihydrofolate reductase: an in silico anti-malarial drug discovery. Indian J. Pharm. 2018;52(1):122–134. doi: 10.5530/ijper.52.1.14. Jan 1. [DOI] [Google Scholar]
- 19.Gupta S., Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Senapati S., Kumar S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1776157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software. 2015;1:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 21.Schüttelkopf A.W., van Aalten D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 22.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52(12):7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 23.Hess B., Bekker H., Berendsen H.J., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18(12):1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:123.0.CO;2-H. [DOI] [Google Scholar]
- 24.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 25.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 26.DeLano W.L. The PyMOL molecular graphics system. http://www.pymol.org.2002
- 27.Bahar I., Atilgan A.R., Demirel M.C., Erman B. Vibrational dynamics of folded proteins: significance of slow and fast motions in relation to function and stability. Phys. Rev. Lett. 1998;80(12):2733–2736. doi: 10.1103/PhysRevLett.80.2733. [DOI] [Google Scholar]
- 28.Kumari R., Kumar R., Open Source Drug Discovery Consortium. Lynn A. g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 29.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017 Mar 3;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frick D.N., Virdi R.S., Vuksanovic N., Dahal N., Silvaggi N.R. Molecular basis for ADP-ribose binding to the Mac 1 domain of SARS-CoV-2 nsp3. Biochemistry. 2020;59(28):2608–2615. doi: 10.1021/acs.biochem.0c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michalska K., Kim Y., Jedrzejczak R., Maltseva N.I., Stols L., Endres M., Joachimiak A. Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes. IUCrJ. 2020;7(Pt 5):814–824. doi: 10.1107/S2052252520009653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debnath P., Debnath B., Bhaumik S., Debnath S. In silico identification of potential inhibitors of ADP-ribose phosphatase of SARS-CoV-2 nsP3 by combining E-pharmacophore- and receptor-based virtual screening of database. Chemistry. 2020;5(30):9388–9398. doi: 10.1002/slct.202001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.https://clinicaltrials.gov/ct2/show/NCT04604704. Assessed on 25-11-2020.
- 34.Cardoso T., Marques C., Dagostin J.L.A., Masson M.L. Lactobionic acid as a potential food ingredient: recent studies and applications. J. Food Sci. 2019;84(7):1672–1681. doi: 10.1111/1750-3841.14686. [DOI] [PubMed] [Google Scholar]
- 35.Hoffhine J.C., inventor, Abbott Laboratories, assignee Aqueous soluble salts of erythromycin. United States patent US. 1956;2:761–859. Sep. 4. [Google Scholar]
- 36.Gandhi G.R., Vasconcelos A.B.S., Wu D.T., Li H.B., Antony P.J., Li H., Geng F., Gurgel R.Q., Narain N., Gan R.Y. Citrus flavonoids as promising phytochemicals targeting diabetes and related complications: a systematic Review of in vitro and in vivo studies. Nutrients. 2020;12(10):2907. doi: 10.3390/nu12102907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin T., Rasul A., Sarfraz A., Sarfraz I., Hussain G., Anwar H., Riaz A., Liu S., Wei W., Li J., Li X. Salvianolic acid A & B: potential cytotoxic polyphenols in battle against cancer via targeting multiple signaling pathways. Int. J. Biol. Sci. 2019;15(10):2256–2264. doi: 10.7150/ijbs.37467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.https://patents.google.com/patent/CN101530409A/en?oq=CN101530409A. Assessed on 25-11-2020.
- 39.Kong J., Wu Z.X., Wei L., Chen Z.S., Yoganathan S. Exploration of antibiotic activity of aminoglycosides, in particular ribostamycin alone and in combination with ethylenediaminetetraacetic acid against pathogenic bacteria. Front. Microbiol. 2020;11:1–9. doi: 10.3389/fmicb.2020.01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gohlke H., Kiel C., Case D.A. Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J. Mol. Biol. 2003;330(4):891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 41.David C.C., Jacobs D.J. Principal component analysis: a method for determining the essential dynamics of proteins. Methods Mol. Biol. 2014;1084:193–226. doi: 10.1007/978-1-62703-658-0_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.