Abstract
Arabi lambs (n =28; body weight = 24 ± 3.7 kg; average age = 120 ± 8 days) were used to investigate the effect of microbial additives on growth performance, microbial protein synthesis and rumen microbial population of fattening lamb based on completely randomized design. Four treatments were studied: (1) control (without additive; CON); (2) Lactobacillus fermentum and L. plantarum (FP); (3) Saccharomyces cerevisiae (SC) plus FP (SCFP); and (4) Megasphaera elsdenii plus SCFP (MSCFP). Lambs were inoculated before morning feeding (daily oral dosed) with a 50 mL microbial suspension as follows: FP, 50 mL bacterial suspension containing 4.5 × 108 colony-forming unit per day (cfu/d) of L. plantarum and L. fermentum (in ratio 50:50); SCFP, 50 mL microbial suspension containing 4.5 × 108 cfu/d FP and 1.4 × 1010 cfu/d SC; MSCFP, 50 mL microbial suspension containing 4.5 × 108 cfu/d Me, 4.5 × 108 cfu/d FP and 1.4 × 1010 cfu/d SC. Feed intake and body weight of lambs were not affected by microbial additives. Average daily gain and feed efficiency were increased on day 0 to 21. The highest concentration of uric acid, total excreted purine derivatives (PD), microbial N, microbial CP, and metabolizable protein were in MSCFP lambs. The ruminal population of Ruminococcus albus and Ruminococcus flavefaciens was higher in MSCFP and SCFP than CON and FP lambs. The highest and the lowest abundance of M. elsdenii and methanogen respectively was observed in lambs fed on microbial additives. The tendency to improve growth performance vs. CON may be due to improvements in microbial protein synthesis and microbial populations, especially fiber-degrading bacteria. The decrease in the population of methanogens as a result of the use of microbial additives is another positive result.
Keywords: lamb, microbial additives, microbial population, microbial protein synthesis, performance
INTRODUCTION
Antibiotics are natural or synthetic compounds that generally kill or inhibit the growth of bacteria. Previous studies show that using low doses of antibiotics improved feed efficiency (FE) and growth performance (Chattopadhyay, 2014). But, the use of antibiotics for growth promotor, treatment, or control of the disease, creates a pool of antibiotic-resistant bacteria that infect animals and products, that it can transmit to humans and animals through the consumption of these animal products (Van der Fels-Klerx et al., 2011). So, global concern about the use of antibiotics has led many countries to ban the use of antibiotics in animal feed (Sarker et al., 2010). Hence, given the prohibition of antibiotics, the use of alternative additives such as probiotics can be useful.
Probiotics or microbial additives (bacterial and yeast additives) are viable organisms that have positive effects on animal health and performance (Arowolo and He, 2018). In a review study of Elghandour et al. (2015) reported that the use of microbial additives leads to changes in the microbial environment, increased weight gains and improved FE. Studies showed that yeast additives such as Saccharomyces cerevisiae lead to an increase in the concentration of ruminal bacteria, especially Fibrobacter spp., due to the equilibrium in rumen pH (Beauchemin et al., 2006). Bacterial additives such as lactate producing bacteria (LAB) (Lactobacillus, Bifidobacterium, Enterococcus, Streptococcus, and Bacillus) and lactate utilizing bacteria (LUB) (Megasphaera elsdenii, Selenomonas ruminantium, and Propionibacterium) are used as microbial additives. The most important hypothesis about LAB is that they are stimulating the growth of LUB leading to stability ruminal pH (Seo et al., 2010). The LUB can influence rumen fermentation in a high grain diet (Mackie and Gilchrist, 1979), change lactate to VFA (e.g., butyrate), increase propionate production than lactic acid, increase feed efficiency, and increase ruminal pH (Seo et al., 2010). Yeast-containing additives have been investigated in lambs (Soren et al., 2013), but there is limited information about the use of combined bacterial and yeast additive on microbial population and microbial protein synthesis in lambs. Since the use of microbial additives based on LAB in dairy cows has become commonplace, our aim was to use treatments based on LAB in growing lambs. So, in the present study, we investigated three treatments based on LAB compared to controls (without additives). It was hypothesized that the mixture of L. fermentum GP10, L. plantarum (FP) (in ratio 50:50) alone or combined M. elsdenii GU1 (Me) and S. cerevisiae (SC) can improve performance, microbial protein synthesis, and rumen microbial population in growing lambs.
MATERIALS AND METHODS
Animals, Diet, and Treatment
Twenty-eight autumn born Arabi lambs (body weight = 24 ± 3.7 kg; average age = 120 ± 8 d) were randomly divided into four groups (n = 7), which were vaccinated against external (Azantole) and internal (Triclabendazole and Levamisole) parasites and were penned individually (1.5 m × 1.3 m). The experimental period was 77 d (14 d adaptation and 63 d trial period) and during this period the complete mixed ration (contained g/kg of dry matter (DM)): alfalfa hay 201; wheat straw 99; barley grain 300; corn grain 210; soybean meal 123.5; wheat bran 55; calcium carbonate 4; NaCl 2.5; vitamin and mineral supplements 5) was provided for each lamb twice daily at 8:00 a.m. and 16:00 p.m. (allowed 5% of orts) (NRC, 2007). The chemical composition of the diet is presented in Table 1. The ration was adjusted to reach 250 g daily weight gain. During the experiment, lambs had free access to fresh water and salt licks.
Table 1.
Chemical composition and metabolizable energy of ration fed to lambs
| Itema, % DM | Total mixed ration |
|---|---|
| Dry matter | 90.3 ± 0.60 |
| Organic matter | 94.8 ± 0.14 |
| Crude protein | 16.1 ± 0.25 |
| Ether extract | 2.7 ± 0.03 |
| Neutral detergent fiber | 29.0 ± 0.15 |
| Acid detergent fiber | 16.5 ± 0.32 |
| Nonfiber carbohydratesa | 47.2 ± 0.52 |
| Metabolizable energy, Mcal/kg DMb | 2.65 ± 0.08 |
aNonfiber carbohydrates (calculated as: 1,000 – (NDF g/kg + CP g/kg + EE g/kg + crude ash g/kg).
bMetabolizable energy (calculated from each feed composition).
Experimental treatments were 1) CON (control, without additive); 2) FP (50 mL bacterial suspension containing 4.5 × 108 colony-forming unit per day (cfu/d) of L. plantarum and L. fermentum (in ratio 50:50); 3) SC + FP (50 mL microbial suspension containing 4.5 × 108 cfu/d FP and 1.4 × 1010 cfu/d SC; SCFP); and 4) Me + SCFP (50 mL microbial suspension containing 4.5 × 108 cfu/d Me, 4.5 × 108 cfu/d FP and 1.4 × 1010 cfu/d SC; MSCFP). For each treatment, the lambs were inoculated with a 50 mL microbial suspension before morning feeding (daily oral administration). The SC was used from Iran Mollasses company (Mashhad, Iran), that each gram of this yeast contains 7×109 cfu/g. The commercial strain of L. plantarum and L. fermentum (GP10) isolated from the rumen of Najdi goat were used as LAB (Mohammadabadi et al., 2018). M. elsdenii (GU1) isolated from the rumen of Najdi goat were used as LUB (Mohammadabadi et al., 2018).
Sample Collection and Chemical Analysis
At the end of adaptation period, the body weights of lambs were recorded before morning feeding as initial weight, and also the lambs were weighted using digital scales before morning feeding on d 21, d 42, and d 63. To determine intake, the amount of feed offered and orts for each lamb was recorded every day before morning feeding. Feed and orts samples were taken for chemical analysis and stored at −20 °C. Finally, average daily gain (ADG) and FE (gain:feed) were calculated for each lamb. On d 50 of experiments, five lambs from each treatment with the same body weight were transferred to the metabolic cages equipped with feces and urine collector for 13 d (7 d adaptation to the metabolic cages and 6 d sampling). During the collection period, samples of feed offered and orts was collected (10%) and stored at −20 °C for subsequent analysis. The total urine volume was collected and weighed during each day of the sampling period in plastic buckets containing 100 mL sulfuric acid (10% v/v) to reduce the pH to less than 3. After that 10% of urine samples were diluted five times with distilled water, to prevent the precipitation of purine derivatives (PD) and stored at −20 °C until subsequent analysis. On d 63 of experiment, ruminal fluid (RF) was obtained with a stomach tube 3 h after morning feeding and frozen at −80 °C until deoxyribonucleic acids (DNA) was extracted.
Chemical Analysis
Feed and orts samples were analyzed after drying at 55 °C for 48 h and grinding (1 mm particle size) to determine crude ash (No. 924.05), crude protein (CP, No. 988.05; N×6.25), ether extract (EE, No. 920.39), and acid insoluble fibers (ADF, No. 973.18) according to AOAC (1998). Neutral detergent fiber (NDF) was analyzed according to Van Soest et al., (1991). Nonfiber carbohydrates (NFC) concentration was calculated by difference as NFC (g/kg DM) = 1000 − (NDF g/kg DM + CP g/kg DM + EE g/kg DM + ash g/kg DM).
Urine Purine Derivatives and Microbial Protein Synthesis
Urinary PD (mmol/d) including allantoin, uric acid, xanthine (X), and hypoxanthine (H) were determined using colorimetric method according to Chen and Gomes (1992). Also, microbial N synthesis (gN/d) was estimated by the following equations of Chen and Gomes (1992): Y = 0.84X + (0.150 BW0.75e−0.25X) and microbial N = 70X/(0.116 × 0.83 × 1,000), where Y (mmol/d) = PD excreted in urine; X (mmol/d) = absorption of microbial purines; and e (mmol/BW0.75 daily) = endogenous PD excretion.
Total DNA Extraction and Real-time Quantitative Polymerase Chain Reaction (qPCR)
The RF was thawed at room temperature and microbial DNA was extracted using a genomic DNA extraction kit (AccuPrep, Bioneer Corporation, Daejeon, South Korea) equipped with spin columns according to the manufacturer’s instructions. To determine the concentration and purity of DNA extracted, Nanodrop Spectrophotometer (NanoDrop ND-1000, USA) was used. Species-specific PCR primers (16S rDNA) used in this study are list in Table 2. The qPCR was performed in a CFX96 optical reaction module (Bio-Rad Laboratories Inc.) and analyzed using software Bio-Rad CFX Manager (version 3.1). Each amplification reaction was run in duplicate with a final volume of 20 μL. Components of the PCR reaction consisted of 1 µL of template DNA, 2 µL of both primers (1 pmol Forward and 1 pmol Reverse), 10 µL of SYBR Green PCR Master Mix Kit and 7 µL of deionized water. SYBR Green qPCR Master Mix contained Taq DNA polymerase, reaction buffer (KCl and (NH4)2SO4), dNTPs, MgCl2, and SYBR Green. The amplification program was one cycle at 95 °C for 5 min (initial denaturation), 35 cycles of 95 °C for 30 s (denaturation), 61°C for 30 s (annealing), and 72 °C for 45 s (extension) and a final elongation at 72 °C for 10 min. The specificity of amplified products was confirmed by melting temperature and dissociation curve after amplification.
Table 2.
PCR primers for real-time PCR assay
| Target species | Primer sequence (5′→3′) | Efficiency (%) | Product size (bp) | Reference |
|---|---|---|---|---|
| Total bacteria | R: GTGSTGCAYGGYTGTCGTCA F: ACGTCRTCCMCACCTTCCTC |
95.3 | 120 | Maeda et al. (2003) |
| Fibrobacter succinogenes | R: GTTCGGAATTACTGGGCGTAAA F: CGCCTGCCCCTGAACTATC |
96.4 | 121 | Zhang et al. (2008) |
| Ruminococcus albus | R: CCCTAAAAGCAGTCTTAGTTCG F: CCTCCTTGCGGTTAGAACA |
93.1 | 175 | Koike and Kobayashi (2001) |
| Ruminococcus flavefaciens | F: CGAACGGAGATAATTTGAGTTTACTTAGG R: CGGTCTCTGTATGTTATGAGGTATTACC |
94.0 | 132 | Zhang et al. (2008) |
| Megasphaera elsdenii | F: AGATGGGGACAACAGCTGGA R: CGAAAGCTCCGAAGAGCCT |
96.7 | 95 | Stevenson and Weimer (2007) |
| Lactobacillus spp. | F: AGCAGTAGGGAATCTTCCA R: CACCGCTACACATGGAG |
100.2 | 341 | Walter et al. (2001) |
| Methanogens | F: TTCGGTGGATCDCARAGRGC R: GBARGTCGWAWCCGTAGAATCC |
95.0 | 140 | Denman et al. (2007) |
| Protozoa | F: GCTTTCGWTGGTAGTGTATT R: CTTGCCCTCYAATCGTWCT |
94.6 | 223 | Denman et al. (2007) |
F, forward; R, reverse.
Changes in targeted populations (fold changes) of Fibrobacter succinogenes, Ruminococcus albus, R. flavefaciens, M. elsdenii, Lactobacillus spp., methanogens, and protozoa were calculated using a relative quantification calculation and the 2−ΔΔCt method (Livak and Schmittgen, 2001), with general bacteria (Denman and Mc Sweeney, 2006) cycle threshold (Ct) values used as the reference and average ΔCt of the CON group as the calibrator value.
Statistical Analysis
The data obtained from assessing feed intake, growth performance, urinary purine derivatives, microbial protein synthesis, and microbial population were analyzed as a randomized complete design using General Linear Models (GLM) procedure in SAS software (SAS Institute, 2008) according to the model: Yij = μ + Ti + eij, where Yij is observation, μ is the general mean, Ti is the effect of treatment, and eij is the standard error of term. Means were compared by the Duncan multiple comparison tests at P < 0.05.
RESULTS
Feed Intake and Growth Performance
Feed intake (DM, OM, CP, NDF, ADF g/kg0.75 body weight (BW) and ME MJ/kg0.75 BW; P = 0.77, P = 0.78, P = 0.80, P = 0.77, P = 0.65, and P = 0.77, respectively) and BW of lambs on d 21, d 42, and d 63 were not affected (P = 0.98, P = 0.95, P = 0.95, respectively) by experimental treatments. Lambs ADG and FE were increased (P = 0.03, P = 0.01, respectively) by dietary microbial additives on d 0 to d 21 compare to CON, but there was no difference between microbial additives and CON on other days of the experiment. However, the total ADG increased (P = 0.04) in MSCFP vs. CON lambs (Table 3).
Table 3.
Effect of microbial feed additives on feed intake and growth performance in lambs at d 0, d 21, d 42, and d 63
| Treatmentsb | ||||||
|---|---|---|---|---|---|---|
| Itema | CON (n = 7) |
FP (n = 7) |
SCFP (n = 7) |
MSCFP (n = 7) |
SEM | P-value |
| Intake, g/kg0.75 BW | ||||||
| DM | 67.9 | 68.4 | 68.6 | 69.6 | 1.15 | 0.77 |
| OM | 64.5 | 65.0 | 65.2 | 66.1 | 1.06 | 0.78 |
| CP | 11.0 | 11.1 | 11.1 | 11.2 | 0.18 | 0.80 |
| NDF | 19.5 | 19.7 | 19.7 | 20.0 | 0.33 | 0.77 |
| ADF | 11.1 | 11.3 | 11.2 | 11.5 | 0.19 | 0.65 |
| ME, MJ/kg0.75 BW | 0.75 | 0.76 | 0.76 | 0.77 | 0.012 | 0.77 |
| Initial BW, kg (0 d) | 24.3 | 23.8 | 24.5 | 24.4 | 1.62 | 0.95 |
| Final BW, kg | ||||||
| 21 d | 28.2 | 28.1 | 28.8 | 28.8 | 1.48 | 0.98 |
| 42 d | 32.7 | 32.3 | 33.2 | 33.5 | 1.51 | 0.95 |
| 63 d | 37.3 | 37.3 | 38.2 | 38.3 | 1.64 | 0.95 |
| ADG, g | ||||||
| 0–21 d | 184 | 204 | 205 | 209 | 6.66 | 0.03 |
| 21–42 d | 213 | 200 | 211 | 225 | 8.77 | 0.33 |
| 42–63 d | 218 | 237 | 238 | 226 | 9.95 | 0.50 |
| 0–63 d | 205 | 214 | 218 | 220 | 6.82 | 0.04 |
| FE | ||||||
| 0–21 d | 0.203 | 0.230 | 0.222 | 0.226 | 0.01 | 0.01 |
| 21–42 d | 0.207 | 0.192 | 0.198 | 0.205 | 0.01 | 0.69 |
| 42–63 d | 0.196 | 0.208 | 0.204 | 0.192 | 0.01 | 0.71 |
| 0–63 d | 0.202 | 0.209 | 0.207 | 0.206 | 0.01 | 0.94 |
aBW, body weight; DM, dry matter; OM, organic matter; CP, crude protein; NDF, neutral detergent fiber; ADF, acid detergent fiber; ME, metabolizable energy ADG, average daily gain; FE, feed efficiency (gain:feed).
bCON, without microbial additive; FP, Lactobacillus fermentum and Lactobacillus plantarum; SCFP, Saccharomyces cerevisiae (SC) plus FP; MSCFP, Megasphaera elsdenii plus SC plus FP.
Urine Purine Derivatives and Microbial Protein Synthesis
Allantoin and X + H concentrations were not affected (P = 0.13, P = 0.06, respectively) by microbial additives. However, the concentration of uric acid was increased (P = 0.03) in MSCFP lambs compare to CON (2.21 vs. 1.97 mmol/d, respectively). Moreover, the concentration of total excreted PD was increased (P = 0.03) in MSCFP vs. FP and CON lambs. The highest levels of microbial N, microbial CP and metabolizable protein was observed in MSCFP lambs (P = 0.01, P = 0.01, P = 0.02, respectively), but there was no difference between MSCFP and SCFP. The amount of microbial N synthesized gN/kg DOMI (digestible organic matter intake) was not affected (P = 0.73) by microbial additives (Table 4).
Table 4.
Effect of microbial feed additives on urinary purine derivatives and microbial protein supply in lambs
| Treatmentsb | ||||||
|---|---|---|---|---|---|---|
| Itema | CON (n = 5) |
FP (n = 5) |
SCFP (n = 5) |
MSCFP (n = 5) |
SEM | P-value |
| DOMI, g/d | 720 | 758 | 791 | 833 | 19.6 | 0.01 |
| Urinary purine derivatives, mmol/d | ||||||
| Allantoin | 10.5 | 10.5 | 10.8 | 11.2 | 0.33 | 0.13 |
| Uric acid | 1.97 | 2.07 | 2.13 | 2.21 | 0.09 | 0.03 |
| X + H | 1.18 | 1.20 | 1.28 | 1.33 | 0.05 | 0.06 |
| TPD | 13.5 | 13.8 | 14.2 | 14.7 | 0.35 | 0.03 |
| Microbial production, g/d | ||||||
| Microbial N, g/d | 11.4 | 11.9 | 12.3 | 12.8 | 0.44 | 0.01 |
| Microbial CP, g/d | 71.1 | 74.6 | 76.7 | 79.8 | 1.89 | 0.01 |
| Metabolizable protein, gN/d | 7.25 | 7.61 | 7.83 | 8.14 | 0.19 | 0.02 |
| Efficiency of microbial N | ||||||
| Microbial N, gN/kg DOMI | 16.2 | 15.7 | 15.5 | 15.3 | 0.52 | 0.73 |
aDOMI, digestible organic matter intake; X + H, xanthine + hypoxanthine; TPD; total purine derivative; microbial CP (g/d), microbial N × 6.25; metabolizable protein (gN/d), microbial N × 0.75 × 0.85 (Alderman and Cottrill, 1993).
bCON, without microbial additive; FP, Lactobacillus fermentum and Lactobacillus plantarum; SCFP, Saccharomyces cerevisiae (SC) plus FP; MSCFP, Megasphaera elsdenii plus SC plus FP.
Rumen Microbial Population
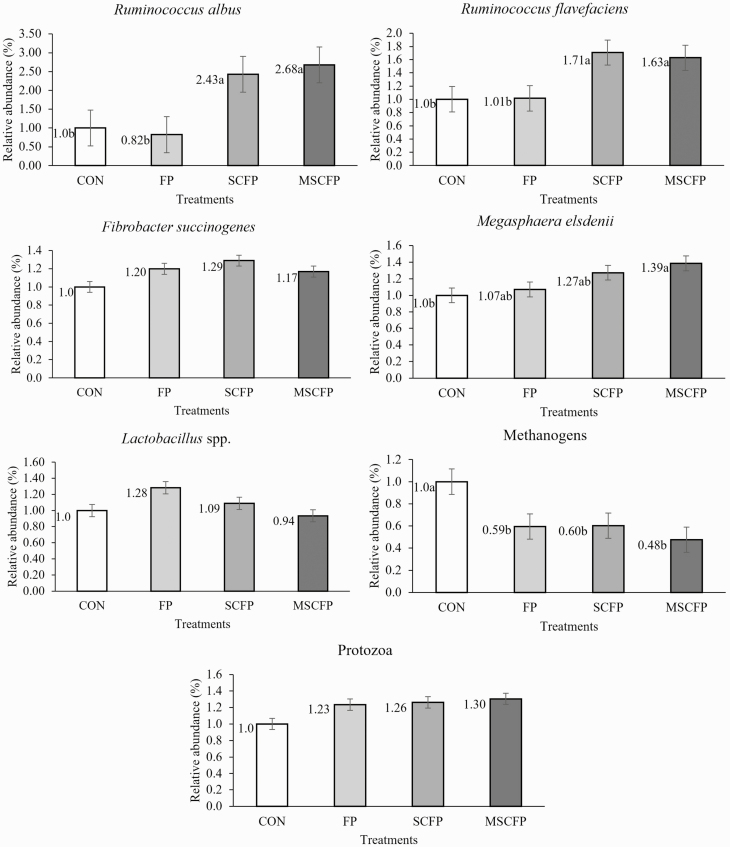
Ruminal microflora of F. succinogenes, Lactobacillus spp., and protozoa were not affected (P = 0.87, P = 0.53, P = 0.78, respectively) by microbial additives. The ruminal population of R. albus and R. flavefaciens were higher (P = 0.01, P = 0.01, respectively) in MSCFP and SCFP lambs than CON and FP. The lowest abundance of M. elsdenii was observed in CON lambs and was differ (P = 0.04) from MSCFP lambs. Moreover, methanogen population decreased (P = 0.01) using microbial additives than CON lambs (Figure 1).
Figure 1.
Effect of microbial feed additives on microbial populations in lambs. Fold change compared to CON and CON was considered as 1. a,bIndicate a differ significantly (P < 0.05). Bar indicates standard error of the mean. CON, without microbial additive; FP, Lactobacillus fermentum and Lactobacillus plantarum; SCFP, Saccharomyces cerevisiae (SC) plus FP; MSCFP, Megasphaera elsdenii plus SC plus FP.
DISCUSSION
Feed Intake and Growth Performance
Feed intake was not affected by the experimental treatments and consistent with our results, the use of microbial additives had no effect on feed intake in lambs (LAB and SC) (Alhidary et al., 2016), goats (LAB and Aspergillus awamori) (Azzaz et al., 2015), and dairy cattle (P. freudenreichii and L. acidophilus) (West and Bernard, 2011). On the other hand, feed intake was improved in young ruminants as a result of the use of live yeast (Lesmeister et al., 2004). Also, dry matter intake (DMI) and organic matter intake (OMI) were improved in the study of Bitencourt et al. (2011) by the yeast dietary supplementation. This may be due to the increased digestibility of the NDF and the DM in their experiment, and also, may be correlated with rumen development (Lesmeister et al., 2004). However, without the effect of experimental treatments on feed intake, ADG was improved on d 0 to d 21 and d 0 to d 63, which consistent with the previous results (Ayala-Monter et al., 2019). They reported that improvement in ADG without altering feed intake was due to improved FE and nutrient digestibility.
Urine Purine Derivatives and Microbial Protein Synthesis
Microbial CP plays an important role in supplying ruminant protein and provides most of the amino acids needed for growth, maintenance and production of the host animal (Vaithiyanathan et al., 2007). The effect of microbial additives on PD and microbial CP synthesis in ruminants has been investigated in limit studies. Yoon and Stern (1995) in a review study also reported microbial CP synthesis increased using probiotics. Bajagai et al. (2016) reported that supplementation with yeast increased microbial CP synthesis. However, in an in vitro experiment, Direkvandi et al. (2019) reported that the use of Me increased the synthesis of microbial protein, which could be due to the increased synthesis of branched-chain amino acids by Me. In present study, the amount of microbial CP synthesis similarly was increased in SC and Me containing treatments (MSCFP and SCFP). This is probably due to the increased use of ruminal ammonia into the microbial CP (Erasmus et al., 1992). Also, the increase in microbial N and CP in the treatments containing microbial additive compared to the CON may be due to the increase in DOMI in this treatment. Pérez et al. (1998) reported a relationship between purine derivatives and DOMI. Other studies have also shown that the amount of urinary PD and microbial CP synthesis are dependent on the amount of DMI and CPI (Puchala and Kulasek, 1992). Balcells et al. (1993) who found a positive relationship between DMI and OMI and the amount of urinary PD excreted in sheep. The amount of feed intake is one of the factors affecting the rate of microbial protein synthesis and by reducing the feed intake, the amount of microbial protein synthesis is reduced. This can be due to the reduction of N and soluble carbohydrates for microbial protein synthesis.
Rumen Microbial Population
The ruminal population of Lactobacillus spp. was not affected by the experimental treatments. Qadis et al. (2014) reported that supplementation with probiotics had no effect on L. plantarum populations and Enterococcus spp. They reported the lack of an increase in LAB was a sensitivity of these bacteria to pH greater than 6. Sari et al. (2019) observed that the total ruminal population of LAB was not affected by L. plantarum TSD-10, L. plantarum MX-16, L. brevis SPCE-39 and a mix of these bacteria. However, they reported although the total ruminal population of LAB was not affected, but populations of some Lactobacillus strains are likely to be significantly different. Also reported that the use of LUB alone or combined with LAB (Propinobacterium and Propinobacterium + E. faecium) had no effect on the ruminal population of LUB and Lactobacillus spp. (Ghorbani et al., 2002). In fact, studies have shown that the use of yeast stimulates the growth of LUB (Newbold et al., 1998), which is consistent with our results.
However, population of ruminal fiber degrading bacteria (Ruminococcus and Fibrobacter) showed no significant difference between FP and CON lambs. Contrary to our results in the in vitro experiment, the use of 0.1% E. faecium (7.0 × 108 cfu/mL) increased the ruminal population of R. flavefaciens and F. succinogenes compared to the control (Mamuad et al., 2019). The increase in the population of fiber-degrading bacteria in treatments SCFP and MSCFP is probably due to the presence of Me and SC. Me increase the population of fiber-degrading bacteria by increasing the synthesis of branched-chain amino acids (a precursor to the synthesis of branched-chain fatty acids in the cell wall of fiber-degrading bacteria) and yeast by reducing the redox potential. Reducing the potential of redox stimulates the attachment of fiber-degrading bacteria to the fiber particles in the rumen. Providing nutrients by SC stimulated the growth of R. albus, R. flavefaciens, and F. succinogenes (Chaucheyras-Durand et al., 2008). Indeed, the positive effect of yeast-containing additives is due to the stabilization of the rumen microbial environment. In an experiment of Malekkhahi et al. (2016) showed that the use of SC (20 × 109 cfu/head per day) increased the ruminal population of F. succinogenes compared to the controls during the adaptation period in dairy cattle.
The results of our study were consistent with other researchers that reported microbial additives had no effect on the ruminal protozoa population (Ghorbani et al., 2002; Galip, 2006; Izuddin et al., 2019). However, Brossard et al. (2006) reported an improvement in the rumen population of protozoa using live yeast in sheep. The effect of microbial additives on the rumen protozoa population varies according to the type of probiotic and the protozoa strain. The decrease in methanogen population may be due to increased propionate production due to the use of microbial additives. Preventing the formation of hydrogen in the rumen or consuming it is a way to prevent it from entering the methane production cycle. In fact, propionate and butyrate production is a pathway for hydrogen utilization and hydrogen is a major precursor to methane production (Jeyanathan et al., 2014). Astuti et al (2018) also reported that the use of the U32 strain of L. plantarum reduced methane production and they reported that this decrease was probably due to the positive effect of LAB on the growth of LUB. The decrease in methanogen population may be due to the increase in F. succinogenes population as a result of microbial additives. Because F. succinogenes is a nonhydrogen producing bacteria, and hydrogen is a substrate for methanogens (Mamuad et al., 2019), therefore, less substrate will be accessible for methanogens.
CONCLUSIONS
The main finding in this study was that the simultaneous use of all three microorganisms in MSCFP treatment compared to control and other treatments had a greater effect to improved nutrient intake, final BW, ADG, and microbial CP production. The tendency to improve growth performance in lambs may be due to improvements in microbial protein synthesis and microbial populations, especially fiber-degrading bacteria (R. flavefaciens and R. albus). The decrease in the population of methanogens (all treatments compared to control) and increased the population M. elsdenii (MSCFP) as a result of the use of microbial additives is another positive result.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the Agricultural Sciences and Natural Resources University of Khuzestan for their valuable assistance.
Conflict of interest statement. No conflict of interest exists.
ETHICS APPROVAL
All animal management and sampling procedures conducted according to The Care and Use of Agricultural Animals in Research and Teaching guidelines (FASS, 2010). All procedures and guidelines involving animals were approved by the Animal Experiment Committee at Agricultural Sciences and Natural Resources University of Khuzestan, Mollasani, Iran.
LITERATURE CITED
- Alderman G., and Cottrill B. R.. . 1993. Energy and protein requirements of ruminants. An advisory manual prepared by the AFRC Technical Committee on Responses to Nutrients. Wallingford, UK: CAB International. [Google Scholar]
- Alhidary I. A., Abdelrahman M. M., and Khan R. U.. . 2016. Comparative effects of direct-fed microbials alone or with a trace minerals supplements on the productive performance, blood metabolites, and antioxidant status in grazing Awassi lambs. Environ. Sci. Pollut. Res. 23(24):25218–25223. [DOI] [PubMed] [Google Scholar]
- AOAC International 1998. Official methods of analysis. 16th ed. Washington, DC: AOAC International. [Google Scholar]
- Arowolo M. A., and He J.. . 2018. Use of probiotics and botanical extracts to improve ruminant production in the tropics: a review. Anim. Nutr. 4:241–249. doi: 10.1016/j.aninu.2018.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti W. D., Wiryawan K. G., Wina E., Widyastuti Y., Suharti S., and Ridwan R.. . 2018. Effects of selected Lactobacillus plantarum as probiotic on in vitro ruminal fermentation and microbial population. Pakistan J. Nutr. 17:131–139. [Google Scholar]
- Ayala-Monter M. A., Hernández-Sánchez D., González-Muñoz S., Pinto-Ruiz R., Martínez-Aispuro J. A., Torres-Salado N., Herrera-Pérez J., and Gloria-Trujillo A.. . 2019. Growth performance and health of nursing lambs supplemented with inulin and Lactobacillus casei. Asian-Australas. J. Anim. Sci. 32:1137–1144. doi: 10.5713/ajas.18.0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzaz H. H., Aziz H. A., Farahat E. S. A., and Murad H. A.. . 2015. Impact of microbial feed supplements on the productive performance of lactating Nubian goats. Glob. Vet. 14(4):567–575. [Google Scholar]
- Bajagai Y. S., Klieve A. V., Dart P. J., and Bryden W. L.. . 2016. Probiotics in animal nutrition. Paper No. 179. Rome: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Balcells J., Fondevila M., Guada J. A., Castrillo C., and Surra J. C. E.. . 1993. Urinary excretions of purine derivates and nitrogen in sheep given straw supplemented with different sources of carbohydrates. Anim. Prod. Sci. 57:287–292. [Google Scholar]
- Beauchemin K. A., Krehbiel C. R., and Newbold C. J.. . 2006. Enzymes, bacterial direct-fed microbials and yeast: principles for use in ruminant nutrition. Biol. Grow. Anim. 4:251–284. [Google Scholar]
- Bitencourt L. L., Silva J. R. M., Oliveira B. M. L. D., Dias Júnior G. S., Lopes F., Siécola S. Jr, Zacaroni O. D. F., and Pereira M. N.. . 2011. Diet digestibility and performance of dairy cows supplemented with live yeast. Sci. Agric. 68(3):301–307. [Google Scholar]
- Brossard L., Chaucheyras-Durand F., Michalet-Doreau B., and Martin C.. . 2006. Dose effect of live yeasts on rumen microbial communities and fermentations during butyric latent acidosis in sheep: new type of interaction. J. Anim. Sci. 82:829–836. [Google Scholar]
- Chattopadhyay M. K. 2014. Use of antibiotics as feed additives: a burning question. Front. Microbiol. 5:334. doi: 10.3389/fmicb.2014.00334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaucheyras-Durand F., Walker N. D., and Bach A.. . 2008. Effects of active dry yeasts on the rumen microbial ecosystem: past, present and future. Anim. Feed Sci. Tech. 145:5–26. [Google Scholar]
- Chen X. B., and Gomes J. M.. . 1992. Estimation of microbial protein supply to sheep and cattle based on urinary excretion of purine derivatives: an overview of the technical details. Aberdeen, UK: International Feed Resources Unit, Rowett Res. Inst., Bucksburn; p. 21 (occasional publication). [Google Scholar]
- Denman S. E., and McSweeney C. S.. . 2006. Development of a realtime PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 58:572–582. [DOI] [PubMed] [Google Scholar]
- Denman S. E., Tomkins N. W., and McSweeney C. S.. . 2007. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol. Ecol. 62:313–322. doi: 10.1111/j.1574-6941.2007.00394.x [DOI] [PubMed] [Google Scholar]
- Direkvandi E., Mohammadabadi T., and Salem A. Z. M.. . 2019. The effect of lactate producing and utilizing bacteria and Saccharomyces cerevisiae on anaerobic biofermentation and digestibility in Arabi sheep. Iranian J. Rumin. Res. 7(4):111–129. [Google Scholar]
- Elghandour M. M. Y., Salem A. Z. M., Castaneda J. S. M., Camacho L. M., Kholif A. E., and Chagoya J. C. V.. . 2015. Direct-fed microbes: a tool for improving the utilization of low-quality roughages in ruminants. J. Integr. Agric. 14:526e33. [Google Scholar]
- Erasmus L. J., Botha P. M., and Kistner A.. . 1992. Effect of yeast culture supplementation on production, rumen fermentation, and duodenal nitrogen flow in dairy cows. J. Dairy Sci. 75:3056–3065. [DOI] [PubMed] [Google Scholar]
- FASS 2010. Guide for the care and use of agricultural animals in agricultural research and teaching. Champaign, IL: Federation of Animal Science Societies. [Google Scholar]
- van der Fels-Klerx H. J., Puister-Jansen L. F., van Asselt E. D., and Burgers S. L.. . 2011. Farm factors associated with the use of antibiotics in pig production. J. Anim. Sci. 89:1922–1929. doi: 10.2527/jas.2010-3046 [DOI] [PubMed] [Google Scholar]
- Galip N. 2006. Effect of supplemental yeast culture and sodium bicarbonate on ruminal fermentation and blood variables in rams. J. Anim. Physiol. Anim. Nutr. (Berl.) 90:446–452. doi: 10.1111/j.1439-0396.2006.00625.x [DOI] [PubMed] [Google Scholar]
- Ghorbani G. R., Morgavi D. P., Beauchemin K. A., and Leedle J. A.. . 2002. Effects of bacterial direct-fed microbials on ruminal fermentation, blood variables, and the microbial populations of feedlot cattle. J. Anim. Sci. 80:1977–1985. doi: 10.2527/2002.8071977x [DOI] [PubMed] [Google Scholar]
- Izuddin W. I., Loh T. C., Samsudin A. A., Foo H. L., Humam A. M., and Shazali N.. . 2019. Effects of postbiotic supplementation on growth performance, ruminal fermentation and microbial profile, blood metabolite and GHR, IGF-1 and MCT-1 gene expression in post-weaning lambs. BMC Vet. Res. 15:315. doi: 10.1186/s12917-019-2064-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyanathan J., Martin C., and Morgavi D. P.. . 2014. The use of direct-fed microbials for mitigation of ruminant methane emissions: a review. Animal 8:250–261. doi: 10.1017/S1751731113002085 [DOI] [PubMed] [Google Scholar]
- Koike S., and Kobayashi Y.. . 2001. Development and use of competitive PCR assays for the rumen cellulolytic bacteria: Fibrobacter succinogenes, Ruminococcus albus and Ruminococcus flavefaciens. FEMS Microbiol. Lett. 204:361–366. doi: 10.1111/j.1574-6968.2001.tb10911.x [DOI] [PubMed] [Google Scholar]
- Lesmeister K. E., Henrich A. J., and Gabler M. T.. . 2004. Effects of supplemental yeast (Saccharomyces cerevisiae) culture on rumen development, growth characteristics and blood parameters in neonatal dairy calves. J. Dairy Sci. 87:1832–1839. [DOI] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D.. . 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mackie R. I., and Gilchrist F. M.. . 1979. Changes in lactate-producing and lactate-utilizing bacteria in relation to pH in the rumen of sheep during stepwise adaptation to a high-concentrate diet. Appl. Environ. Microbiol. 38:422–430. doi: 10.1128/AEM.38.3.422-430.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Fujimoto C., Haruki Y., Maeda T., Kokeguchi S., Petelin M., Arai H., Tanimoto I., Nishimura F., and Takashiba S.. . 2003. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med. Microbiol. 39:81–86. doi: 10.1016/S0928-8244(03)00224-4 [DOI] [PubMed] [Google Scholar]
- Malekkhahi M., Tahmasbi A. M., Naserian A. A., Danesh-Mesgaran M., Kleen J. L., AlZahal O., and Ghaffari M. H.. . 2016. Effects of supplementation of active dried yeast and malate during sub-acute ruminal acidosis on rumen fermentation, microbial population, selected blood metabolites, and milk production in dairy cows. Anim. Feed Sci. Tech. 213:29–43. [Google Scholar]
- Mamuad L. L., Kim S. H., Biswas A. A., Yu Z., Cho K. K., Kim S. B., Lee K., and Lee S. S.. . 2019. Rumen fermentation and microbial community composition influenced by live Enterococcus faecium supplementation. AMB Express 9:123. doi: 10.1186/s13568-019-0848-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadabadi T., Bakhtiari M. A., and Alimirzaei P.. . 2018. Isolation and identification of lactate-producing and utilizing bacteria from the rumen of Najdi goats. Indian J. Small Rumin. 24(2):276–280. [Google Scholar]
- Newbold C. J., McIntosh F. M., and Wallace R. J.. . 1998. Changes in the microbial population of a rumen simulating fermenter in response to yeast culture. Can. J. Anim. Sci. 78:241–244. [Google Scholar]
- NRC (National Research Council) 2007. Nutrient requirements of small ruminants. Washington, DC: National Academy Press. [Google Scholar]
- Pérez J. F., Balcells J., Cebrián J. A., and Martín-Orúe S. M.. . 1998. Excretion of endogenous and exogenous purine derivatives in sheep: effect of increased concentrate intake. Br. J. Nutr. 79:237–240. doi: 10.1079/bjn19980040 [DOI] [PubMed] [Google Scholar]
- Puchala R., and Kulasek G.. . 1992. Estimation of microbial protein flow from the rumen of sheep using microbial nucleic acid and urinary excretion of purine derivatives. Can. J. Anim. Sci. 72:821–830. [Google Scholar]
- Qadis A. Q., Goya S., Ikuta K., Yatsu M., Kimura A., Nakanishi S., and Sato S.. . 2014. Effects of a bacteria-based probiotic on ruminal pH, volatile fatty acids, and bacterial flora of Holstein calves. J. Vet. Med. Sci. 76:877–885. doi: 10.1292/jvms.14-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sari N. F., Ridwan R., Fidriyanto R., Astuti W. D., and Widyastuti Y.. . 2019. The Effect of probiotics on high fiber diet in rumen fermentation characteristics. IOP Conf. Ser.: Earth Environ. Sci. 251:012057. doi: 10.1088/1755-1315/251/1/012057 [DOI] [Google Scholar]
- Sarker M. K., Lee S. M., Kim G. M., Choi J. K., and Yang C. J.. . 2010. Effects of different feed additives on growth performance and blood profiles of Korean Hanwoo calves. Asian Austral J. Anim. 23:52–60. [Google Scholar]
- SAS (Statistical Analysis System) 2008. SAS/STAT 9.2 user’s guide. Cary, NC: SAS Institute Inc. [Google Scholar]
- Seo J. K., Kim S. W., Kim M. H., Upadhaya S. D., Kam D. K., and Ha J. K.. . 2010. Direct-fed microbials for ruminant animals. Asian Australas. J. Anim. Sci. 23:1657–1667. [Google Scholar]
- Soren N. M., Tripathi M. K., Bhatt R. S., and Karim S. A.. . 2013. Effect of yeast supplementation on the growth performance of Malpura lambs. Trop. Anim. Health. Pro. 45:547–554. [DOI] [PubMed] [Google Scholar]
- Stevenson D. M., and Weimer P. J.. . 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75:165–174. doi: 10.1007/s00253-006-0802-y [DOI] [PubMed] [Google Scholar]
- Vaithiyanathan S., Bhatta R., Mishra A. S., Prasad R., Verma D.L., and Singh N. P.. . 2007. Effect of feeding graded levels of prosopis cineraria leaves on rumen ciliate protozoa, nitrogen balance and microbial protein synthesis in lambs and kids. Anim. Feed Sci. Tech. 133:177–191. [Google Scholar]
- Van Soest P. V., Robertson J. B., and Lewis B. A.. . 1991. Methods for dietary fiber, neutral detergent fiber, and non-starch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74(10):3583–3597. [DOI] [PubMed] [Google Scholar]
- Walter J., Hertel C., Tannock G. W., Lis C. M., Munro K., and Hammes W. P.. . 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578–2585. doi: 10.1128/AEM.67.6.2578-2585.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J. W., and Bernard J. K.. . 2011. Effects of addition of bacterial inoculants to the diets of lactating dairy cows on feed intake, milk yield, and milk composition. Prof. Anim. Sci. 27(2):122–126. [Google Scholar]
- Yoon I. K., and Stern M. D.. . 1995. Influence of direct-fed microbials on ruminal microbial fermentation and performance of ruminants: a review. Asian Australas. J. Anim Sci. 8:533–555. [Google Scholar]
- Zhang C., Guo Y., Yuan Z., Wu Y., Wang J., Liu J., and Zhu W.. . 2008. Effect of octadeca carbon fatty acids on microbial fermentation, methanogenesis and microbial flora in vitro. Anim. Feed Sci. Technol. 146:259–269. [Google Scholar]