Abstract
We recently showed that dietary grape powder (GP) imparts considerable protection against ultraviolet B (UVB)-mediated skin carcinogenesis in SKH-1 mice. To determine molecular mechanisms of this response, we employed tandem mass tag (TMT) quantitative global proteomics approach on skin tumors from mice exposed to 180 mJ/cm2 UVB twice per week and fed control or 5% GP diet. We found 2,629 proteins modulated by GP feeding, with 34 identified using stringent cutoffs (false discovery rate (FDR) q-value ≤0.1, fold change ≥1.2, p-value ≤0.05, ≥3 unique peptides). Ingenuity Pathway Analysis helped identify seven proteins involved in protein ubiquitination, including the deubiquitinase UCHL5 and 6 subunits of the 20S proteasome (PSMA1,3,4,6 and PSMB4,7). A second data set without the FDR q-value identified 239 modulated proteins, seven of which are involved in protein ubiquitination. Further, 14 proteins involved in acute phase response signaling were modulated >1.5-fold, including acute phase proteins APCS, FGA, FGB, HP, HPX, and RBP1. Evaluation of upstream regulators found inhibition of ERK1/2 phosphorylation and NF-κB p65, and an increase in IκBα in GP-treated tumors. Overall, our data suggested that GP consumption may mitigate tumorigenesis by enhancing protein ubiquitination and degradation caused by oxidative stress, and manipulates an otherwise tumor-promoting anti-inflammatory environment.
Keywords: grape, non-melanoma skin cancers, chemoprevention, proteomics
Graphical Abstract
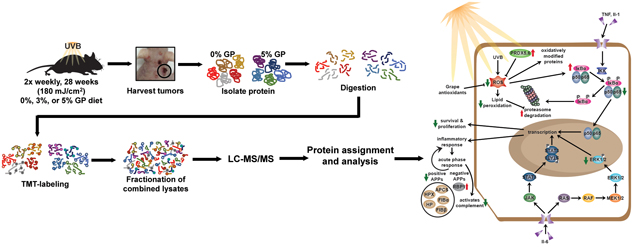
INTRODUCTION
Ultraviolet (UV) radiation is the most prevalent carcinogen in our environment, and can be divided into three categories: UVA, UVB, and UVC (reviewed in (1)). Of these, UVB is commonly thought to cause the most damage to the skin, although UVA and UVC can be harmful as well. While UVB light is required to stimulate the skin to produce vitamin D that is essential for calcium homeostasis and metabolism (2, 3), excessive UVB can lead to oxidative stress within the skin via disproportionate generation of reactive oxygen species (ROS), if not balanced by antioxidant defenses to maintain redox homeostasis (4). UVB radiation also directly causes DNA damage by formation of cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6–4)-pyrimidone photoproduct lesions, which have been shown to be carcinogenic (1, 5). Collectively, oxidative stress, DNA damage, and inflammation, as well as mutation(s) in regulatory genes by UV radiation have been linked to cellular dysfunction and the development of greater than 90% of skin cancer cases (1). As the most common neoplasms in the United States, non-melanoma skin cancers (NMSC) are morbid, but can often be treated by surgery, chemotherapy, photodynamic therapy, and/or radiation therapy unless left untreated for too long or present in hard-to-treat areas (6). However, after a primary diagnosis of NMSC, most commonly basal cell carcinoma (BCC) or squamous cell carcinoma (SCC) of the skin, patients are at an increased risk of developing subsequent NMSCs and other cancers (7, 8). Further, the existing approaches have not been sufficient in curtailing the increasing incidence of NMSCs, emphasizing the need for newer preventative and/or therapeutic strategies for the management of these cancers.
Recently, the use of nutritional supplements and/or antioxidants for the management of diseases, including cancer, has increased dramatically. Emerging research strongly supports the beneficial effects of strategically combining two or more natural agents or consuming whole foods, as these combinations may provide synergistic response over the individual constituents (9–11). We recently demonstrated that a grape powder (GP)-supplemented diet reduces UVB-mediated skin tumorigenesis in SKH-1 hairless mice, which was accompanied by reduced oxidative stress, increased repair of DNA damage, reduced cell proliferation, and enhanced apoptotic response and ROS metabolism (12). The study rationale for the use of GP was based on the fact that grapes contain hundreds of polyphenols, and their combination may provide a synergistic chemopreventive response via enhancing their cumulative bioavailability and actions against multiple oncogenic signaling pathways. As an extension of this study, here we seek to identify the molecular mechanisms of the observed skin cancer chemopreventive effect of GP by employing a comparative quantitative proteomics approach. Employing TMT LC-MS/MS for peptide quantification and analysis, we identified multiple pathways linked to the observed biological response of GP. These pathways included acute phase response signaling, response to inflammation, and protein ubiquitination, which regulate proteins in many ways including degradation via the proteasome. Our study provides insights into the molecular targets of GP-mediated skin cancer chemoprevention of UVB-mediated skin carcinogenesis.
MATERIALS AND METHODS
Materials
TMT-10plex Isobaric Label Reagent Sets were purchased from Thermo Scientific. GP was received from the California Table Grape Commission (CTGC). Resveratrol, catechin, epicatechin, peonidin, cyanidin, malvidin, kaempferol, isorhamnetin, taxifolin and quercetin were the top 10 polyphenols present in the GP used in this study (detailed in (12)).
Mice, treatments, tissue collection, and protein isolation
For this study, we used tissue samples collected during our recently published study where we determined the chemopreventive efficacy of dietary grape against UVB-mediated skin carcinogenesis in SKH-1 hairless mice (12). Briefly, we employed a UVB initiation-promotion protocol, where mice were exposed to 180 mJ/cm2 UVB twice weekly for 28 weeks. The mice were given GP-fortified diet at a dose of 3% or 5%. All diets were matched to the natural sugar content of the 5% GP diet. Details of mice and treatment protocol are provided in our previous publication (12). At the termination of the experiment, mice were euthanized and skin and tumor tissue were collected and flash-frozen in liquid nitrogen and stored at −80°C until further use. For protein isolation, skin tissues were pulverized into a powder using a mortar and pestle on liquid nitrogen. Powdered tissue was lysed in 1X RIPA lysis buffer (MilliporeSigma) with freshly added PMSF (Amresco) and protease inhibitor cocktail (Thermo Scientific). Total protein concentration was determined using Pierce BCA Protein Assay (Thermo Scientific) per manufacturer’s protocol.
Protein precipitation and digestion
For this study, we used the tumor lysates obtained from five randomly selected mice, each from the control diet and 5% GP diet. Equal amounts of protein were aliquoted into tubes, and sample volumes were adjusted with water to 20 μL. Proteins were precipitated by adding 180 μL 8:1 acetone:trichloroacetic acid (TCA) for final concentrations of 80% acetone and 10% TCA and incubated at −20°C for 1 h. Precipitated proteins were washed twice with cold neat acetone followed by cold 80% methanol in water. Proteins were resolubilized in 25 μL 8 M urea dissolved in 50 mM ammonium bicarbonate (ABC, pH 8) in water containing 5 mM Tris. Proteins were held overnight at 4°C to resolubilize. To each sample was added 125 μL ABC to dilute urea to 1.33 M. Protein disulfides were reduced using dithiothreitol (DTT) at a solution concentration of 2 mM by incubating at 55°C 30 min. Free sulfhydryls were then alkylated by reaction with iodoacetamide (IAA) at 5 mM final concentration with incubation at room temperature in the dark for 45 min. A second addition of DTT to final concentration of 2 mM was made to quench the alkylation reaction. Proteins were simultaneously digested with lys-C (Wako) and trypsin (Promega) proteases at an enzyme to substrate ratio of 1:25 and 1:20, respectively. Digestion was done for 16 h at 37°C with gentle shaking and the digests were acidified with heptafluorobutyric acid to pH 2, then cleaned up by solid-phase extraction using 100 μL Omix tips (Agilent Technologies, Inc.) and eluting with 75% acetonitrile, 0.1% formic acid in water.
Tandem mass tag (TMT) labeling
Samples were labeled using the TMT-10plex Isobaric Label Reagent Set (Thermo Scientific) generally according to manufacturer’s instructions with some minor changes as follows. Samples were dried by vacuum centrifugation and resolubilized in 40 μL 100 mM triethylammonium bicarbonate (TEAB) pH 8 (Thermo Scientific). To each 8 mg vial of TMT labeling reagent was added 100 μL dry acetonitrile. Then, 41 μL dissolved TMT reagent was added to each sample (131-fold excess) and the reaction was carried out for 1 h at room temperature. The reaction was quenched by the addition of hydroxylamine to 0.44%.
A trial pool of digested samples individually labeled with the 10 different TMT reagents was created by pooling 2 μL from each reaction. This trial pool was acidified and subjected to solid-phase extraction, followed by an analysis of 1.1 μg total labeled digest by Orbitrap LC-MS/MS. Reporter ion abundances for all identified peptides were summed to determine an approximate protein abundance ratio among all the samples in a 10-plex experiment. Inputs for each TMT-labeled sample were then adjusted to compensate for lower- and higher-abundance samples as shown in Supplementary Table S1.
High pH fractionation
TMT-labeled pooled samples were subjected to reverse-phase fractionation at pH 10. Solvent A consisted of 10 mM ammonium formate in water (pH adjusted to 10 with ammonium hydroxide) and solvent B consisted of 10 mM ammonium formate (pH 10), 80% acetonitrile in water. The column used was a Gemini C18 column (Phenomenex), 4.6 mm × 250 mm packed with 5 μm particles. The gradient is shown in Supplementary Table S2a.
The flow rate was 1 mL/min. Absorbance was monitored by a photodiode array detector at 214 nm and 280 nm. Labeled, pooled samples (described above) were dried by vacuum centrifugation and resolubilized in 200 μL solvent A. The entire sample was loaded into a 500 μL sample loop and fractions were collected every 1 min from the start of the gradient program. Fractions were pooled together so as to reduce LC-MS/MS analyses while preserving chromatographic distribution. Every sixth fraction starting from fraction 6 and ending at fraction 31 were pooled together, yielding 5 overall fractions (e.g. fractions 6, 11, 16, 21, 26, and 31 are combined to make fraction 1, while fractions 7, 12, 17, 22, and 27 are combined to make fraction 2, etc.). This results in a reduction in complexity of the individual fractions compared to the unfractionated material while preserving a broad distribution of peptide hydrophobicities within a fraction. Pooled fractions were dried and resolubilized in 0.2% formic acid in water at a concentration of 1 μg/μL.
LC-MS/MS
LC-MS/MS was performed on an Orbitrap Elite (Thermo Scientific) fitted with the Easy-Spray source and coupled to Agilent 1100 Nanopump and temperature-controlled autosampler. Chromatography was performed using an Easy-Spray column with integrated emitter and heater, 15 cm × 75 μm, packed with Pepmap RSLC C18, 3 μm, 100 Å stationary phase (Thermo Scientific). The HPLC method used 0.1% formic acid in water as solvent A and 0.1% formic acid in acetonitrile as solvent B. The gradient is shown in Supplementary Table S2b.
The autosampler was held at 6°C. The Orbitrap Elite mass spectrometer was operated in data-dependent mode using higher-energy collisional dissociation (HCD) activation for MS/MS. The Orbitrap acquisition parameters are as follows: 230 min acquisition time with 1 MS scan collected in profile mode at 60,000 resolving power over the m/z range 350–1800 followed by 10 HCD MS/MS scans collected in centroid mode at 30,000 resolving power. MS/MS spectra used a 2.0 m/z isolation window, 38% normalized collision energy, 0.1 millisec activation time, and a fixed start m/z of 100. Dynamic exclusion was enabled with exclusion duration of 30 s and exclusion window of −0.51 Da to +1.1 Da. Charge state exclusion was also enabled with unassigned and singly-charged ions excluded from precursor selection. Nano-electrospray ionization was performed at 1.8kV, capillary temperature of 205°C, and S-lens RF level at 45%. Roughly, 38,000 spectra were collected across 230 min of acquisition time for each of the five high pH fractions per TMT experiment.
Data analysis
Raw mass spectral data were assigned to peptides and proteins using MaxQuant (13) and searching with a mouse protein database from Uniprot containing 50,961 sequences. All five high pH fractions from a single TMT experiment were searched together. Parameters for MaxQuant searching included trypsin specificity with up to two missed cleavages, fixed carbamidomethylation of cysteines and peptide N-termini and lysines with the TMT-10plex reagent, variable oxidation of methionine, variable acetylation of protein N-termini, variable deamidation of asparagine and glutamine residues, and variable carbamylation of peptide N-termini. TMT quantitation acceptance required a precursor intensity fraction of 0.75 to limit co-isolation interference. The first search precursor tolerance was 20 ppm with main search precursor tolerance of 4.5 ppm. Fragment ion tolerance was 20 ppm. Identifications were controlled to 1% false discovery rate at the peptide spectrum match (PSM) and protein levels. Proteins with only 1 unique or razor peptide were permitted. The post-search analysis was performed using Perseus (14). Quantitative analysis was performed at the protein group level by importing the MaxQuant results file. Reverse-decoy and contaminant proteins were removed and protein groups were eliminated that did not have at least one unique peptide. Entries were required to have at least 3 reporter ion abundances (of 10 possible) greater than zero. Reporter ion abundances were normalized within a TMT channel across all proteins by dividing by the mean abundance for that TMT channel. TMT channels were then assigned to groups based on the sample type and analyzed by two-tailed Student’s T-test. Resultant p-values were converted to q-values to correct for multiple hypothesis testing using the Permutation-FDR approach. Finally, fold-change heat maps were created for protein expression by dividing each TMT channel abundance by the mean of all channels for that protein and subjecting the result to log2 transformation. Hierarchical clustering was performed on the total protein matrix, with increasing TMT abundance assigned to red and decreasing TMT abundance assigned to green. The proteomics data generated in this study has been deposited to the NIH public repository MassIVE and has assigned a data set identifier MSV000084316 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=6b8d04475e734ebeb0c8874f32f3c895).
In silico pathway and gene ontology analysis
After post-search analysis, the 2,629 proteins were subjected to additional cut-off criteria of i) peptides with a permutation-FDR calculated q-value of ≤0.1, ii) a fold change ≥1.2, iii) 3 or more unique peptide hits, and iv) significant change in expression (T-test p-value of ≤0.05) between 5% GP and control diets. A secondary data set was created excluding the FDR-calculated q-value. The molecular function and biological process of the remaining proteins were assessed using PANTHER (Protein ANalysis THrough Evolutionary Relationships) software (15). Additionally, the remaining protein aliases and fold change were uploaded into Qiagen’s Ingenuity Pathway Analysis (IPA) software (16). The predicated canonical pathways were generated and evaluated.
Immunoblot analysis
Immunoblot analyses were performed using standard protocols, described previously (12) using lysates from control, 3% GP, and 5% GP treatment groups. Details of antibodies used are provided in Supplementary Table S3 Densitometry analysis was performed using Adobe Photoshop CC 2018. Band intensity was transformed against the loading control, then normalized to the band intensity in lane one. The mean ± standard error of the three normalized lanes are represented graphically using GraphPad Prism version 7 for Windows (GraphPad Software). Statistical analysis was performed using two way ANOVA followed by Tukey’s multiple comparisons test.
20S Proteasome activity assay
For determining the 20S proteasome activity, tumor lysates in RIPA buffer from control, 3% GP, and 5% GP tumors were pooled for 2 mice per sample for a final concentration of 20 μg protein per well in technical duplicate. The 20S Proteasome Activity Assay (MilliporeSigma) was performed per the manufacturer’s protocol and incubated at 37°C for 2 h. The 20S proteasome activity was quantified using fluorescence intensity (λex = 380/λem = 460) on the BioTek Synergy H1 Hybrid Multi-Mode Microplate reader. The assay was repeated twice with 3–6 biological replicates that were averaged. To account for assay variability within each set, all values were transformed Y/K, where Y = sample readout and K = lowest sample readout within the control group. Sets were then combined, represented graphically, and one way ANOVA followed by Tukey’s multiple comparisons test was performed using GraphPad Prism version 7 for Windows (GraphPad Software).
RESULTS AND DISCUSSION
Identification of GP-modulated proteins using TMT-10plex LC-MS/MS
We have previously demonstrated the chemopreventive effects of dietary GP (3% and 5% GP in diet) feeding against UVB-mediated skin tumorigenesis in SKH-1 hairless mice. This is an ideal model for UVB-mediated skin carcinogenesis due to the following: 1) the loss of hair cycle reduces variability of tumorigenesis based on the modifying effects of hair cycle, 2) tumors are independent, displaying significant differences in the rate of development and aggressiveness, and 3) the developed tumors model UVR-induced tumors in human (17). Therefore, tumors can range from pre-malignant papilloma growths to malignant SCCs and spindle cell carcinomas (17). In both treatment groups of 3% and 5% GP diet, we demonstrated reductions in tumorigenesis through a decrease in proliferative markers, oxidative stress, and skin damages, and increase in apoptosis (12). To determine downstream molecular mechanisms of the reduction of tumorigenesis from GP supplementation, we used the tumor lysates obtained from five randomly selected mice, each from control diet and 5% GP diet, and subjected them to tandem mass tagged (TMT-10plex) LC-MS/MS (Figure 1a). A total of 2,629 differentially modulated proteins were quantified across all tumor replicates (Supplementary Table S4). Fold-change heat maps were created using Perseus and hierarchical clustering was performed with increasing TMT abundance assigned to red and decreasing TMT abundance assigned to green (Figure 1b). Distribution of unique peptides recognized and distribution of the reporter ion intensity ratios (5% GP/control) in the identified 2,629 proteins are outlined in Figures 1c and 1d. Interestingly, Ctl3 and Ctl4 clustered with 5GP samples, possibly due to the resemblance of tumor biology. We also speculate this is due to the tumor heterogeneity of the SKH-1 hairless mouse.
Figure 1. Summary of quantitative proteomics approach in protocol and analysis.
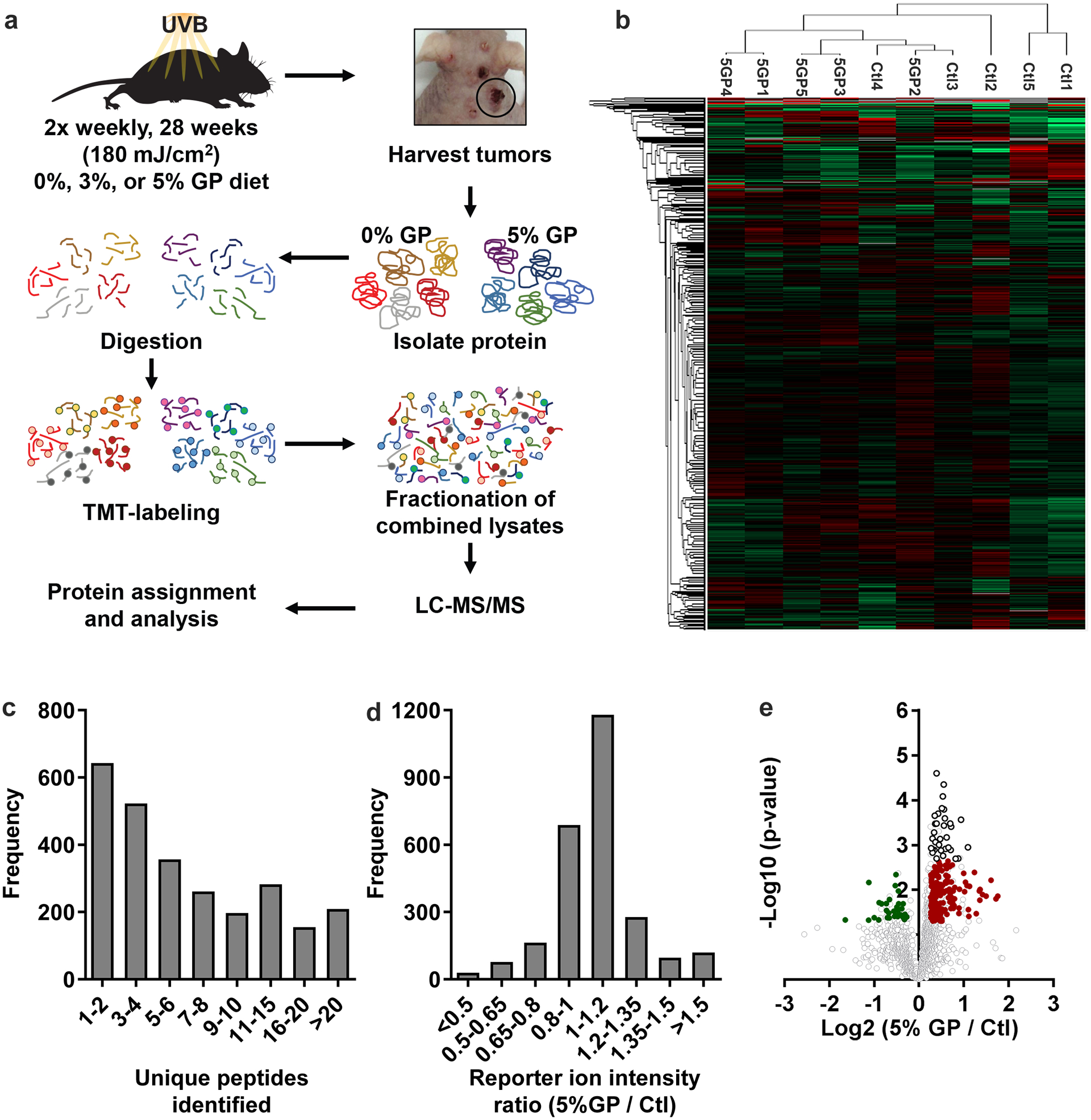
(a) Overview of TMT 10-plex mass tagging experimental design from large tumor protein lysates isolated from control mice (n=5) and 5% GP mice (n=5). (b) Heat map of log 2 transformed fold change of reporter ion intensity mean of all channels, including hierarchical clustering using MaxQuant software based upon the tumor sample ID (Ctl or 5GP). (c) Histogram of unique peptide sequences and (d) distribution of the reporter ion intensity ratios (5% GP / Ctl) in the identified 2,629 proteins. (e) Volcano plot of log 2 transformed reporter ion intensity ratios (x-axis) against log 10 transformed p-values calculated by a student’s t-test (y-axis). Significant, differentially expressed proteins were determined by two sets of cut-off parameters. Primary analysis involved proteins with a FDR q-value ≤0.1, fold change ≥1.2, p-value ≤0.05, and ≥3 unique peptides, whereas secondary analysis excluded the FDR q-value cut-off. Proteins with the primary cut-offs are indicated by black circles. Colored dots indicate decreased (green) and increased (red) protein levels within the secondary cut-off parameters. Ctl, control diet. 5GP, 5% GP diet. TMT, tandem mass tag. FDR, false discovery rate.
To identify proteins of interest, our initial cut-off parameters included i) peptides with a permutation-FDR calculated q-value of ≤0.1, ii) a fold change ≥1.2, iii) 3 or more unique peptide hits, and iv) significant change in expression (T-test p-value of ≤0.05) between 5% GP and control diets, resulting in 34 upregulated proteins (shown as circle with black outline in Figure 1e). A second data set was generated to help us elucidate modified targets of GP consumption, which had the same cut-off parameters, except we excluded the permutation FDR cut-off. As shown in the volcano plot (Figure 1e), this resulted in 239 total proteins (32 down-regulated (green) and 207 up-regulated (red)). A list of the modified proteins can be found in Supplementary Table S5. Due to the nature of isobaric mass tagging, ratio compression is a bias in the quantitative output leading to an underestimation or compression of the actual protein amount when co-eluting peptides (18). To compensate for this, we have lowered our fold-change threshold to 1.2-fold to allow us to identify the most modified proteins of interest and generate further hypotheses on downstream targets of GP consumption.
Gene ontology and pathway analysis
We utilized PANTHER software to assess cellular processes affected by GP feeding. Within the secondary data set of 239 proteins, 48% of the proteins are involved in catalytic activity (Figure 2a). Interestingly 15% of these catalytically involved proteins, including peroxiredoxins 5 (PRDX5) and 6 (PRDX6), are associated with oxidoreductase activity, supporting our previous claim that GP feeding modulates oxidative stress in skin. Furthermore, using PANTHER to identify biological processes, a large number of the affected proteins appear to be involved in metabolic or cellular processes (Figure 2b). A small fraction of the proteins (~7%) were found to be related to response to the stimulus, involving responses to stress, external stimulus, and immune response. Next, we attempted to identify the molecular pathways that are affected by GP consumption in tumorigenesis by employing IPA, a knowledge base that uses algorithms to predict upstream regulators, canonical pathways, and regulatory effects of data sets such as proteomics or gene expressions (16). The secondary data set was uploaded to IPA with the calculated q-value, the p-value, the number of peptides identified, and the corresponding ratio (5% GP/control). Interestingly, disease and function prediction analysis in IPA found that the modulation of 204 of the 239 proteins were correlated to slightly decrease cancer (Supplementary Figure S1), which supported our previous findings (12). Moreover, we also found the involvement of GP-modulated proteins in enhancing the metabolism of ROS and inhibiting hydrogen peroxide (Figure 2c). This supports previous evidence suggesting resveratrol (a major antioxidant constituent of GP) acts as an anti-inflammatory agent (19) and the notion that GP modulates oxidative stress. Upon further evaluation of the data sets, we found that several other canonical pathways were affected by GP (Figure 2d), with the top two hits being protein ubiquitination pathway (in the first data set) and APR signaling (in the second data set). The identification of APR signaling by IPA, as a top affected pathway matches with the identification of response to the stimulus by PANTHER, which includes stress response. These findings led us to explore this pathway further along with the effects of GP on protein ubiquitination and the 20S proteasome.
Figure 2. Functional analysis of the significant differentially expressed proteins.
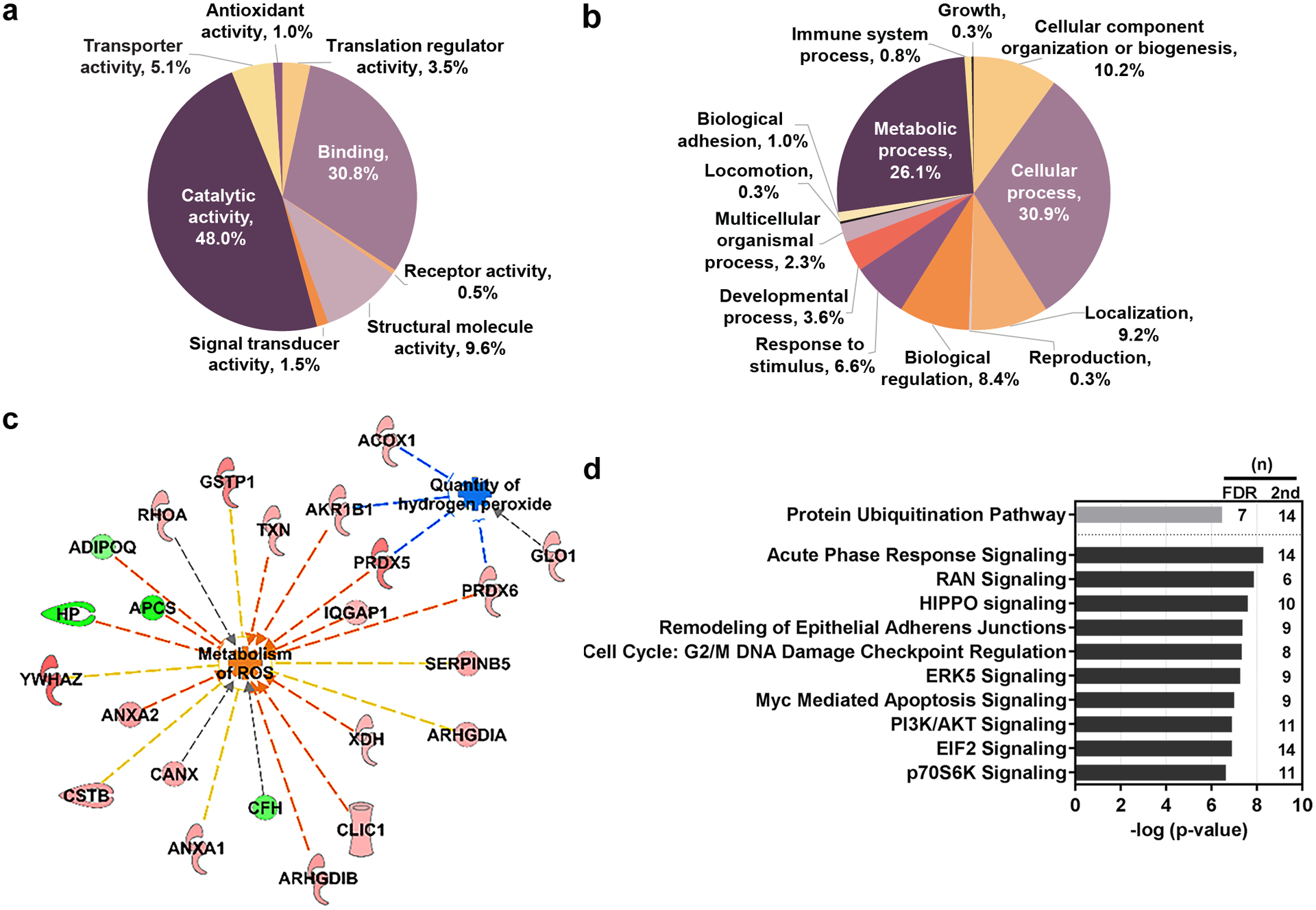
Proteins of interest were classified by PANTHER reporting (a) molecular function (b) and biological processes. (c) Functional analysis of the proteins with the secondary cut-off parameters utilizing IPA shows interactions of proteins suppressing oxidative stress through increased metabolism of ROS and reduced quantity of hydrogen peroxide. Upregulated proteins appear red in color and downregulated proteins are green. Indirect interactions are denoted by dashed lines. Blue lines suggest inhibition; orange suggest activation; yellow indicates inconsistent findings; grey indicates that an effect is not predicted. (d) Pathway analysis was conducted using IPA. Primary FDR analysis (gray bar) and secondary cut-off analysis (black bars). Bars represent the –log-transformed p-value calculated using a right-tailed Fisher Exact test of the number of focus genes in the dataset and the total number of genes known to be associated with the process. (n) indicates the number of proteins within our dataset that are involved in the corresponding pathway. IPA, Ingenuity Pathway Analysis. FDR, false discovery rate. ROS, reactive oxygen species.
Dietary GP modulates multiple 20S proteasomal subunits
Our analysis revealed that GP consumption results in the modulation of proteins involved in the Protein Ubiquitination Pathway (grey bar in Figure 2d). Due to the increased cut-off stringency, this was the only pathway identified by IPA to be significantly modulated in this data set. The 26S proteasome (Figure 3a) is a proteolytic complex, which plays an integral role in cellular homeostasis and is responsible for the degradation of poly-ubiquitinated proteins (20, 21). This complex consists of 19S regulatory cap structures surrounding the catalytic 20S core particle. The 19S cap recognizes the poly-ubiquitinated chain, then unfolds and translocates the protein to the 20S catalytic core for degradation. The 20S core is composed of four stacked rings containing seven α (PSMA1–7) or β (PSMB1–7) proteins each with an α7β7β7α7 configuration. The β proteins PSMB6, PSMB7, and PSMB5 (β1, β2, and β5, respectively) are responsible for caspase-like, trypsin-like, and chymotrypsin-like protein cleavage, whereas α subunits act as a docking site for the 19S cap and prevent proteins from random degradation (22, 23).
Figure 3. Proteomics analysis showing proteins associated with the Protein Ubiquitination Pathway.
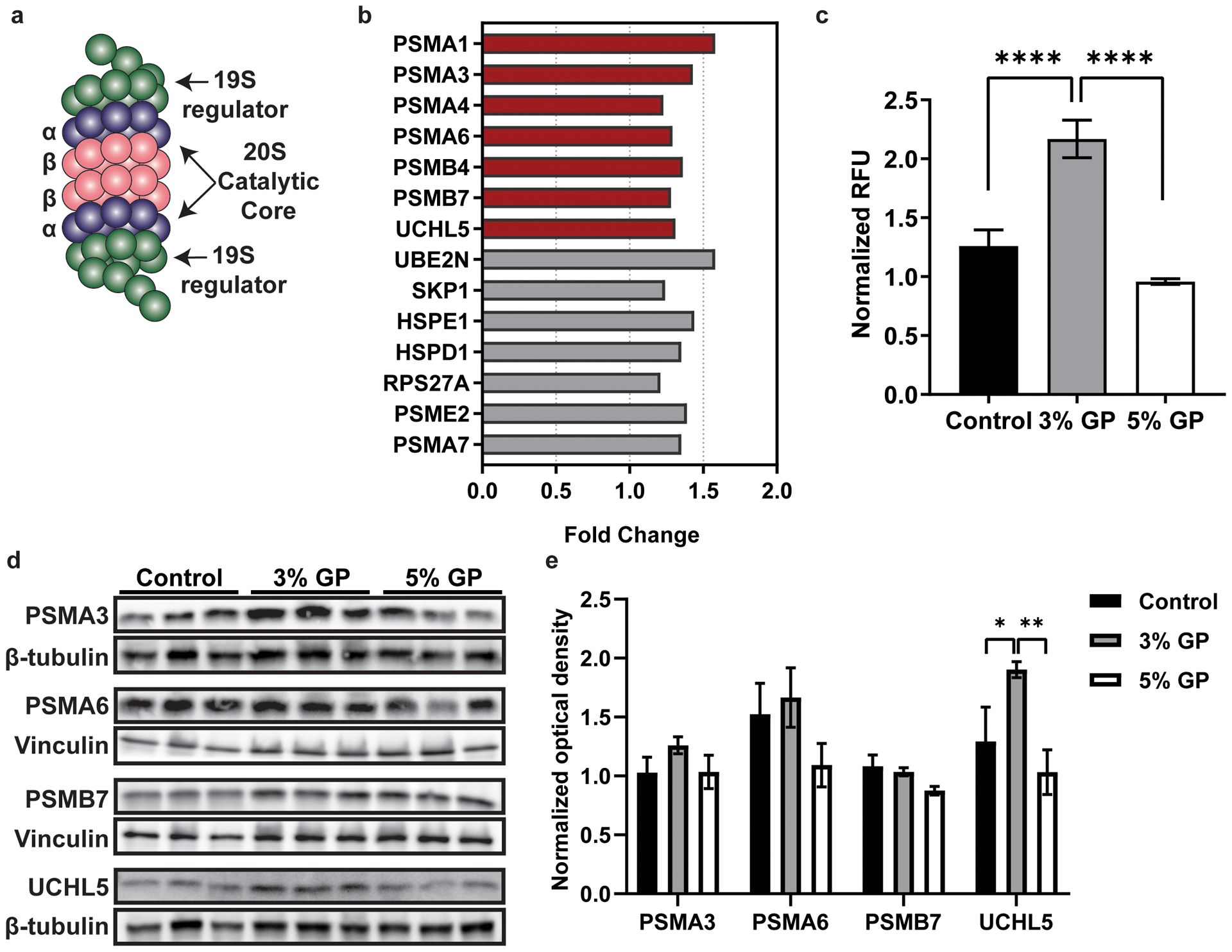
(a) Basic structure of the 26S proteasome, a complex composed of two 19S regulatory particles capping the barrel-shaped 20S catalytic core. The 20S core consists of four stacked rings with an α7β7β7α7 configuration. The α subunits are predominantly structural while the β subunits are primarily catalytic. (b) Proteins identified in our dataset that are involved in the Protein Ubiquitination Pathway identified by IPA. Red bars represent modulated proteins that are within primary cut-off criteria, while grey bars are within secondary cut-off. (c) 20S proteasome activity assay (**** p < 0.0001), with statistical significance determined using one way ANOVA with Tukey’s multiple comparisons. (d) Immunoblot analysis of modulated 20S subunits PSMA3, PSMA6, and PSMB7 and deubiquitinating enzyme, UCHL5. β-tubulin and vinculin are loading controls for the respective immunoblots. (e) Densitometry analysis for quantification of the ratios of the proteasome proteins to the loading control. After normalization to the loading control, all densitometry bars are normalized to the first lane of the control group. Data are represented as the mean ± standard error of the mean of the normalized three bands. Two-way ANOVA with Tukey’s multiple comparisons was performed to determine significance (** p < 0.01). RFU, relative fluorescent units. GP, grape powder diet.
Within the primary data set created using stringent cutoffs, we identified 6 of the 39 proteins to be subunits of the 20S proteasome, and 1 involved enzyme (UCHL5) (red, Figure 3b). When compared to the tumors of mice on a control diet, the tumors from the 5% GP treated mice indicated a 1.23–1.58 fold change in α subunits 6,7,3, and 1 (PSMA1, 3, 4, 6), and β subunits 7 and 2 (PSMB4, 7). Additional analysis using the secondary data set found that the protein ubiquitination pathway was scored at #11 in terms of significance in the secondary data set, and revealed 7 additional proteins involved in this pathway (grey bars, Figure 3b). Because these data suggest that GP affects many of the subunits within the 20S proteasome, we sought to assess if the observed protein modulations affect proteasome cleavage. To do this, we quantified 20S proteasome activity in the tumors of all treatment groups (control, 3% GP, and 5% GP). Our data suggested that the 3% GP enhanced proteasome activity, whereas 5% GP diet had no effect (Figure 3c), although the proteomics data set showed protein expression differences between the 5% GP and control diets (Figure 3b). We also performed immunoblot analysis of all treatment groups evaluating two of the α subunits, PSMA3 and 6, the trypsin-like, catalytically active β2 subunit, PSMB7, and UCHL5, an enzyme responsible for deubiquitination prior to proteasomal degradation of proteins (Figure 3d) (24, 25). By performing density analysis, we observed significant increases in UCHL5 expression within the 3% GP group, but not the 5% GP group consistent with the proteasome assay. This increase was accompanied by slight increases in the 3% GP group of PSMA3 and PSMA6, yet no change in PSMB7 (Figure 3e). Interestingly, these four proteins (UCHL5, PSMA3, PSMA6 and PSMB7) were significantly upregulated in proteomics analysis utilizing 5% GP samples.
Although proteasomes play a pivotal role in regulating the redox balance of cells by degrading oxidized proteins and is susceptible to oxidative modifications, their regulation is poorly understood. Proteasome dysfunction can lead to the accumulation of oxidized proteins, which in turn can feedback to further inhibition of the proteasome and lead to cytotoxicity. Unfortunately, dysregulation can also contribute to pathologies and oxidative stress-associated disorders including neurodegenerative disorders and various cancers (26). In our previous study, we demonstrated that GP reduced the lipid peroxidation product 4-hydroxynoneal (HNE) (12). A number of studies have suggested that proteins cross-linked by HNE can inhibit proteasome function, thereby altering the balance of ubiquitination and degradation of oxidized proteins (27–29). This disturbance can then lead to the accumulation of cellular damages which contribute to skin aging. Furthermore, the antioxidant capacity of natural compounds should enhance the proteasome activity against these damage by balancing homeostasis. Consumption of the Mediterranean diet (a balanced diet of non-starchy vegetables, fruits, legumes, etc) in elderly populations has demonstrated the efficacy of natural compounds against aging (30). Therefore, we reason that the natural components within GP may enhance proteasome activity, leading to the proper removal of damaged proteins.
Dietary GP reduces chronic acute phase response
Within the secondary canonical pathway analysis (Figure 2d), acute phase response (APR) signaling was the top predicted pathway, with 14 proteins identified. This pathway is linked with UV radiation, as UV induces inflammation in the skin, leading to the influx of cytokines by surrounding cells. This response prompts the liver to produce acute-phase proteins (APPs), a set of early responses by the body to acute damage or trauma (Figure 4a) (31, 32). APR results in the changes of many proteins grouped as either positive or negative acute-phase proteins. Positive acute-phase proteins, such as fibrinogen and haptoglobin, appear to be increased during inflammation, whereas negative APPs, including albumin, retinol-binding protein, and transferrin, are decreased. Although APR response aids in initial inflammatory mediation, prolonged expression of acute-phase proteins can support a constitutively active inflammatory environment (33–35). Within our comparative proteomic analysis, we found that GP altered 14 proteins involved in the APR signaling pathway (Figure 2b). Following our analysis, we identified five positive APPs (haptoglobin (HP), fibrinogen α (FGA), fibrinogen β (FGB), serum amyloid P-component (APCS) and (hemopexin) HPX; green bars in Figure 4b) as being more than 1.5-fold downregulated and one negative APP (retinol-binding protein 1 (RBP1); red bar in Figure 4b) as 2.14-fold upregulated. The other seven APPs identified during additional analysis using the secondary data set included AMBP, C1RA, C1S1, IL-36γ, ITIH3, RALB, and RAP1B (grey bars in Figure 4b). Because these data suggest a change of positive APP between the 5% GP and the control diet tumors, we performed immunoblot and densitometry analysis to determine if the changes applied to 3% GP treated mice as well. We confirmed significant decreases in positive APPs- HP, FGA and FGB, as well as slight decreases in APCS and alpha-1-microglobulin/bikunin precursor (AMBP) in both treatment groups (Figure 4c–d). This data suggests an anti-inflammatory response for GP in the skin.
Figure 4. Proteomics analysis showing proteins associated with the Acute Phase Response.

(a) Overview of APR, which is induced upon cellular damage from UV. After UV damage, the surrounding cells will secrete cytokines, such as Il-6 and TNFα, which signal the liver to increase the production of APPs to aid in regeneration and repair of the tissue. Upon repair, the signal is abated. However, a chronic tumor inflammatory response can lead to the constitutive activation on upstream inflammatory regulators, and therefore APPs. (b) Proteins identified in the dataset involved in APR, including positive (green) and negative (red) APPs. Green and red bars indicate proteins identified during primary analysis, and grey bars indicate the proteins found during secondary analysis. (c) Immunoblot evaluation of APPs. (d) Densitometry analysis for quantification of the ratios of the APPs to the loading control. (e) Immunoblot analysis of the activated (pERK) and total ERK1/2 (p44/42). (f)) Densitometry analysis for quantification of the ratios of activated to total ERK units p44 and p42. Bands were normalized to loading control prior to calculating the ratio. (g) Immunoblot evaluation of NF-κB and IκBα. (h) Densitometry analysis for quantification of the ratios of NF-κB and IκBα to the loading control. After normalization to the loading control, all densitometry bars are normalized to the first lane of the control group. Data are represented as the mean ± standard error of the mean of the normalized three bands. Two-way ANOVA with Tukey’s multiple comparison was performed to determine significance in all densitometry graphs (* p < 0.05, ** p < 0.01, *** p < 0.001). β-tubulin, β-actin and vinculin are loading controls for the respective immunoblots. APR, acute phase response. APP, acute-phase protein. GP, grape powder diet.
Upon inflammatory response modulating the expression of APPs, C-reactive protein upregulation can lead to the activation of the complement system (C1S and C1R, modulated −1.43 and −1.38, respectively), which further recruit inflammatory cells (36). Because the feedback loop of chronic inflammation in tumor environments, many of these positive APPs, including SAA and HP, have been proposed as potential biomarkers in patients with colorectal (37–39), lung (40, 41), hepatocellular (42), breast (43), and endometrial (44) cancers. In addition to involvement in coagulation, fibrinogen (FG) appears to have a direct role in the inflammatory response. In a wound-healing model, FG-deficient mice exhibited a deficiency in wound repair through the inability of cells to efficiently organize, although wound closure times were similar (45). Elevated FG levels have been associated with various malignancies (46–49) and can indicate the metastatic potential of circulating tumor cells in models of lung carcinoma and melanoma (50). Our proteomics data also suggests that GP preserves the functions of negative APPs, such as RBP1, which is responsible for the binding and transport of retinol (vitamin A), from the liver to cells (51). Further, as described below, due to the increased levels of APPs within our data set, we sought to evaluate the expression of upstream regulators that are linked to chronic inflammation.
Dietary GP reduces the chronic inflammatory response
Although neither of our proteomics data sets picked up these proteins, we were interested in assessing if the increased positive APP expression in control tumors was due to upstream regulators including nuclear factor kappa B (NF-κB) and mitogen-activated protein kinase (MAPK) signaling pathways. UV-induced photodamage activates multiple protective signaling cascades involved in inflammation and ROS, which are known to enhance each other (52). To maintain the redox balance within the cell, antioxidant response systems scavenge the ROS produced by the photodamage. With time, the overabundance of ROS caused by UV radiation can deplete antioxidants and decrease the efficiency of the response systems, leading to cumulative DNA damages and stress (53). Oxidative stress caused by UV radiation has been shown to induce many cascades leading to the activation of MAPKs, which regulate multiple transcriptional factors, such as NF-κB (54). ERKs 1 and 2, members of the MAPK family, can be activated by UV radiation and oxidative stress to mediate multiple cellular functions, including proliferation. Activation of ERK is dysregulated in many cancers, including skin cancer (55–58). Upon assessment of ERK1/2 activation in our tumor lysates, we observed a significant inhibition in the activation of ERK2 (p42) in the 3% GP group, with marked reduction in the ERK2 (p42) 5% GP and ERK1 (p44) in the 3% GP group upon immunoblot and densitometry analysis (Figure 4e–f).
Next, we sought to explore changes in the NF-κB signaling pathway as MAPK proteins (ERK1/2, p38 kinase, and JNK) are known to be mediators of NF-κB signaling (59, 60). UV light activates p38 and inhibitory kappa kinase (IKK) to activate NF-κB, which serves as a key regulator in the pro-inflammatory response upon activation and localization to the nucleus (61). NF-κB is a key transcription factor required for the induction of pro-inflammatory genes including Il-6 and TNF-α (62). STAT3, another protein that has been shown to be constitutively active in cancers, the activation occurs upon an accumulation of Il-6. Additionally, activated STAT3 and NF-κB can interact within the nucleus leading to further inflammatory response (63). Upon their upregulation, both STAT3 and NF-κB p65 have been linked to upregulation of HP (64) and SAA (65). Therefore, we evaluated the expression of NF-κB to determine if upstream activation is leading to constitutively active positive acute-phase proteins within UVB-mediated tumors. As demonstrated in Figure 4g–h, the 3% GP had a significant reduction in expression of NF-κB p65 and significant increase in IκBα, a protein known to tether non-phosphorylated subunits of the NF-κB complex (p65 and p50) to the cytosol. Although the slight increase in IκBα was confounding in the 5% GP group, it was accompanied by a marked reduction in NF-κB p65 expression. Interestingly, we have previously demonstrated that the grape antioxidant resveratrol can inhibit the inflammatory and oncogenic effector NF-κB in the skin (66). Therefore, we believe that the combined polyphenol effects of the GP diet inhibit the phosphorylation of ERK signaling by oxidative stress, therefore inhibiting activation of downstream NF-κB signaling.
CONCLUSIONS
As the most common malignancy in the United States, NMSC cases are at an all-time high and continuing to rise (6). Therefore, we need additional measures for the management of these cancers. In recent years, the beneficial effects of commonly consumed, naturally occurring dietary agents and supplements are being widely investigated for the prevention of cancer. Moreover, the consumption of these compounds in their natural matrix, as whole foods, in a polyphenol-rich diet has been repeatedly linked to additive/synergistic responses against a variety of diseases, including cancer (10, 67–69). Therefore, whole foods as part of a manageable diet are gaining considerable attention for better health and disease prevention. However, the molecular mechanisms of action of dietary supplements need to be carefully evaluated. Our study has sought to evaluate the mechanisms of the beneficial effects of dietary grape against skin tumorigenesis. Our data suggest that grape powder acts as an anti-inflammatory agent and enhances the activity of the 20S proteasome for the disposal of ubiquitinated proteins. Based on our findings, we have proposed a mechanism of GP-meditated action against skin cancer in Figure 5. Further studies are needed to determine upstream regulators and identify the interactions among different pathways and proteins, providing an overall skin cancer chemopreventive response.
Figure 5. Proposed mechanisms of the cutaneous biological response to UV damage with GP consumption.
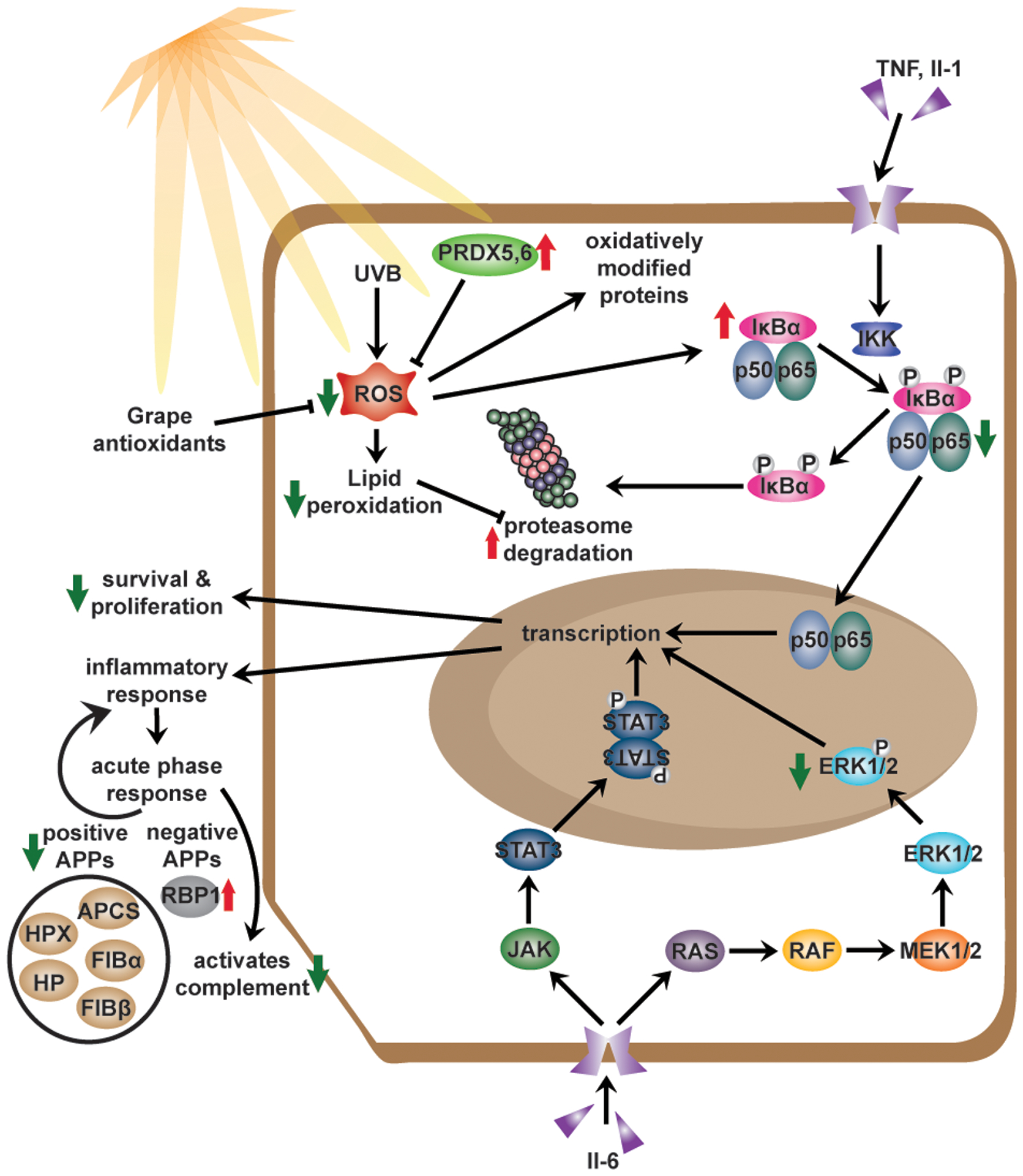
Summary of the mechanisms explored in our previous study (12) and based on data obtained in this study. Previously we reported that the chemoprotective effects of GP consumption were associated with reductions in oxidative stress, survival, and proliferation. The current study demonstrated the anti-inflammatory response of GP was associated with the modulation of APPs and therefore, upstream regulators (NF-κB and MAPK). GP consumption also modulated the proteasome activity, which activity is suggestively decreased upon oxidative stress. Red arrows indicate an upregulation of the protein or process. Green arrows indicate downregulation of protein or process. APP, acute-phase protein.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by funding from the California Table Grape Commission, as well as the National Institutes for Health (grant numbers R01AR059130 and R01CA176748 to NA), and the Department of Veterans Affairs (VA Merit Review Awards I01CX001441 and I01BX004221; and a Research Career Scientist Award IK6BX003780 to NA). We also acknowledge the core facilities supported by the Skin Diseases Research Center (SDRC) Core Grant P30AR066524 from NIH/NIAMS. The mass spectrometry work reported here was performed in the Mass Spectrometry/Proteomics Facility in the Biotechnology Center, UW-Madison.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Mancebo SE; Wang SQ, Skin cancer: role of ultraviolet radiation in carcinogenesis. Rev. Environ. Health 2014, 29 (3), 265–273. [DOI] [PubMed] [Google Scholar]
- 2.Wacker M; Holick MF, Sunlight and Vitamin D: A global perspective for health. Dermatoendocrinol. 2013, 5 (1), 51–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacLaughlin JA; Anderson RR; Holick MF, Spectral character of sunlight modulates photosynthesis of previtamin D3 and its photoisomers in human skin. Science 1982, 216 (4549), 1001–1003. [DOI] [PubMed] [Google Scholar]
- 4.Heck DE; Vetrano AM; Mariano TM; Laskin JD, UVB light stimulates production of reactive oxygen species: unexpected role for catalase. J. Biol. Chem 2003, 278 (25), 22432–22436. [DOI] [PubMed] [Google Scholar]
- 5.Ichihashi M; Ueda M; Budiyanto A; Bito T; Oka M; Fukunaga M; Tsuru K; Horikawa T, UV-induced skin damage. Toxicology 2003, 189 (1–2), 21–39. [DOI] [PubMed] [Google Scholar]
- 6.Rogers HW; Weinstock MA; Feldman SR; Coldiron BM, Incidence Estimate of Nonmelanoma Skin Cancer (Keratinocyte Carcinomas) in the U.S. Population, 2012. JAMA Dermatol. 2015, 151 (10), 1081–1086. [DOI] [PubMed] [Google Scholar]
- 7.Nugent Z; Demers AA; Wiseman MC; Mihalcioiu C; Kliewer EV, Risk of second primary cancer and death following a diagnosis of nonmelanoma skin cancer. Cancer Epidemiol. Biomarkers Prev 2005, 14 (11 Pt 1), 2584–2590. [DOI] [PubMed] [Google Scholar]
- 8.Kahn HS; Tatham LM; Patel AV; Thun MJ; Heath CW Jr., Increased cancer mortality following a history of nonmelanoma skin cancer. JAMA 1998, 280 (10), 910–912. [DOI] [PubMed] [Google Scholar]
- 9.Singh P; Arora D; Shukla Y, Enhanced chemoprevention by the combined treatment of pterostilbene and lupeol in B[a]P-induced mouse skin tumorigenesis. Food Chem. Toxicol 2017, 99, 182–189. [DOI] [PubMed] [Google Scholar]
- 10.Singh CK; Liu X; Ahmad N, Resveratrol, in its natural combination in whole grape, for health promotion and disease management. Ann. N. Y. Acad. Sci 2015, 1348 (1), 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh CK; Siddiqui IA; El-Abd S; Mukhtar H; Ahmad N, Combination chemoprevention with grape antioxidants. Mol. Nutr. Food Res 2016, 60 (6), 1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh CK; Mintie CA; Ndiaye MA; Chhabra G; Dakup PP; Ye T; Yu M; Ahmad N, Chemoprotective Effects of Dietary Grape Powder on UVB Radiation-Mediated Skin Carcinogenesis in SKH-1 Hairless Mice. J. Invest. Dermatol 2019, 139 (3), 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox J; Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 2008, 26 (12), 1367–1372. [DOI] [PubMed] [Google Scholar]
- 14.Tyanova S; Temu T; Sinitcyn P; Carlson A; Hein MY; Geiger T; Mann M; Cox J, The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13 (9), 731–740. [DOI] [PubMed] [Google Scholar]
- 15.Thomas PD; Campbell MJ; Kejariwal A; Mi H; Karlak B; Daverman R; Diemer K; Muruganujan A; Narechania A, PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003, 13 (9), 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramer A; Green J; Pollard J Jr.; Tugendreich S, Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30 (4), 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benavides F; Oberyszyn TM; VanBuskirk AM; Reeve VE; Kusewitt DF, The hairless mouse in skin research. J. Dermatol. Sci 2009, 53 (1), 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rauniyar N; Yates JR, Isobaric Labeling-Based Relative Quantification in Shotgun Proteomics. J. Proteome Res 2014, 13 (12), 5293–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das S; Das DK, Anti-inflammatory responses of resveratrol. Inflamm Allergy Drug Targets 2007, 6 (3), 168–173. [DOI] [PubMed] [Google Scholar]
- 20.Shen M; Schmitt S; Buac D; Dou QP, Targeting the ubiquitin-proteasome system for cancer therapy. Expert Opin. Ther. Targets 2013, 17 (9), 1091–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayter JR; Doherty MK; Whitehead C; McCormack H; Gaskell SJ; Beynon RJ, The subunit structure and dynamics of the 20S proteasome in chicken skeletal muscle. Mol. Cell. Proteomics 2005, 4 (9), 1370–1381. [DOI] [PubMed] [Google Scholar]
- 22.Sahara K; Kogleck L; Yashiroda H; Murata S, The mechanism for molecular assembly of the proteasome. Adv. Biol. Regul 2014, 54, 51–58. [DOI] [PubMed] [Google Scholar]
- 23.Voutsadakis IA, Proteasome expression and activity in cancer and cancer stem cells. Tumour Biol. 2017, 39 (3), 1010428317692248. [DOI] [PubMed] [Google Scholar]
- 24.Arpalahti L; Hagstrom J; Mustonen H; Lundin M; Haglund C; Holmberg CI, UCHL5 expression associates with improved survival in lymph-node-positive rectal cancer. Tumour Biol. 2017, 39 (7), 1010428317716078. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka K, The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci 2009, 85 (1), 12–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aiken CT; Kaake RM; Wang X; Huang L, Oxidative stress-mediated regulation of proteasome complexes. Mol. Cell. Proteomics 2011, 10 (5), R110 006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulteau AL; Lundberg KC; Humphries KM; Sadek HA; Szweda PA; Friguet B; Szweda LI, Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J. Biol. Chem 2001, 276 (32), 30057–30063. [DOI] [PubMed] [Google Scholar]
- 28.Keller JN; Huang FF; Zhu H; Yu J; Ho YS; Kindy TS, Oxidative stress-associated impairment of proteasome activity during ischemia-reperfusion injury. J. Cereb. Blood Flow Metab 2000, 20 (10), 1467–1473. [DOI] [PubMed] [Google Scholar]
- 29.Friguet B; Szweda LI, Inhibition of the multicatalytic proteinase (proteasome) by 4-hydroxy-2-nonenal cross-linked protein. FEBS Lett. 1997, 405 (1), 21–25. [DOI] [PubMed] [Google Scholar]
- 30.Athanasopoulou S; Chondrogianni N; Santoro A; Asimaki K; Delitsikou V; Voutetakis K; Fabbri C; Pietruszka B; Kaluza J; Franceschi C; Gonos ES, Beneficial Effects of Elderly Tailored Mediterranean Diet on the Proteasomal Proteolysis. Front. Physiol 2018, 9, 457–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cray C; Zaias J; Altman NH, Acute Phase Response in Animals: A Review. Comp. Med 2009, 59 (6), 517–526. [PMC free article] [PubMed] [Google Scholar]
- 32.D’Orazio J; Jarrett S; Amaro-Ortiz A; Scott T, UV Radiation and the Skin. Int. J. Mol. Sci 2013, 14 (6), 12222–12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabay C; Kushner I, Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med 1999, 340 (6), 448–454. [DOI] [PubMed] [Google Scholar]
- 34.Jain S; Gautam V; Naseem S, Acute-phase proteins: As diagnostic tool. J. Pharm. Bioallied Sci 2011, 3 (1), 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krzystek-Korpacka M; Matusiewicz M; Diakowska D; Grabowski K; Blachut K; Kustrzeba-Wojcicka I; Terlecki G; Gamian A, Acute-phase response proteins are related to cachexia and accelerated angiogenesis in gastroesophageal cancers. Clin. Chem. Lab. Med 2008, 46 (3), 359–364. [DOI] [PubMed] [Google Scholar]
- 36.Korkmaz HI; Krijnen PAJ; Ulrich MMW; de Jong E; van Zuijlen PPM; Niessen HWM, The role of complement in the acute phase response after burns. Burns 2017, 43 (7), 1390–1399. [DOI] [PubMed] [Google Scholar]
- 37.Holm M; Saraswat M; Joenväärä S; Ristimäki A; Haglund C; Renkonen R, Colorectal cancer patients with different C-reactive protein levels and 5-year survival times can be differentiated with quantitative serum proteomics. PLoS One 2018, 13 (4), e0195354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glojnaric I; Casl MT; Simic D; Lukac J, Serum amyloid A protein (SAA) in colorectal carcinoma. Clin. Chem. Lab. Med 2001, 39 (2), 129–133. [DOI] [PubMed] [Google Scholar]
- 39.Sun L; Hu S; Yu L; Guo C; Sun L; Yang Z; Qi J; Ran Y, Serum haptoglobin as a novel molecular biomarker predicting colorectal cancer hepatic metastasis. Int. J. Cancer 2016, 138 (11), 2724–2731. [DOI] [PubMed] [Google Scholar]
- 40.Cho WC; Yip TT; Cheng WW; Au JS, Serum amyloid A is elevated in the serum of lung cancer patients with poor prognosis. Br. J. Cancer 2010, 102 (12), 1731–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang Y-K; Lai Y-H; Chu Y; Lee M-C; Huang C-Y; Wu S, Haptoglobin is a serological biomarker for adenocarcinoma lung cancer by using the ProteomeLab PF2D combined with mass spectrometry. Am. J. Cancer Res 2016, 6 (8), 1828–1836. [PMC free article] [PubMed] [Google Scholar]
- 42.Tai C-S; Lin Y-R; Teng T-H; Lin P-Y; Tu S-J; Chou C-H; Huang Y-R; Huang W-C; Weng S-L; Huang H-D; Chen Y-L; Chen WL, Haptoglobin expression correlates with tumor differentiation and five-year overall survival rate in hepatocellular carcinoma. PLoS One 2017, 12 (2), e0171269–e0171269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabassum U; Reddy O; Mukherjee G, Elevated serum haptoglobin is associated with clinical outcome in triple-negative breast cancer patients. Asian Pac. J. Cancer Prev 2012, 13 (9), 4541–4544. [DOI] [PubMed] [Google Scholar]
- 44.Cocco E; Bellone S; El-Sahwi K; Cargnelutti M; Buza N; Tavassoli FA; Schwartz PE; Rutherford TJ; Pecorelli S; Santin AD, Serum amyloid A: a novel biomarker for endometrial cancer. Cancer 2010, 116 (4), 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drew AF; Liu H; Davidson JM; Daugherty CC; Degen JL, Wound-healing defects in mice lacking fibrinogen. Blood 2001, 97 (12), 3691–3698. [DOI] [PubMed] [Google Scholar]
- 46.Pichler M; Hutterer GC; Stojakovic T; Mannweiler S; Pummer K; Zigeuner R, High plasma fibrinogen level represents an independent negative prognostic factor regarding cancer-specific, metastasis-free, as well as overall survival in a European cohort of non-metastatic renal cell carcinoma patients. Br. J. Cancer 2013, 109 (5), 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeuchi H; Ikeuchi S; Kitagawa Y; Shimada A; Oishi T; Isobe Y; Kubochi K; Kitajima M; Matsumoto S, Pretreatment plasma fibrinogen level correlates with tumor progression and metastasis in patients with squamous cell carcinoma of the esophagus. J. Gastroenterol. Hepatol 2007, 22 (12), 2222–2227. [DOI] [PubMed] [Google Scholar]
- 48.Fan S; Guan Y; Zhao G; An G, Association between plasma fibrinogen and survival in patients with small-cell lung carcinoma. Thorac. Cancer 2018, 9 (1), 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seebacher V; Aust S; D’Andrea D; Grimm C; Reiser E; Tiringer D; Von Mersi H; Polterauer S; Reinthaller A; Helmy-Bader S, Development of a tool for prediction of ovarian cancer in patients with adnexal masses: Value of plasma fibrinogen. PLoS One 2017, 12 (8), e0182383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palumbo JS; Kombrinck KW; Drew AF; Grimes TS; Kiser JH; Degen JL; Bugge TH, Fibrinogen is an important determinant of the metastatic potential of circulating tumor cells. Blood 2000, 96 (10), 3302–3309. [PubMed] [Google Scholar]
- 51.Quadro L; Hamberger L; Colantuoni V; Gottesman ME; Blaner WS, Understanding the physiological role of retinol-binding protein in vitamin A metabolism using transgenic and knockout mouse models. Mol. Aspects Med 2003, 24 (6), 421–430. [DOI] [PubMed] [Google Scholar]
- 52.Pillai S; Oresajo C; Hayward J, Ultraviolet radiation and skin aging: roles of reactive oxygen species, inflammation and protease activation, and strategies for prevention of inflammation-induced matrix degradation - a review. Int. J. Cosmet. Sci 2005, 27 (1), 17–34. [DOI] [PubMed] [Google Scholar]
- 53.Takashima A; Bergstresser PR, Impact of UVB radiation on the epidermal cytokine network. Photochem. Photobiol 1996, 63 (4), 397–400. [DOI] [PubMed] [Google Scholar]
- 54.Kyriakis JM; Avruch J, Mammalian Mitogen-Activated Protein Kinase Signal Transduction Pathways Activated by Stress and Inflammation. Physiol. Rev 2001, 81 (2), 807–869. [DOI] [PubMed] [Google Scholar]
- 55.Yang G; Fu Y; Malakhova M; Kurinov I; Zhu F; Yao K; Li H; Chen H; Li W; Lim DY; Sheng Y; Bode AM; Dong Z; Dong Z, Caffeic acid directly targets ERK1/2 to attenuate solar UV-induced skin carcinogenesis. Cancer Prev. Res. (Phila.) 2014, 7 (10), 1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russo AE; Torrisi E; Bevelacqua Y; Perrotta R; Libra M; McCubrey JA; Spandidos DA; Stivala F; Malaponte G, Melanoma: molecular pathogenesis and emerging target therapies (Review). Int. J. Oncol 2009, 34 (6), 1481–1489. [DOI] [PubMed] [Google Scholar]
- 57.Einspahr JG; Calvert V; Alberts DS; Curiel-Lewandrowski C; Warneke J; Krouse R; Stratton SP; Liotta L; Longo C; Pellacani G; Prasad A; Sagerman P; Bermudez Y; Deng J; Bowden GT; Petricoin EF 3rd, Functional protein pathway activation mapping of the progression of normal skin to squamous cell carcinoma. Cancer Prev. Res. (Phila.) 2012, 5 (3), 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moriyama M; Moriyama H; Uda J; Kubo H; Nakajima Y; Goto A; Morita T; Hayakawa T, BNIP3 upregulation via stimulation of ERK and JNK activity is required for the protection of keratinocytes from UVB-induced apoptosis. Cell Death Dis. 2017, 8 (2), e2576–e2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Somensi N; Brum PO; de Miranda Ramos V; Gasparotto J; Zanotto-Filho A; Rostirolla DC; da Silva Morrone M; Moreira JCF; Pens Gelain D, Extracellular HSP70 Activates ERK1/2, NF-kB and Pro-Inflammatory Gene Transcription Through Binding with RAGE in A549 Human Lung Cancer Cells. Cell. Physiol. Biochem 2017, 42 (6), 2507–2522. [DOI] [PubMed] [Google Scholar]
- 60.Bonvin C; Guillon A; van Bemmelen MX; Gerwins P; Johnson GL; Widmann C, Role of the amino-terminal domains of MEKKs in the activation of NFκB and MAPK pathways and in the regulation of cell proliferation and apoptosis. Cell. Signal 2002, 14 (2), 123–131. [DOI] [PubMed] [Google Scholar]
- 61.Bickers DR; Athar M, Oxidative stress in the pathogenesis of skin disease. J. Invest. Dermatol 2006, 126 (12), 2565–2575. [DOI] [PubMed] [Google Scholar]
- 62.Liu T; Zhang L; Joo D; Sun S-C, NF-κB signaling in inflammation. Signal Transduct. Target. Ther 2017, 2, 17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chung SS; Vadgama JV, Curcumin and epigallocatechin gallate inhibit the cancer stem cell phenotype via down-regulation of STAT3-NFkappaB signaling. Anticancer Res. 2015, 35 (1), 39–46. [PMC free article] [PubMed] [Google Scholar]
- 64.Uskokovic A; Dinic S; Mihailovic M; Grdovic N; Arambasic J; Vidakovic M; Bogojevic D; Ivanovic-Matic S; Martinovic V; Petrovic M; Poznanovic G; Grigorov I, STAT3/NF-kappaB interactions determine the level of haptoglobin expression in male rats exposed to dietary restriction and/or acute phase stimuli. Mol. Biol. Rep 2012, 39 (1), 167–176. [DOI] [PubMed] [Google Scholar]
- 65.Hagihara K; Nishikawa T; Sugamata Y; Song J; Isobe T; Taga T; Yoshizaki K, Essential role of STAT3 in cytokine-driven NF-kappaB-mediated serum amyloid A gene expression. Genes Cells 2005, 10 (11), 1051–1063. [DOI] [PubMed] [Google Scholar]
- 66.Adhami VM; Afaq F; Ahmad N, Suppression of Ultraviolet B Exposure-Mediated Activation of NF-κB in Normal Human Keratinocytes by Resveratrol. Neoplasia 2003, 5 (1), 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shah R, The Role of Nutrition and Diet in Alzheimer Disease: A Systematic Review. J. Am. Med. Dir. Assoc 2013, 14 (6), 398–402. [DOI] [PubMed] [Google Scholar]
- 68.Godic A; Poljšak B; Adamic M; Dahmane R, The role of antioxidants in skin cancer prevention and treatment. Oxid. Med. Cell. Longev 2014, 2014, 860479–860479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scalbert A; Manach C; Morand C; Remesy C; Jimenez L, Dietary polyphenols and the prevention of diseases. Crit. Rev. Food Sci. Nutr 2005, 45 (4), 287–306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


