To the Editor,
Acute respiratory infections including rhinovirus (RV) infections are a major cause of asthma exacerbations.1 Recent studies suggest that eosinophils play important roles in the development of asthma exacerbation.2 Not only neutrophils but also eosinophils increase in asthmatic airways during viral infection,3 suggesting that eosinophils are indeed recruited to and activated in the airways during virus-related asthma exacerbations.
Cadherin-related family member 3 (CDHR3), a member of the cadherin superfamily, is a transmembrane protein with six extracellular cadherin domains. However, the biological function of CDHR3 is still unknown. Recently, Bønnelykke et al. reported that a coding single nucleotide polymorphism (SNP) (rs6967330; C529Y) in CDHR3 is associated with severe exacerbations of childhood asthma.4 Furthermore, we reported that CDHR3 is a receptor for rhinovirus C (RV-C) species,5 which is more virulent and more likely to cause exacerbations of childhood asthma as compared with RV-B.1, 6 CDHR3 rs6967330; C529Y increases the expression of CDHR3 protein on the cell surface 4, 5 resulting in increased RV-C binding and progeny yields.5 Moreover, this SNP is associated with increased RV-C illnesses in vivo.7 Therefore, CDHR3-Y529 variant is likely to promote the development of asthma exacerbations by increasing susceptibility to RV-C illnesses. CDHR3 is mainly expressed on the apical surface of ciliated epithelial cells, but the natural ligands and function are unknown.
In this study, we examined whether CDHR3 binds to and activates eosinophils. We first examined the effect of CDHR3 on eosinophil adhesion. Eosinophils were obtained from healthy volunteers, and their adhesion to CDHR3 or intercellular adhesion molecule (ICAM)-1 was measured using residual eosinophil peroxidase (EPO) assays. Compared to control or ICAM-1, CDHR3 induced significantly higher spontaneous adhesion of eosinophils (Figure 1A). More than 10 μg/ml of CDHR3 augmented eosinophil adhesion, and 100 µg/ml of CDHR3 induced a maximal response (Figure 1B). IL-5 further enhanced eosinophil adhesion to CDHR3 (Figure 1B). Eosinophil adhesion to CDHR3 was inhibited by anti-αM integrin or anti-β2 integrin mAb, but not by anti-α4 integrin mAb (Figure S1), suggesting that eosinophils can adhere to CDHR3 via αMβ2 integrin. The inhibition of adhesion by anti-αM integrin or anti-β2 integrin mAb could be due to steric hindrance. CDHR3-induced eosinophil adhesion of asthmatic patients was higher than that of healthy volunteers (Figure 1C).
FIGURE 1.
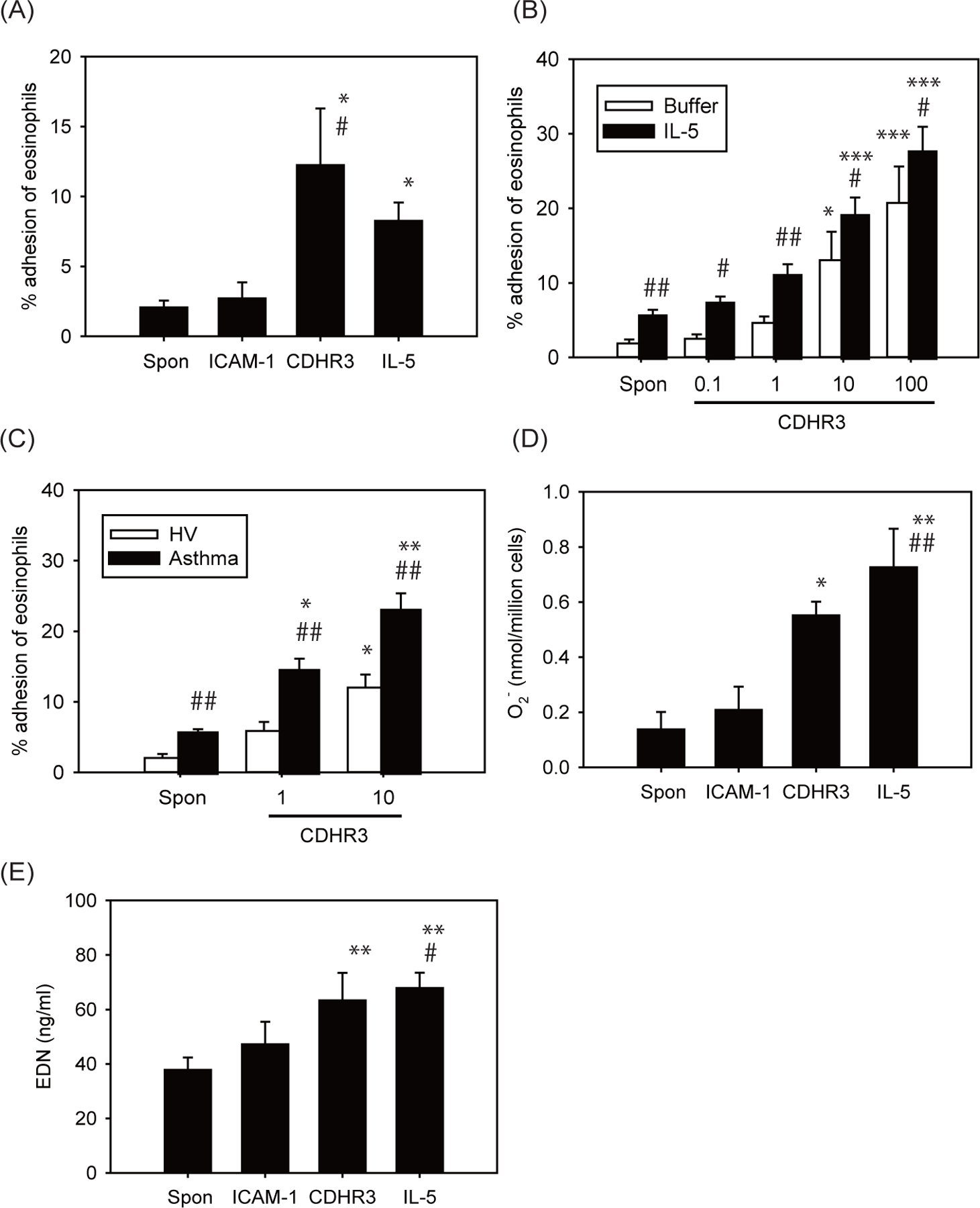
Effect of CDHR3 on eosinophil functions. (A and B) Effect of CDHR3 on eosinophil adhesion. A, Eosinophils (1×105 cells/ml) collected from non-allergic donors were incubated in CDHR3 or ICAM-1-coated plates (10 μg/ml) or in medium (HBSS/gel)-coated plates, and their adhesion was measured. As for IL-5, eosinophils (1×105 cells/ml) were incubated with IL-5 (100 pM) in medium-coated plates and their adhesion was measured. Data are shown as means ± SEM of 6 experiments using cells from different donors. * P < 0.05 vs. spontaneous adhesion (Spon). # P < 0.05 vs. ICAM-1. B, Eosinophils (1×105 cells/ml) collected from non-allergic donors were incubated with or without IL-5 (100 pM), and their adhesion to various concentrations of rh-CDHR3 (0.1–100 µg/mL) was measured (n = 6). * P < 0.05 and *** P < 0.001 vs. spontaneous adhesion (Spon). # P < 0.05 and ## P < 0.01 vs. buffer (without IL-5). C, Effect of asthma on CDHR3-induced eosinophil adhesion. Eosinophils (1×105 cells/ml) collected from healthy volunteers or asthmatic patients were incubated in CDHR3-coated plates (1 or 10 μg/ml) and their adhesion was measured (n = 6). * P < 0.05 and ** P < 0.01 vs. spontaneous adhesion (Spon). ## P < 0.01 vs. healthy volunteers. D, Effect of CDHR3 on eosinophil O2− generation. Eosinophils (1 × 106 cells/ml) from non-allergic donors were incubated in CDHR3 or ICAM-1-coated plates (10 μg/ml) or in medium-coated plates. As for IL-5, eosinophils (1 × 106 cells/ml) were incubated with IL-5 (100 pM) in medium-coated plates. The generation of eosinophil O2− was examined (n = 6). Maximum value of eosinophil O2− generation over the next 240 min were shown. * P < 0.05 and ** P < 0.01 vs. spontaneous O2− generation (Spon). ## P < 0.01 vs. ICAM-1. E, Effect of CDHR3 on EDN release. Eosinophils (1 × 106 cells/ml) from non-allergic donors were incubated in CDHR3 or ICAM-1-coated plates (10 μg/ml) or in medium-coated plates for the 240 min. As for IL-5, eosinophils (1 × 106 cells/ml) were incubated with IL-5 (100 pM) in medium-coated plates for the 240 min. Levels of EDN in cell-free supernatants were quantified using ELISA (n = 6). ** P < 0.01 vs. spontaneous EDN release (Spon). # P < 0.05 vs. ICAM-1.
We next examined the effect of CDHR3 on superoxide anion (O2−) generation or release of eosinophil-derived neurotoxin (EDN) by eosinophils. Compared to medium control, CDHR3 induced significantly greater eosinophil O2− generation, to a level similar to that induced by IL-5 (Figure 1D). Eosinophil O2− generation induced by CDHR3 was suppressed by anti-αM integrin or anti-β2 integrin mAb but not by anti-α4 integrin mAb (Figure S1). In addition, CDHR3 induced significantly higher EDN release (Figure 1E) compared to control. Anti-αM integrin or anti-β2 integrin mAb, but not anti-α4 integrin mAb, suppressed EDN release induced by CDHR3 (Figure S1).
We next tested whether CDHR3 C529Y mutation affects eosinophil functions. Eosinophils adhesion to plates coated with the wild-type CDHR3-C529 or the CDHR3-Y529 variant protein were similar (Figure S2). Eosinophil O2− generation by CDHR3 variant protein did not differ from that by wild-type CDHR3 protein (Figure S2). Therefore, we transfected HeLa cells with these CDHR3 variants, and then examined effects of CDHR3 C529Y mutation on eosinophil adhesion and functions. We previously demonstrated that expression of CDHR3-Y529 variant after transfection of HeLa-H1 cells lead to enhanced surface expression of protein as compared to CDHR3-C529,5 whereas levels of overall cellular expression of CDHR3 were similar,5 and confirmed these finding in this study (data not shown). Along with increased CDHR3 expression on the cell surface, transfection with CDHR3-Y529 also increased eosinophil adhesion compared to CDHR3-C529 or the negative control (Figure 2A). Similar results were obtained when we used fluorescently-labeled eosinophils (Figure 2B, 2C) or performed major basic protein (MBP) staining (Figure S3). Furthermore, CDHR3-Y529 transfection induced greater eosinophil O2− generation (Figure 2D) and EDN release (data not shown).
FIGURE 2.
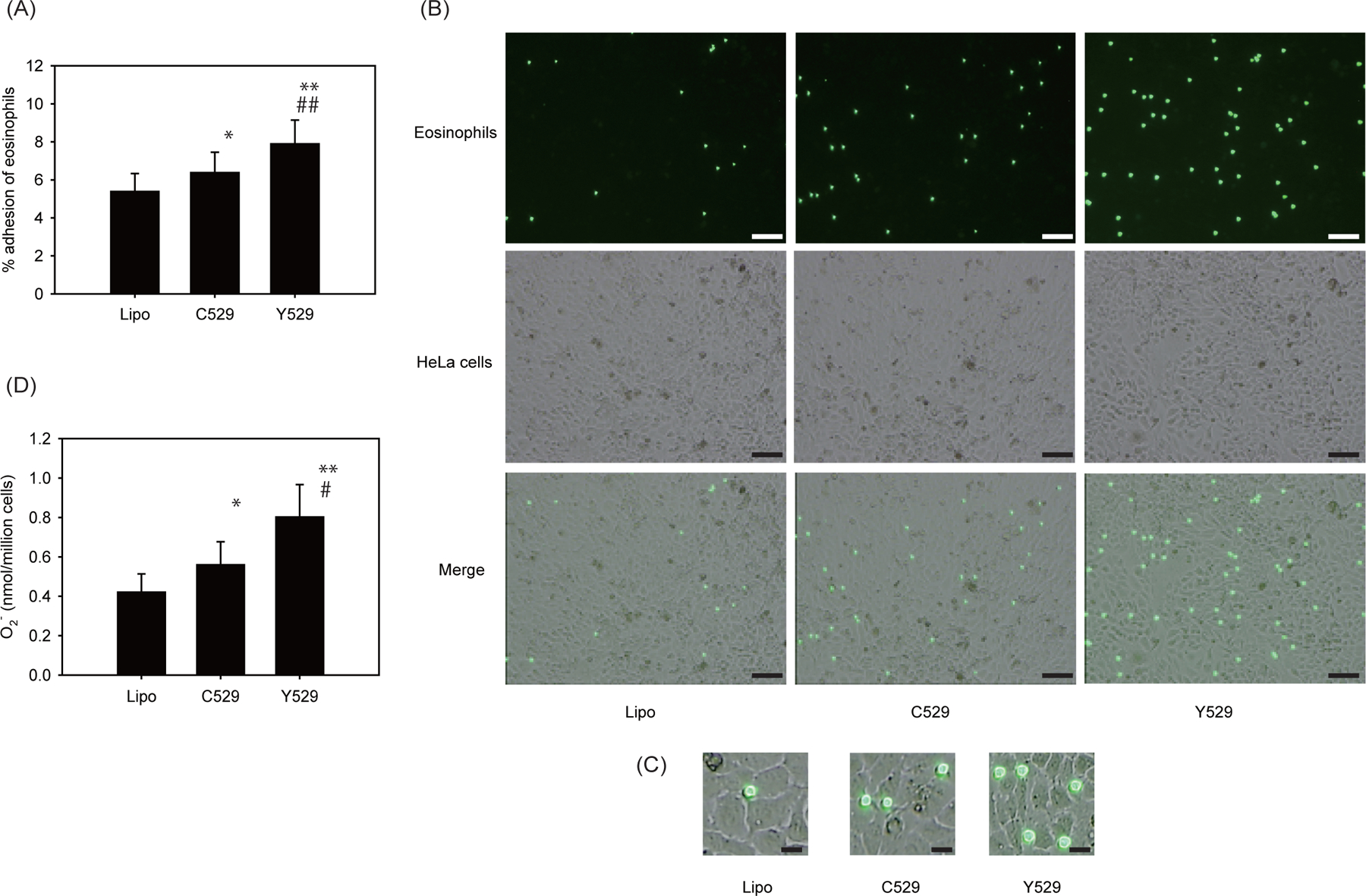
Effect of CDHR3 C529Y gene mutation on eosinophil functions. A, Effect of CDHR3 C529Y gene mutation on eosinophil adhesion to HeLa cells transfected with plasmid DNA. HeLa cells were transfected with plasmids encoding wild-type CDHR3 (C529) or CDHR3 variant (Y529) or lipofectamine only (Lipo), and eosinophil adhesion to transfected cells was measured (n = 6). * P < 0.05 and ** P < 0.01 vs. Lipo. ## P < 0.01 vs. C529. B, Fluorescently-labeled eosinophil adhesion to transfected HeLa cells. HeLa cells were transfected with plasmids encoding wild-type CDHR3 (C529) or CDHR3 variant (Y529) or lipofectamine only (Lipo). Eosinophils were labeled with calcein-AM and then incubated with the transfected HeLa cells for 20 min. After the plates were washed, cells were observed under a fluorescence microscope. Representative figures of 3 independent experiments with different donors were shown. Scale bar, 100 μm. C, Fluorescently-labeled eosinophil adhesion to transfected HeLa cells at higher magnification. Scale bar, 20 μm. D, Effect of CDHR3 C529Y gene mutation on eosinophil O2− generation. HeLa cells were transfected with plasmids encoding wild-type CDHR3 (C529) or CDHR3 variant (Y529) or lipofectamine only (Lipo), and eosinophils were incubated with transfected HeLa cells for 240 min. The generation of eosinophil O2− was examined (n = 6). The maximum value of eosinophil O2− generation was shown. * P < 0.05 and ** P < 0.01 vs. Lipo. # P < 0.05 vs. C529.
In this study, we found that CDHR3 activated eosinophil function such as adhesion, O2− generation and degranulation. This was true for plate-bound CDHR3 (Figure 1) and for CDHR3 expressed in transfected HeLa cells (Figure 2). However, the effects of genetic variation at rs6967330 on eosinophil adhesion and function depended on whether the proteins were plate bound (Figure S2; no difference) or expressed in HeLa cells (Figure 2; Y529 induced increased eosinophil activation). These findings suggest that the difference in eosinophil adhesion was due to the level of surface expression of CDHR3 protein, but not due to the different ability of the CDHR3 variants to induce eosinophil adhesion. The plate-bound experiments demonstrate that if the display of CDHR3 protein on a surface is similar, the effects on eosinophil adhesion are also similar.
Given the close association between eosinophilic inflammation and asthma exacerbation,2 CDHR3-Y529 could promote eosinophil adhesion and proinflammatory functions linked to asthma exacerbations. Recent studies suggested that CDHR3-Y529 increases the risk for developing chronic rhinosinusitis or severe adult asthma with early onset, suggesting that CDHR3 variant may contribute to eosinophilic inflammation in chronic rhinosinusitis as well as bronchial asthma.
Whether CDHR3 expression is increased in the airway of patients with asthma is incompletely understood. For example, Bai et al. reported that CDHR3 mRNA expression in airway epithelial cells cultured at air-liquid interface was slightly lower in cells obtained from donors with asthma.8 Furthermore, Everman et al. reported that CDHR3 is exclusively expressed on ciliated cells, and that expression is greatest in cells undergoing ciliation as compared to mature ciliated cells,9 suggesting that eosinophil adhesion to ciliated cells could be greatest in airways with active cell differentiation or repair. In addition, Jones et al. reported that CDHR3 protein expression of bronchial epithelial cells increased in atopic asthma as compared with that in non-atopic controls.10 Additional studies are needed to test for cell surface expression of CDHR3 in vivo with regard to CDHR3 genotype at rs6967330 and asthma status.
We found that CDHR3-induced eosinophil adhesion of asthmatic patients was higher than that of healthy volunteers (Figure 1C), although the mechanism is unknown. In this study, eosinophil adhesion to CDHR3 in allergic asthma did not differ from that in non-allergic asthma (data not shown). Furthermore, in allergic asthma, the titer of specific IgE or total IgE did not affect the degree of eosinophil adhesion (data not shown), suggesting that IgE-mediated mechanisms are unlikely to be involved.
One limitation of this study is that the experiments were performed in vitro and these findings need to be confirmed in nontransformed airway epithelial cells and in clinical studies. Another limitation is that we tested for interactions between CDHR3 and a limited number of integrins. It is possible that other integrins in addition to αM integrin or β2 integrin may interact with CDHR3.
In conclusion, CDHR3 upregulated eosinophil functions such as adhesion, O2− generation and EDN release. Furthermore, eosinophil activation by CDHR3 was at least partially mediated through αMβ2 integrin. These findings suggest that genetic variation in CDHR3 can modify the risk for developing asthma by activating eosinophils, as well as by increasing the risk of RV-C induced illnesses.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Ms. Akemi Yokote and Dr. Hitoshi Miyazawa for their excellent technical assistance.
FUNDING
This study was funded by a grant from the Ministry of Education, Culture, Sports, Science and Technology (15K09228).
Footnotes
CONFLICTS OF INTEREST
J.E.G received consultant’s fees from Regeneron, Ena Therapeutics and Meissa Vaccines Inc. The rest of the authors have no conflicts of interest to disclose.
SUPPORTING INFORMATION
Additional supporting information may be found in the Supporting Information section at the end of the article
REFERENCES
- 1.Bochkov YA, Gern JE. Rhinoviruses and Their Receptors: Implications for Allergic Disease. Curr Allergy Asthma Rep. 2016;16:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green RH, Brightling CE, McKenna S, Hargadon B, Parker D, Bradding P, Wardlaw AJ, Pavord ID. Asthma exacerbations and sputum eosinophil counts: a randomised controlled trial. Lancet. 2002;360:1715–1721. [DOI] [PubMed] [Google Scholar]
- 3.Pizzichini MM, Pizzichini E, Efthimiadis A, Chauhan AJ, Johnston SL, Hussack P, Mahony J, Dolovich J, Hargreave FE. Asthma and natural colds. Inflammatory indices induced sputum; a feasibility study. Am J Respir Crit Care Med 1998;158: 1178–1184. [DOI] [PubMed] [Google Scholar]
- 4.Bønnelykke K, Sleiman P, Nielsen K, Kreiner-Møller E, Mercader JM, Belgrave D, den Dekker HT, Husby A, Sevelsted A, Faura-Tellez G, Mortensen LJ, Paternoster L, Flaaten R, Mølgaard A, Smart DE, Thomsen PF, Rasmussen MA, Bonàs-Guarch S, Holst C, Nohr EA, Yadav R, March ME, Blicher T, Lackie PM, Jaddoe VW, Simpson A, Holloway JW, Duijts L, Custovic A, Davies DE, Torrents D, Gupta R, Hollegaard MV, Hougaard DM, Hakonarson H, Bisgaard H. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–55. [DOI] [PubMed] [Google Scholar]
- 5.Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, Palmenberg AC, Gern JE. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci USA. 2015;112:5485–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakagome K, Bochkov YA, Ashraf S, Brockman-Schneider RA, Evans MD, Pasic TR, Gern JE. Effects of rhinovirus species on viral replication and cytokine production. J Allergy Clin Immunol. 2014;134:332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bønnelykke K, Coleman AT, Evans MD, Thorsen J, Waage J, Vissing NH, Carlsson CJ, Stokholm J, Chawes BL, Jessen LE, Fischer TK, Bochkov YA, Ober C, Lemanske RF Jr., Jackson DJ, Gern JE, Bisgaard H. Cadherin-related Family Member 3 Genetics and Rhinovirus C Respiratory Illnesses. Am J Respir Crit Care Med. 2018;197:589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai J, Smock SL, Jackson GR Jr, MacIsaac KD, Huang Y, Mankus C, Oldach J, Roberts B, Ma YL, Klappenbach JA, Crackower MA, Alves SE, Hayden PJ. Phenotypic responses of differentiated asthmatic human airway epithelial cultures to rhinovirus. PLoS One. 2015;10:e0118286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everman JL, Sajuthi S, Saef B, Rios C, Stoner AM, Numata M, Hu D, Eng C, Oh S, Rodriguez-Santana J, Vladar EK, Voelker DR, Burchard EG, Seibold MA. Functional genomics of CDHR3 confirms its role in HRV-C infection and childhood asthma exacerbations. J Allergy Clin Immunol. 2019;144:962–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones AC, Troy NM, White E, Hollams EM, Gout AM, Ling KM, Kicic A, Stick SM, Sly PD, Holt PG, Hall GL, Bosco A. Persistent activation of interlinked type 2 airway epithelial gene networks in sputum-derived cells from aeroallergen-sensitized symptomatic asthmatics. Sci Rep. 2018;8:1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


