Abstract
Complementary (c)DNA clones corresponding to the full‐length genome of T36CA (a Californian isolate of Citrus tristeza virus with the T36 genotype), which shares 99.1% identity with that of T36FL (a T36 isolate from Florida), were made into a vector system to express the green fluorescent protein (GFP). Agroinfiltration of two prototype T36CA‐based vectors (pT36CA) to Nicotiana benthamiana plants resulted in local but not systemic GFP expression/viral infection. This contrasted with agroinfiltration of the T36FL‐based vector (pT36FL), which resulted in both local and systemic GFP expression/viral infection. A prototype T36CA systemically infected RNA silencing‐defective N. benthamiana lines, demonstrating that a genetic basis for its defective systemic infection was RNA silencing. We evaluated the in planta bioactivity of chimeric pT36CA‐pT36FL constructs and the results suggested that nucleotide variants in several open reading frames of the prototype T36CA could be responsible for its defective systemic infection. A single amino acid substitution in each of two silencing suppressors, p20 (S107G) and p25 (G36D), of prototype T36CA facilitated its systemic infectivity in N. benthamiana (albeit with reduced titre relative to that of T36FL) but not in Citrus macrophylla plants. Enhanced virus accumulation and, remarkably, robust systemic infection of T36CA in N. benthamiana and C. macrophylla plants, respectively, required two additional amino acid substitutions engineered in p65 (N118S and S158L), a putative closterovirus movement protein. The availability of pT36CA provides a unique opportunity for comparative analysis to identify viral coding and noncoding nucleotides or sequences involved in functions that are vital for in planta infection.
Keywords: citrus tristeza virus, heat shock protein 70 homolog, infectious clone, p65, silencing suppressor, systemic movement
Differential bioactivity of citrus tristeza virus (CTV), T36CA, in two plant hosts is delineated by specific amino acid residues in two viral suppressors of RNA silencing and a putative CTV movement protein.
1. INTRODUCTION
Citrus tristeza virus (CTV), a species of the genus Closterovirus (family Closteroviridae), is one of the most complex plant RNA viruses on the basis of its large genome size and complicated gene expression strategies. As with all RNA viruses, CTV exists as a population of sequence variants that are distributed around a predominant consensus genomic sequence (Harper, 2013; Rubio et al., 2001). Based on comparison of consensus genomic sequences, at least seven genetically diverse groups (genotypes) or strains of CTV with moderate (10%–20%) nucleotide variability have been distinguished (Harper, 2013; Harper et al., 2015a). Depending on the particular infecting strain, the citrus species or cultivar, and the combination of scion–rootstock used for grafting, CTV infection can be symptomless or cause distinctive symptoms, such as quick decline, stem pitting, and seedling yellowing (Harper, 2013). In nature, it is common for more than one CTV strain to infect a single host and for the infectivity of one strain in a specific host to be modulated by another genetically diverse, coinfecting strain, suggesting the existence of complex strain–strain interactions within an infected host (Harper et al., 2015b). In contrast to the moderate nucleotide variability among CTV strains, the nucleotide divergence among different isolates of the same strain is relatively low (less than 4%) and can be maintained over time even in hosts that are located at different geographical regions (Albiach‐Martí et al., 2000; Chen et al., 2018; Harper, 2013). It is unclear if, or how, diverse sequence variants within the population of a given CTV isolate are reflected in the biology of the virus in the context of infection, for example fitness, virulence, and pathogenesis.
The single‐stranded, monopartite 19.3‐kb genome of CTV has 12 open reading frames (ORFs). The first two ORFs, ORF1a and ORF1b, encode proteins associated with viral replication. ORF1a translates into a polyprotein that contains the helicase, methyltransferase, and leader protease domains, while the ORF1b product functions as an RNA‐dependent RNA polymerase (RdRP). The remaining ORFs encode proteins involved in other functions, including encapsidation, movement, suppression of RNA silencing, and host interaction (Albiach‐Martí et al., 2010; Lu et al., 2004; Satyanarayana et al., 2004; Tatineni et al., 2011). The long flexuous, filamentous virion (2,000 × 10–12 nm) of CTV consists of two types of coat proteins: the minor coat protein (CPm), which encapsidates c.630 nucleotides at the 5′ terminus of the genomic RNA, and the major coat protein (CP), which encapsidates the remainder of the RNA genome (Satyanarayana et al., 2004). In addition, p65 (a heat shock protein 70 homolog) and p61 enhance virion assembly and restrict the encapsidation of the CPm to the c.630 nucleotides at the 5′ end of the genomic RNA (Satyanarayana et al., 2000, 2004). To counteract plant antiviral defence, CTV encodes three viral suppressors of RNA silencing (VSRs), CP and p23, which act intercellularly and intracellularly, respectively, as well as p20, which functions at both cellular levels (Lu et al., 2004). p33, p18, and p13 are not required for systemic infection in many susceptible Citrus species, but are essential to extend the host range of CTV (Tatineni et al., 2011). Potential CTV proteins that function in systemic infection have been investigated in several studies. For example, p33 is required for CTV systemic infection in sour orange and lemon. However, CTV engineered without a p33 can still infect grapefruit or calamondin in the presence of a functional p18 or p13, respectively (Tatineni et al., 2011). Furthermore, deletion of p6 or p20 impairs the systemic infectivity of CTV in Citrus macrophylla and Mexican lime (Tatineni et al., 2008). The functional basis for the defect in systemic infection of these engineered CTV mutants remains to be determined.
Most of the studies on viral gene functions in the preceding section were performed using the infectious complementary (c)DNA clones of an isolate of the T36 strain that originated from Florida. One of the cDNA clones was later also developed into a bioactive vector, named C86 (referred to from here on as pT36FL), capable of expressing green fluorescent protein (GFP) in Nicotiana benthamiana (the laboratory host) and C. macrophylla plants (El‐Mohtar and Dawson, 2014). Recently, we obtained and sequenced a Californian isolate of the T36 strain, T36CA, and a comparison of its consensus genomic sequence with that of T36FL showed that they share a high degree (99.1%) of nucleotide identity. Of the 178 nucleotide variants between the two genomes, 68 are nonsynonymous differences. An analysis of the sequences in the genomic cDNA library of T36CA revealed 71 nucleotide variants, of which 50 are nonsynonymous (Chen et al., 2018).
This study began with the construction of two prototype versions of a T36CA‐based vector, pT36CA (made using cDNA clones of the T36CA genome), followed by an investigation of their performance in planta. The T36CA isolate used for vector construction was originally obtained from an infected Madam Vinous sweet orange (Citrus sinensis) in Ventura county, California (Chen et al., 2018). The pT36CA prototypes showed bioactivity in the local but not the systemic leaves of agroinfiltrated N. benthamiana plants. We sought to investigate the basis of this defect using a combination of genetic and molecular approaches. First, the defective systemic infection of a T36CA prototype could be overcome in RNA silencing‐defective N. benthamiana lines ago2 and RDR6i (Ludman et al., 2017; Schwach et al., 2005), demonstrating that a genetic basis for its failure to systemically infect N. benthamiana plants was RNA silencing. Next, the analysis of chimeric constructs of pT36CA and pT36FL revealed a potential association of the defective systemic infection phenotype with nucleotide variants in nine CTV ORFs (p6, p65, p61, CPm, CP, p18, p13, p20, and p23). Finally, comparative analysis of sequence variants in the T36CA population and the consensus sequence of T36FL, combined with site‐directed mutagenesis, allowed us to identify four deduced amino acids (encoded by nucleotides present in the T36CA population)—one each in the VSRs p25(CP) and p20, and two in the putative movement protein p65—key to establishing a bioactive pT36CA. The implications of how these few, yet crucial, amino acids can profoundly affect virus bioactivity and systemic infection in plants are discussed.
2. RESULTS
2.1. T36CA prototypes are defective in systemic infection in N. benthamiana plants
pT36FL was essentially constructed in a binary plasmid backbone, with a truncated p33 coding sequence, and in which the GFP coding sequence was engineered immediately downstream of the p23 ORF (El‐Mohtar and Dawson, 2014). Using pT36FL as a working template for vector construction, we systematically replaced all T36FL sequences with those obtained from cDNA clones of the T36CA population (Chen et al., 2018), resulting in the generation of two prototype versions of pT36CA, pT36CA‐ori and pT36CA‐V0 (Figures 1a and S1). Results of the sequence analysis of both prototypes are provided in the Text S1 and Figure S1. Given the complexity and sheer size of the CTV genome, it was necessary to construct and test more than one prototype version of the vector as there was a chance that at least one would be infectious. However, when the prototypes were agroinfiltrated to transgenic N. benthamiana plants expressing potyvirus HC‐Pro, both showed bioactivity in the infiltrated leaves but not the systemic leaves (Text S1 and Figure S2). These results offered the first clue that nucleotide variants in the T36CA prototypes negatively impacted the systemic infectivity of the viruses. Both prototypes proved to be useful tools in subsequent studies (see below) aimed at understanding the mechanisms underlying the systemic infection of T36CA.
FIGURE 1.
T36CA and its infectivity in RNA silencing‐defective Nicotiana benthamiana plants. Two prototype vectors, pT36CA‐ori and pT36CA‐V0, were constructed and inoculated to RDR6i and ago2 N. benthamiana plants by agroinfiltration and evaluated for bioactivity. (a) A schematic map of the T36CA binary plasmid (pT36CA) illustrating the region between the cauliflower mosaic virus 35S promoter and nopaline synthase (NOS) terminator. White boxes represent open reading frames (ORFs) encoding a polyprotein (ORF1a), the RNA‐dependent RNA polymerase (RdRP), the minor coat protein (CPm), the major coat protein (CP), and proteins named according to their predicted molecular weights (numerals preceded by the letter “p”). A gene encoding green fluorescent protein (GFP), expressed under the control of a controller element of T36CA CP (dark green), is inserted between p23 and the 3′ noncoding region (NCR). The dashed line represents the region in p33 deleted (Δp33) in pT36CA. (b) and (c) GFP fluorescence (left panels) was observed in the infiltrated (attached) and systemic (detached) leaves of N. benthamiana lines RDR6i (in the presence of exogenous HC‐Pro) (b) and ago2 (c) following the agroinfiltration of pT36CA‐V0 or pT36FL. Virus accumulation was determined by double antibody sandwich‐ELISA (right panels) at 9 weeks postinfiltration. The A405 value for each virus/treatment indicated on the x axis is the average for four individual samples from a representative experiment (with error bar representing SE)
2.2. The defect in systemic infection of prototype T36CA can be overcome in RNA silencing‐defective N. benthamiana lines
A barrier to successful systemic infection of the T36CA prototypes could be RNA silencing, a ubiquitous plant antiviral defence that inhibits the systemic spread of viruses (Bayne et al., 2005; Kasschau and Carrington, 2001; Qu and Morris, 2005). This consideration was further fuelled by the presence of two nonconservative amino acid differences between the T36CA‐V0 and T36FL encoded p20, a known VSR (Figure S1). To determine if T36CA‐V0 was inhibited by RNA silencing in N. benthamiana plants, agroinfiltration was performed using two RNA silencing‐defective lines, RDR6i and ago2, in which the expression of RNA‐dependent RNA polymerase 6 (RDR6) and Argonaute (AGO) 2 were down‐regulated (Ludman et al., 2017; Schwach et al., 2005), respectively. The RDR6i plants coinfiltrated with pT36CA‐V0 and an HC‐Pro‐expressing vector showed GFP expression in the local leaves and, beginning at 5 weeks postinoculation (wpi), in the systemically infected leaves (Figure 1b), while no systemic GFP expression was observed in plants (total 10 individuals) infiltrated with pT36CA‐V0 alone (data not shown). T36CA‐V0 accumulated in the systemically infected leaves of RDR6i plants at a lower viral titre compared to that of T36FL (Figure 1b). Infiltration of pT36CA‐V0 to N. benthamiana ago2 mutant plants resulted in GFP expression in both the infiltrated and systemically infected leaves and virus accumulation in the systemically infected leaves was also observed (Figure 1c). The systemic GFP expression and virus accumulation in both RDR6i and ago2 plants strongly suggested that T36CA‐V0 was silenced in N. benthamiana plants.
To determine whether T36CA‐V0 could also infect C. macrophylla plants, we performed virion purification using RDR6i and ago2 tissues systemically infected with T36CA‐V0 and delivered the resulting virion preparations to C. macrophylla plants by bark flap inoculation. However, neither local nor systemic GFP expression was observed in the target C. macrophylla plants (data not shown).
2.3. Mapping prototype T36CA genomic regions associated with defective systemic infectivity
We posited that the defective systemic infection phenotype of the T36CA prototypes could be mapped to specific regions and/or ORFs in the T36CA genome. To this end, mapping was performed using chimeric pT36CA‐pT36FL vectors, pHyb1 to pHyb7, constructed essentially with specific genomic regions of pT36CA‐ori and pT36FL (Figure 2). The in planta bioactivity of chimeric viruses was determined by one or more of the following analyses/assays following the agroinfiltration of chimeric vectors: visualization of GFP expression, double antibody sandwich (DAS)‐ELISA using crude sap extracts, and immunocapture‐transmission electron microscopy (TEM) of virion preparations. Hyb1 showed bioactivity in both the local and systemic leaves (Figure 2) as GFP signals were seen in those leaves (Figure 3a). Infected systemic leaves developed symptoms—epinasty, vein chlorosis, stunting, and wrinkling—and GFP distribution patterns that resembled those seen in plants infected with T36FL (Figure 3a,b) (El‐Mohtar and Dawson, 2014). Hyb1 increased over time to reach titres comparable to that of T36FL (Figure 3c) and produced CTV virion‐like particles (Figure 3d). Local (data not shown) and systemic GFP expression was observed in C. macrophylla plants inoculated with Hyb1 virion preparations (Figures 2 and 3e). Thus, region I (nt 1–8,106), containing most of the ORF1a of T36CA‐ori, supported Hyb1’s bioactivity in planta, suggesting that the determinants of defective systemic infection of T36CA‐ori might be located in region II (nt 8,107–19,292) corresponding to the 3′‐moiety of the T36CA genome (Figure 2).
FIGURE 2.
A summary of the chimeric pT36CA‐pT36FL constructs pHyb1 to pHyb7, and the infectivity of the resulting chimeric viruses. Chimeric constructs with sequences corresponding to specific genomic regions of T36CA and T36FL were constructed and inoculated to transgenic (HC‐Pro) Nicotiana benthamiana plants by agroinfiltration. Virus preparations purified from specific chimeric construct‐inoculated N. benthamiana plants were inoculated to Citrus macrophylla plants by bark flap inoculation. Left panel: The schematic map of pT36CA‐ori (the same as in Figure 1a) showing the two regions (I and II), subregions (IIa, b, and c), and restriction enzyme sites (SmaI, PmeI, PstI, and SwaI) used to make the chimeric constructs. pHyb1 contains region I and II originated from pT36CA‐ori (white box) and pT36FL (grey box), respectively. Other constructs (pHyb2 to pHyb7) were generated by swapping the corresponding subregions (IIa, IIb and IIc) of pHyb1 and pT36CA‐ori. Right panel: Bioactivity of the chimeric viruses was determined by one or more of the following analyses/assays: visualization of green fluorescent protein (GFP) expression in treated plants, double antibody sandwich (DAS)‐ELISA of sap extracts, and transmission electron microscopy analysis of virus preparations. L and S represent local or systemic tissues, respectively; + and − represent the presence of absence of bioactivity. Note that for Hyb4 to Hyb7, although bioactivity in the systemic leaves of N. benthamiana plant is indicated by +, only sporadic GFP‐expressing foci were observed (accompanied by lower DAS‐ELISA absorbance readings (see Figure 4) relative to the abundant GFP signals (and high DAS‐ELISA absorbance values) observed for those of Hyb1 and Hyb2. NT denotes not tested
FIGURE 3.

The infectivity of chimeric T36CA‐T36FL virus (Hyb1). Chimeric vector pHyb1 (Figure 2) and pT36FL were inoculated to transgenic (HC‐Pro) Nicotiana benthamiana plants by agroinfiltration and evaluated for bioactivity. (a) Green fluorescent protein (GFP) expression was observed in both the infiltrated and systemic leaves, at 9 days postinoculation (dpi) and 6 weeks postinoculation (wpi), respectively, under UV illumination with the leaves attached to the plants. (b) Representative images showing systemic symptoms characterized by leaf epinasty (left) and vein chlorosis in Hyb1‐infected N. benthamiana plants at 10 wpi. (c) Virion accumulation in the systemic leaves of infected N. benthamiana plants over time was determined by double antibody sandwich‐ELISA. The average absorbance at 405 nm (A405) is calculated from a total of six individual samples from two repeated experiments and error bars represent the SE. (d) A transmission electron micrograph (with scale bar) showing virion‐like particles of Hyb1 purified from infected N. benthamiana. (e) A representative wide‐field fluorescence microscopy image of GFP signals observed in the peeled bark of a newly developed branch of a Citrus macrophylla plant inoculated with Hyb1 virion preparation at 10 wpi
Region II, demarcated by three subregions, IIa, IIb, and IIc, based on four unique restriction enzyme sites, SmaI, PmeI, PstI, and SwaI, was the focus of subsequent mapping studies involving six chimeric vectors (Figure 2). The chimeric vectors were constructed by swapping the corresponding subregions of pT36CA‐ori and pHyb1as follows: subregion IIa (resulting in pHyb2 and pHyb3), subregion IIb (resulting in pHyb4 and pHyb5), and subregion IIc (resulting in pHyb6 and pHyb7) (Figure 2). Hyb2 showed bioactivity in both the local and systemic leaves (Figure 2) as GFP signals were seen in both the local (data not shown) and systemic leaves (Figure 4a). Hyb2 reached titres that were comparable to that of Hyb1 (Figure 4b) and, like Hyb1, also formed filamentous virion‐like particles (Figure S3). Local and systemic GFP expression was also observed in C. macrophylla plants inoculated with Hyb2 virion preparations (Figure S3). These data suggest that subregion IIa of Hyb2, which contained the ORF1b of T36CA‐ori, supported virus systemic infection. GFP signals were observed in pHyb3 infiltrated leaves (data not shown), suggesting the presence of Hyb3 bioactivity (Figure 2). However, no GFP expression was observed in the systemic leaves, consistent with the absence of systemic infection as determined by DAS‐ELISA (Figure 4). These results suggest that subregion IIa of Hyb3, with the ORF1b of T36FL, failed to support virus systemic infection in N. benthamiana plants. Taken together, the Hyb2 and Hyb3 data suggest that subregions IIb and IIc of T36CA‐ori probably contain the determinants responsible for its defective systemic infectivity (Figure 2).
FIGURE 4.
The infectivity of chimeric T36CA‐T36FL viruses (Hyb2 to Hyb7). Chimeric vectors, pHyb2 to pHyb7 (Figure 2), were inoculated to transgenic Nicotiana benthamiana plants expressing HC‐Pro by agroinfiltration and evaluated for bioactivity. (a) Representative images showing the presence (or absence) and distribution of green fluorescent protein signals in the systemic leaves of plants infiltrated with pHyb2, pHyb3, pHyb4, pHyb5, pHyb6, and pHyb7 at 8 weeks postinfiltration (wpi). (b) Assessment of chimeric virus accumulation in the systemic leaves of agroinfiltrated plants by double antibody sandwich‐ELISA at 8 wpi. The A405 value for each virus/treatment indicated on the x axis is the average for six individual samples from a representative experiment (with error bar representing SE)
The local and systemic leaves of N. benthamiana plants infiltrated with pHyb4 to pHyb7 all displayed GFP signals, indicating that the resulting chimeric viruses exhibited bioactivity (Figure 2; data not shown). However, the systemic infection of Hyb4 to Hyb7 was associated with varying degrees of suboptimal GFP expression (Figure 4a) and corresponded with considerably reduced virus titres in the systemic leaves relative to that of Hyb1 (Figure 4b). Considered together, the data for Hyb4 and Hyb5 suggest that the reduced titre of these chimeric viruses was probably associated with subregions IIb and IIc of T36CA‐ori, respectively (Figure 2). The data for Hyb6 also suggest that its reduced titre was associated with subregion IIc of T36CA‐ori. This inference was further supported by the Hyb7 data, suggesting that substituting subregion IIc of T36CA‐ori with that of T36FL allowed the resulting chimeric virus (Hyb7) to overcome, albeit with limited effect, an otherwise defective systemic infectivity of T36CA‐ori (Figure 2). Furthermore, Hyb7 formed virion‐like particles (Figure S3) but failed to infect C. macrophylla plants. This clearly showed that subregion IIc of T36FL alone was insufficient to support Hyb7’s systemic infectivity in its native (citrus) host.
Thus, the mapping studies revealed that nearly half the genome (region II) of T36FL was required to overcome/repair the defective systemic infection of T36CA‐ori in N. benthamiana plants, with genomic regions IIb and IIc appearing essential.
2.4. A single amino acid substitution in each of two VSRs encoded by prototype T36CA renders it capable of systemically infecting N. benthamiana plants
Regions IIb and IIc of the T36CA genome contain nine ORFs (p6, p65, p61, CPm, CP, p18, p13, p20, and p23) interspersed with several noncoding sequences, including the 273‐nucleotide long 3′ noncoding region (NCR) (Figure 2). With respect to these regions, the consensus genomic sequence of T36CA differs from that of T36FL by 63 nucleotides, of which 22 are nonsynonymous, 37 are synonymous, three are in the noncoding regions between ORFs, and one is in the 3′ NCR. Because RNA silencing was identified as a genetic basis for the defect in systemic infection of T36CA‐V0 (Figure 1), it seemed a reasonable first hypothesis that the defect was associated with some aspects of impaired anti‐RNA silencing function by the CTV‐encoded VSRs: CP, p20, and p23. An alignment of the CP, p20, and p23 sequences from a cDNA library of the T36CA population (Chen et al., 2018) identified two unique nonsynonymous nucleotide variants in pT36CA‐V0. The first variant encodes a glycine at position 36 of the CP, while a conserved aspartic acid is encoded at that same position based on the reference sequences of T36CA (and that of T36FL). The second variant encodes a serine residue at position 107 of p20, while a conserved glycine residue is encoded at that same position based on the reference sequences (T36CA and T36FL) (Figure S1b). The p23 sequence of pT36CA‐V0 contains two nonsynonymous nucleotides compared to that of T36FL. However, these nucleotides are conserved in the T36CA population.
We performed site‐directed mutagenesis to substitute the two variant nucleotides in pT36CA‐V0 with the conserved nucleotides in the T36CA population to determine if this would remedy the defect in systemic infection of the virus. When the resulting constructs pT36CA‐V1.0 (with a S107G modification) and pT36CA‐V1.1 (with a S107G and G36D modification) were each infiltrated with in transgenic (HC‐Pro) N. benthamiana plants, GFP expression was observed in infiltrated leaves (Figure 5a). Unlike for T36CA‐V0, which did not support GFP expression in the systemic leaves (Figure S2a), systemic GFP expression was observed in the V1.0 and V1.1 infected plants, although the signal intensities were much lower than that observed in T36FL‐infected plants (Figure 5a). The titres of both viruses in the systemic leaves of infected plants were also significantly lower relative to that of T36FL (p < .05, Student's t test; Figure 5b). Moreover, the coinfiltration of TBSV p19 did not significantly affect the titres of both T36CA‐V1.0 and T36CA‐V1.1 (data not shown). Virion‐like particles of both viruses were seen in virion preparations obtained using the systemic leaves (Figure 5c). However, when the virion preparations were inoculated to C. macrophylla plants, no GFP signal was observed in the bark tissues of apical branches up to 16 wpi (data not shown), suggesting that systemic infection could not be established. An examination of the V1.1 virion preparation‐inoculated bark tissues at 10 wpi, however, revealed the presence of GFP signals, suggesting that the virus exhibited bioactivity in those tissues but failed to move to the systemic tissues (Figure 5c). In the case of the V1.0 virion preparation‐inoculated bark tissues, no GFP signal was observed up to 15 wpi.
FIGURE 5.
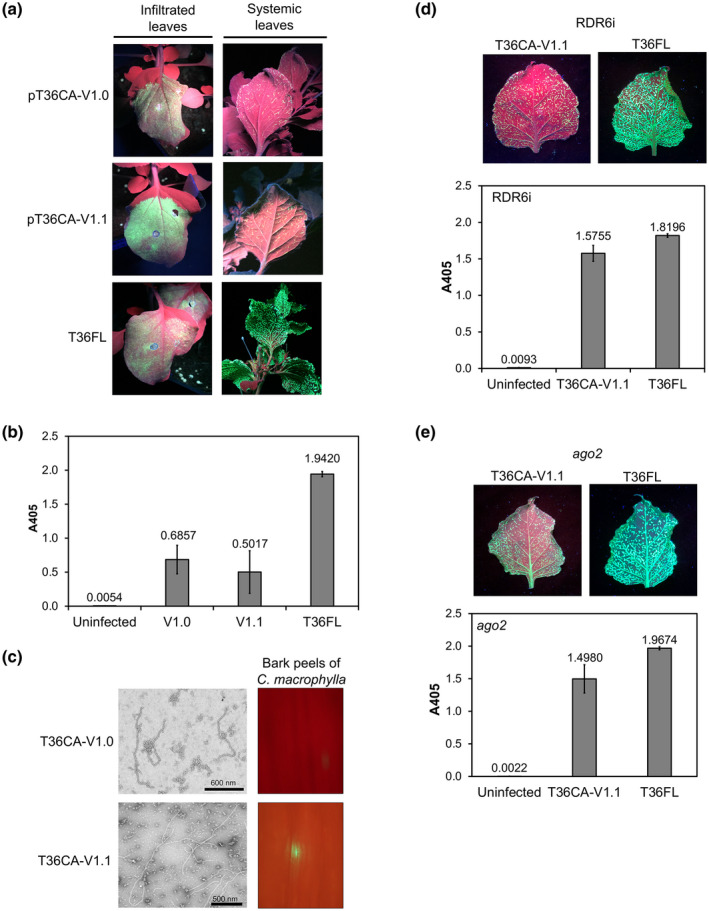
The infectivity of T36CA with a single amino acid substitution introduced into each of two silencing suppressors. A conserved nucleotide from the T36CA population was engineered into the p20 gene, or in both the coat protein (CP [p25]) and p20 genes, of the prototype vector pT36CA‐V0 to replace a nonconserved/nonsynonymous variant identified in each of these genes. The resulting constructs, pT36CA‐V1.0 and pT36CA‐V1.1, and pT36FL (positive control) were each inoculated to transgenic (HC‐Pro‐expressing) Nicotiana benthamiana plants by agroinfiltration. pT36CA‐V1.0 encodes a p20 containing a serine to glycine substitution at position 107 (S107G), while pT36CA‐V1.1 encodes a CP containing a glycine to aspartic acid substitution at position 36 (G36D) in addition to a p20 with the S107G substitution. (a) Representative images showing the presence of green fluorescent protein (GFP) fluorescence in the infiltrated leaves and systemic leaves (attached on the plants) at 8 days postinfiltration and 8 weeks postinfiltration (wpi), respectively. (b) Virus accumulation in the systemic leaves was determined by double antibody sandwich (DAS)‐ELISA at 8 wpi. The A405 value for each virus/treatment indicated on the x axis is the average for three individual plant samples from a representative experiment (with error bar representing SE). (c) Left panels: transmission electron micrographs with scale bars (as indicated), showing the presence of virion‐like particles of T36CA‐V1.0 and T36CA‐V1.1. Right panels: Citrus macrophylla bark peels inoculated with the virion preparations of T36CA‐V1.0 and T36CA‐V1.1 observed under wide‐field fluorescence microscopy. (d) and (e) The systemic infectivity of T36CA‐V1.1 was examined in N. benthamiana RDR6i (coinfiltrated with an HC‐Pro‐expressing vector) (d) and ago2 (e) lines. Top panels: the distribution of GFP signals in the detached systemic leaves of pT36CA‐V1.1‐ versus pT36FL‐infiltrated plants at 9 wpi. Bottom panels: DAS‐ELISA results (for the viruses/treatment indicated on the x axes) following the agroinfiltration of N. benthamiana RDR6i (d) and ago2 (e) lines, with each A405 value being the average for four individual samples from a representative experiment (and error bar representing SE)
We next infiltrated V1.1 in the RDR6i and ago2 N. benthamiana lines to determine if this would improve virus systemic infection and increase the virus titre in tissues used for virion preparation. Indeed, based on DAS‐ELISA data, infiltrating V1.1 to RDR6i N. benthamiana lines in the presence of exogenous HC‐Pro, or to ago2 lines alone, improved virus systemic infection and increased the virus titre to about 86% and 76%, respectively, of the T36FL titre (Figure 5d,e). However, when the resulting virion preparations were inoculated to C. macrophylla plants, none of them became infected (data not shown), suggesting that virus inoculum concentration was not a reason for the defect in systemic infection of T36CA‐V1.1 in citrus plants.
2.5. The differential systemic infection of N. benthamiana and C. macrophylla plants by prototype T36CA maps to two amino acids of a putative closterovirus movement protein
The systemic infection of N. benthamiana but not C. macrophylla plants by T36CA‐V1.1 led to the next hypothesis that other nucleotide variations in the T36CA‐V1.1 genome might contribute to its differential systemic infection of these two host plant species. In subregions IIb and IIc of T36CA‐V0 and T36CA‐V1.1, nonsynonymous nucleotide variants were also identified in the ORFs encoding p6, p65 (a heat shock protein 70 homolog), p61, and p18 (besides the ones already described for CP, p20, and p23 above). p65 has been identified as a movement protein of the closterovirus beet yellows virus (Peremyslov et al., 1999). Among five nucleotide variants in the p65‐coding region identified in the T36CA population, two were unique to pT36CA‐V0 and pT36CA‐V1.1—one encoding an asparagine and the other encoding a serine at positions 118 (N118) and 158 (S158), respectively—whereas nucleotides encoding a serine (S118) and a leucine (L158) were conserved among the reference T36CA sequences and that of T36FL. We substituted the two variant nucleotides in pT36CA‐V1.1, N118S and S158L, and infiltrated the resulting construct, pT36CA‐V1.3, in transgenic (HC‐Pro) N. benthamiana. The V1.3‐infected plants showed bright GFP signals along the veins of the systemic leaves and developed vein chlorosis and wrinkling like that in T36FL‐infected plants (Figure 6a). The systemic infection of V1.3 was outstanding in that its titre was much higher than that of T36FL (at 233% that of T36FL) (Figure 6b). Virion preparations obtained from the systemic N. benthamiana leaves contained virion‐like particles (Figure 6c) and were inoculated to C. macrophylla plants. Remarkably, 8 out of 16 (50%) of the inoculated plants were found to harbour GFP signals in the peeled bark tissues of new branches that emerged weeks after the inoculation, and the signals became increasingly stronger and more abundant in tissues examined at 10 wpi versus 5 wpi (Figure 6d).
FIGURE 6.
The infectivity of citrus tristeza virus (T36CA‐V1.3) with two amino acid substitutions introduced into the T36CA‐V1.1 encoded p65. Conserved nucleotides from the T36CA population were engineered into the p65 gene of pT36CA‐V1.1 to replace two nonconserved/nonsynonymous nucleotide variants. The resulting construct, pT36CA‐V1.3, encoding a p65 that contained two amino acid substitutions, N118S and S158L, was agroinfiltrated to transgenic (HC‐Pro‐expressing) Nicotiana benthamiana plants. (a) Representative images of green fluorescent protein (GFP) fluorescence observed in the infiltrated and systemic leaves of pT36CA‐V1.3‐inoculated plants at 10 days postinoculation (dpi) and 4 weeks postinoculation (wpi), respectively. A systemic leaf with vein chlorosis at 5 wpi, like those observed in Hyb1‐infected plants (see Figure 3b), is shown to the right of the GFP‐fluorescing systemic leaf. (b) Assessment of virus accumulation in the systemic leaves of transgenic (HC‐Pro) N. benthamiana plants at 5 wpi. The A405 value for each virus/treatment indicated on the x axis is the average for eight individual samples in total from two representative experiments (with error bar representing SE). (c) A transmission electron micrograph with scale bar (as indicated), showing a virion‐like particle of T36CA‐V1.3. (d) Examination of bark peels obtained from the new branches of T36CA‐V1.3 virion preparations‐inoculated Citrus macrophylla plants by wide‐field fluorescence microscopy, revealing the presence of GFP fluorescence that increased in intensity and abundance from 5 to 10 wpi
3. DISCUSSION
Evidence for nucleotide heterogeneity in the T36CA population has been documented previously using conventional cDNA cloning and Sanger sequencing methods (Chen et al., 2018). The results obtained here are consistent with those findings, and have contributed to genetic and molecular insights on how specific nucleotide variants in the cDNA clones incorporated into the prototype vectors, pT36CA‐ori and pT36CA‐V0, have negatively affected the bioactivity and systemic infection of the resulting viruses. While the presence of aberrant nucleotides introduced during PCR amplification remains a possibility, there is also evidence to support the suggestion that the nucleotide variants are constituents of a mutant spectrum in the T36CA population. First, the cloned cDNA templates used in this study were generated using a large amount of double‐stranded (ds)RNA (approximately 200 ng) to minimize the possibility of aberrant nucleotide incorporation (Chen et al., 2018; Rubio et al., 2001). Secondly, the same nucleotide variants were seen in multiple independently generated cDNA clones (Chen et al., 2018) including, for example, the nonsynonymous nucleotide difference (i.e., G vs. A) at position 52 of the p20 ORF between the two T36CA prototypes and T36FL, or the synonymous nucleotide differences in the p13 ORF between the two T36CA prototypes and T36FL (i.e., U vs. C and U vs. G in positions 147 and 219, respectively; Figure S1). Thirdly, while some T36CA nucleotide variants, such as those in the terminal sequences of the genome, are different from the ones deemed conserved within the T36CA population, they are identical to the nucleotides in exactly the same genomic locations in T36FL (Chen et al., 2018). Such co‐occurrences of nucleotide variants cannot simply be attributed to coincidence associated with PCR errors alone. The four nonsynonymous nucleotide variants that we altered in the T36CA prototype vectors are not present in the T36FL genotype, and it is clear from results of the biological assays that they are unique to, and important for the fitness of, viruses with the T36CA genotype. It remains to be seen whether these nucleotides would also negatively affect a virus with another genotypic background such as T36FL. We also cannot rule out if nucleotide variants (other than the ones we have tested) in regions IIb and IIc of the T36CA genome, especially those located in potential regulatory regions, could negatively impact the virus. However, if they do, our studies suggest that it may be overcome or compensated by the four nucleotide replacements.
Our results have clearly demonstrated that a genetic basis underlying the compromised fitness (impaired systemic infection in N. benthamiana plants) of T36CA‐V0 is RNA silencing. RNA silencing involves multiple plant components such as DICER‐LIKE proteins (DCLs) that process dsRNA into small interfering RNAs (siRNAs), Argonaute proteins (AGOs) that associate with siRNAs and assemble the RNA‐induced silencing complexes (RISCs) responsible for degrading the targeted sequences or for translation inhibition, and RNA‐dependent RNA polymerases (RDRs) that generate secondary siRNAs for the amplification of the silencing response (Csorba and Burgyán, 2016; Llave, 2010; Machado et al., 2017; Yang and Li, 2018). Some of these components have been identified in N. benthamiana plants and their antiviral functions have been investigated (Gursinsky et al., 2015; Ludman et al., 2017; Odokonyero et al., 2015; Scholthof et al., 2011; Schwach et al., 2005). In our study, the association of the RDR6‐ and/or AGO2‐dependent silencing with the systemic infection of T36CA in N. benthamiana plants was demonstrated by results showing the rehabilitation of T36CA‐V0 systemic infection in the RDR6i and ago2 lines (Figure 1b,c). In addition, T36CA‐V1.1 showed enhanced systemic infectivity in both RDR6i and ago2 plants, as the increase in T36CA‐V1.1 titre (in reference to the T36FL titre) was notably more pronounced in the two silencing‐defective N. benthamiana lines than in HC‐Pro‐expressing N. benthamiana plants (Figure 5b,d,e).
To counteract RNA silencing, plant viruses encode diverse VSRs (Csorba et al., 2015; Kubota and Ng, 2016) that target different steps of the RNA silencing pathway (Burgyán and Havelda, 2011; Csorba et al., 2015; Yang and Li, 2018). In the case of CTV, three viral‐encoded proteins exhibit VSR activity at the intercellular (CP and p20) and intracellular (p20 and p23) levels (Lu et al., 2004). The CP and p20 ORFs are in regions IIb and IIc, respectively, of the CTV genome based on the restriction enzyme sites marked out in the chimeric constructs (Figure 2). Results from the mapping studies of the chimeric (T36CA‐T36FL) viruses clearly support the conclusion that determinants that influence the systemic infectivity of T36CA are present in these two genomic regions (Figure 2). Indeed, our studies using individual point mutations have shown that the defective systemic infection of T36CA‐V0 in N. benthamiana plants was overcome with a single amino acid substitution (S107G) in T36CA‐V1.0 p20, as well as both S107G and G36D substitutions in T36CA‐V1.1 p20 and CP, respectively (Figure 5b). This strongly suggests an association of the two intercellular VSRs with T36CA systemic infection, and that the amino acids at positions 107 (of p20) and 36 (of CP) are very important for VSR activity. It is difficult to interpret the functions of these amino acids without a structure of the proteins. However, we know that an S to G substitution will clearly make the protein more flexible, while the G to D substitution represents a change from an uncharged to charged amino acid.
The effects of HC‐Pro and TBSV p19 on the suppression of silencing of other unrelated plant viruses have been well documented (Lakatos et al., 2006; Siddiqui et al., 2008; Yelina et al., 2002; Yoon et al., 2011). Because the suppression mechanisms of both VSRs involve targeting different steps of the RNA silencing pathway (Csorba et al., 2015; Ivanov et al., 2016; Kontra et al., 2016; Qiao and Falk, 2018; Valli et al., 2018), we initially expected either or both VSRs to exhibit at the least some discernible effects on the suppression of silencing associated with the VSR (p20/CP)‐defective T36CA‐ori or T36CA‐V0. However, both viruses failed to move systemically in HC‐Pro‐expressing N. benthamiana plants, even in the presence of exogenous TBSV p19 (Figure S2). In addition, as mentioned in the results, the presence of exogenous TBSV p19 in HC‐Pro‐expressing N. benthamiana plants did no better in raising the titre of T36CA‐V1.0 and T36CA‐V1.1 than when exogenous TBSV p19 was absent. Taken together, these results indicate that the suppression activity/function of TBSV p19 is ineffective against the RDR6‐ and/or AGO2‐mediated anti‐T36CA defence in transgenic (HC‐Pro‐expressing) N. benthamiana plants. Adding to the complexity is the observation that exogenous HC‐Pro was needed for the systemic infection of T36CA‐V0 and T36CA‐V1.1 in RDR6i plants, but was dispensable for both local and systemic infection of the virus in ago2 plants (Figures 1b,c and 5d,e; data not shown for T36CA‐V1.1). It is possible that although the spread of the mobile silencing signals is restricted in RDR6i lines (Melnyk et al., 2011; Schwach et al., 2005), the presence of HC‐Pro (administered by coinfiltration) is still required to suppress the intracellular silencing triggered by these viruses (Valli et al., 2018). In the absence of an active AGO2 in ago2 plants, a key component of RNA silencing is inhibited, thus allowing the VSR‐defective T36CA‐V0 to infect systemically, and for T36CA‐V1.1 to increase its titre, without the need for exogenous VSRs. N. benthamiana has a number of DCLs, AGOs, and RDRs associated with the RNA silencing process (Muhammad et al., 2019); therefore, it is not surprising that more host components may be involved in the antiviral defence mechanism against T36CA.
The fact that GFP signals were observed in the initial (bark flap) inoculation site of T36CA‐V1.1 virions in C. macrophylla plants suggests that viral replication‐driven gene expression had occurred in the local tissues. However, the absence of systemic infection in C. macrophylla plants, even when much higher virus inoculum concentrations were used (Figure 5d,e), brings to light the potential involvement of another/other viral component(s). That component, as determined in our study, was p65 (a HSP70 homolog), a putative CTV movement protein encoded by the p65 ORF located in genomic region IIb (Figure 2). The HSP70 homolog in beet yellows virus (BYV), BYV p65, is one of at least five viral proteins encoded by the so‐called “quintuple gene block”—p65, p64 (an ortholog of CTV p61) as well as the two capsid proteins, CP and CPm, and a small hydrophobic protein, p6—involved in the cell‐to‐cell movement of BYV, and is conserved among all closterovirus genomes (Agranovsky, 2016; Alzhanova et al., 2000, 2001; Dolja et al., 2006). In our study, the high titre of T36CA‐V1.3 in N. benthamiana plants and its robust systemic infectivity in C. macrophylla have provided clear evidence that the amino acids in positions 118 and 158 of p65 are critical determinants of systemic T36CA movement in C. macrophylla plants. This finding represents new experimental proof for the association of p65 in the systemic movement of CTV. The proteins, along with their specific amino acids identified in this study and the roles they play in facilitating interactions with the host machineries involved in RNA silencing and long‐distance virus transport should and will be further investigated in future studies.
4. EXPERIMENTAL PROCEDURES
4.1. Construction of T36CA‐based binary plasmids
The T36CA binary plasmid, pT36CA, was constructed by replacing all regions corresponding to the CTV genome in C86 (El‐Mohtar and Dawson, 2014) with the equivalent regions from each of two cDNA clones, containing DNA corresponding to nucleotide (nt) positions 1 to 8,391 and 7,800 to 19,292 (for simplicity referred to as the 5′ and 3′ fragments, respectively), of the full‐length (19,292 nt) T36CA genome (Chen et al., 2018). Multiple cloned DNA and PCR‐amplified products encompassing specific regions of the 5′ or 3′ fragments were obtained and used for systematically constructing two prototype vectors, pT36CA‐ori and pT36CA‐V0. In both prototype vectors, regions corresponding to the T36CA genome from nt position 1 to 8,106 originated from one cDNA clone of the 5′ fragment, while those from nt positions 8,107 to 17,204 and the 3′ NCR were from a single 3′ fragment cDNA clone. Nucleotides 17,205 to 19,019 in pT36CA‐ori and pT36CA‐V0 were from two different cDNA clones of the 3′ fragment, clones #8 and #12, respectively.
Detailed steps and key intermediates involved in the construction of pT36CA‐ori are laid out in the Text S2 and Figure S4. Nucleotide sequences of T36CA origin incorporated in pT36CA‐ori are available in the NCBI database (accession number MH279617). The procedures for constructing pT36CA‐V0 were similar to that of pT36CA‐ori with three exceptions. First, steps leading to the generation of pGemT36#12p23GFP (data not shown), a cDNA clone equivalent to pGemT36p23GFP (Figure S4b), involved PCR amplification using the 3′ fragment cDNA clone #12 as the template (data not shown), rather than using pT36CAori‐full (Figure S4a), which was generated, in part, using 3′ fragment cDNA clone #8 (data not shown). Secondly, steps leading to the generation of pGEMT36p23#12GFPnos (data not shown), a cDNA clone equivalent to pGEMT36p23GFPnos (Figure S4b), involved replacing the ApaI/HindIII fragment excised from pGEMT36p23GFPnos with a similarly cut fragment from pGemT36#12p23GFP (data not shown). Thirdly, the PstI/SwaI fragment excised from pGEMT36p23#12GFPnos was ligated into the vector backbone of similarly digested pT36CA‐ori (Figure S4b) to generate the final product pT36CA‐V0 (data not shown).
Procedures for making the pT36CA‐pT36FL chimeric construct, pHyb1, are described in Text S2 and Figure S4a. pHyb2, pHyb4, and pHyb6 were generated by excising subregions IIa, IIb, and IIc with SmaI/PmeI, PmeI/PstI, and PstI/SwaI, respectively, of pT36CA‐ori and ligating the excised fragments with the vector backbone of similarly cut pHyb1. pHyb3, pHyb5, and pHyb7 were generated in a reciprocal manner, that is, restriction enzymes‐excised fragments from pHyb1 were ligated into the vector backbone of similarly cut pT36CA‐ori (Figure 2a).
The site‐directed mutagenesis of specific nonconserved and nonsynonymous nt variants in the p20, p25 (CP), and p65 coding regions of T36CA‐V0 was achieved by overlapping PCRs using pT36CA‐V0 as a template and specific oligonucleotide primers (Table S1). An A to G modification at nt position 18,078 that would result in a deduced S107G amino acid change in p20 was introduced into a PCR product generated by oligonucleotide primers CTV206, CTV422, CTV421, and CTV27. A G to A modification at nt position 16,257 that would result in a deduced G36D amino acid change in the CP was incorporated in a PCR product generated by oligonucleotide primers CTV110, CTV416, CTV415, and CTV89. Both PCR products were each cloned into the pGemT Easy vector (Promega) and then subcloned into pT36CA‐V0 in two sequential steps. First, the DNA fragment with the A/G nt change was excised with PstI/SwaI and ligated into the vector backbone of the similarly digested pT36CA‐V0, resulting in pT36CA‐V1.0. Next, the DNA with the G/A nt modification was excised with PmeI/PstI and ligated into the vector backbone of the similarly digested pT36CA‐V1.0 to generate pT36CA‐V1.1. To modify the two nt variants—from A to G at position 12,397 and C to T at position 12,517 that would result in a deduced N118S and S158L amino acid change, respectively—in the p65‐coding region, two sequential overlapping PCRs were performed using pT36CA‐V1.1 as a template and the following oligonucleotide primers: CTV110, CTV426, CTV425, and CTV89 (for the A/G nt change) and CTV110, CTV428, CTV427, and CTV89 (for the C/T nt change). The resulting product was cloned into the pGemT Easy vector (Promega), excised with PmeI/PstI, and subcloned into the vector backbone of similarly digested pT36CA‐V1.1 to generate pT36CA‐V1.3.
All PCRs were performed using the high‐fidelity Herculase II Fusion DNA polymerase (Agilent Technologies), and products were cloned and sequenced to confirm the introduced nt changes as well as to ensure that no spurious mutations were present. DNA purification, using the MiniElute Gel Extraction kit (Qiagen) or the Zymoclean Gel DNA Recovery kit (Zymo Research), and DNA ligation by T4 ligase (Promega) were performed according to the manufacturers’ instructions. Plasmids were transformed into appropriate Escherichia coli strains and selected by growth on antibiotic‐containing agar media.
4.2. Agroinfiltration of N. benthamiana plants
Transgenic N. benthamiana plants overexpressing a potyvirus, Turnip mosaic virus, HC‐Pro, a viral suppressor of RNA silencing (VSR) (kindly provided by Bryce Falk, UC Davis) has been shown to enhance the infectivity of lettuce infectious yellows virus, a member of the family Closteroviridae (Qiao and Falk, 2018). Therefore, HC‐Pro‐expressing N. benthamiana plants were used for the agroinfiltration of CTV constructs unless indicated specifically. All N. benthamiana plants, including RDR6i (kindly provided by David Baulcombe, University of Cambridge) and ago2 lines, were grown in a greenhouse until five or six fully expanded leaves were formed before they were used for infiltration. The binary plasmids were transformed into Agrobacterium tumefaciens EHA105 using the freeze‐thaw method (Ambrós et al., 2011). For infiltration, A. tumefaciens cultures each transformed with a vector construct were prepared following the procedure described in Ambrós et al. (2011). Briefly, a single colony of transformed A. tumefaciens was inoculated into 5 ml Luria‐Bertani (LB) medium with 50 µg/ml kanamycin (the starter culture) and grown for 48 hr at 28°C. The starter culture was transferred to an induction LB medium, supplemented with 10 mM 2‐(N‐morpholino)ethanesulfonic acid (MES), pH 5.85, and 20 µM acetosyringone, in a 1:80 (vol/vol) ratio, and incubated at 28°C overnight with shaking at 240 rpm. For A. tumefaciens containing p35S:TBSV p19, the starter culture was incubated for 24 hr before being transferred to the induction medium in a 1:200 (vol/vol) ratio. A. tumefaciens was collected by centrifugation at 2,000 × g for 10 min at 24°C, and washed with buffer (10 mM MES, pH 5.85. 10 mM MgCl2). After another round of centrifugation at 2,000 × g for 10 min at 24°C, A. tumefaciens was resuspended in induction buffer (10 mM MES, pH 5.85, 10 mM MgCl2, 150 µM acetosyringone) to a final OD600 of 0.5 and kept in the dark for at least 3 hr before use. The leaves of N. benthamiana plants were infiltrated with an A. tumefaciens suspension using a 3‐ml needleless syringe and the plants were maintained at 20°C with a 16 hr light/8 hr dark cycle. Eight to 16 plants were used for each infiltration event.
4.3. RT‐PCR, ELISA, and statistical analysis
Total RNA of N. benthamiana plants was extracted from leaves using TRIzol reagent (Thermo Fisher Scientific) with modifications as described previously (Gehrig et al., 2000). The first strand cDNA was synthesized by Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega) using oligonucleotide primer CTV30‐AC, and a 1274‐bp product was subsequently PCR‐amplified by Taq DNA polymerase using oligonucleotide primers CTV351‐AC and CTV30‐AC (Table S1). DAS‐ELISA was performed to detect CTV in N. benthamiana plants according to the user's guide for the ELISA reagent set for CTV identification (SRA78900; Agdia) with some modifications. For example, the antibody 1052‐2LA specific to CTV virion (kindly provided by Bill Dawson, University of Florida) was diluted in a carbonate coating buffer at a 1:1,500 ratio for coating a 96‐well plate (Fisherbrand) placed in a humidity chamber overnight at 4°C. Plant extracts were prepared by grinding 0.1 g of leaf tissues in 1 ml of phosphate‐buffered saline with 0.05% Tween‐20 (PBST) using a mortar and pestle (Garnsey and Cambra, 1991). The preparations of antibody and substrate for detection followed the manufacturer's instructions. The absorbance at 405 nm was measured using an Emax precision microplate reader (Molecular Devices). Paired Student t tests were performed using the GraphPad program (https://www.graphpad.com/).
4.4. Virion purification, transmission electron microscopy analysis, and inoculation of citrus plants
Virion purification and inoculation of citrus were carried out using the methods described previously (El‐Mohtar and Dawson, 2014). Systemic leaves of N. benthamiana plants with GFP fluorescence were harvested for virion purification. Nine‐ to 12‐month‐old C. macrophylla plants were inoculated by a bark flap inoculation technique (Robertson et al., 2005). Each virion preparation was inoculated to between four and 16 plants. Following virion inoculation, citrus plants were kept indoors for 1–5 days before being moved to and maintained in a greenhouse for observation.
A formvar/carbon‐coated copper grid was floated on 8 µl of CTV capture antibodies (Agdia CAB 78900/5000, diluted 1:10 in phosphate buffer [10 mM potassium phosphate, pH 7.0]) for 10 min. The grid was rinsed with phosphate buffer and transferred to 6 µl of a virion preparation. After 15 min, the grid was rinsed with phosphate buffer and stained with 0.5% phosphotungstic acid, pH 7.0, with 50 µg/ml bacitracin. After blotting, the grid was examined under a Hitachi 7500 transmission electron microscope.
4.5. Visualization of GFP expression in plants
GFP fluorescence in N. benthamiana leaves was observed using a Blak‐Ray longwave UV lamp (model B‐100A; UVP). To visualize GFP fluorescence in C. macrophylla plants, individual strips of bark (each measuring approximately 3 × 20 mm) were peeled off the branch using a scalpel. The exposed inner layer of each bark peel, which contained the vascular tissues, was observed under wide‐field fluorescence microscopy using a Nikon Labophot fluorescence microscope fitted with fluorescein isothiocyanate/cyanine 3 (FITC/CY3) double bandpass filters, and a Canon EOS T5i digital single‐lens reflex camera.
Supporting information
FIGURE S1 Sequence comparison of the p13 and p20 coding regions between T36CA‐ori and T36CA‐V0. Nucleotide (nt) sequences of the p13 and p20 coding regions in T36CA‐ori and T36CA‐V0 were aligned with those of T36FL using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/). (a) The alignment of nt sequences of p13. (b) The alignment of nt sequences of p20. Nucleotide variants in T36CA‐ori and/or T36CA‐V0 are indicated in red. Nonsynonymous nt variants are highlighted in blue; indicated below them are the resulting change in deduced amino acids (in bold)
FIGURE S2 The infectivity of citrus tristeza virus (T36CA). Two prototype vectors, pT36CA‐ori and pT36CA‐V0, or pT36FL (positive control), were inoculated to transgenic Nicotiana benthamiana plants expressing HC‐Pro by agroinfiltration either alone (no p19) or coinfiltrated with a vector expressing tomato bushy stunt virus p19 and evaluated for bioactivity. (a) GFP fluorescence was observed in the leaves infiltrated with each of the three vectors at 10 days postinfiltration (dpi), and only in the systemic leaves of pT36FL‐infiltrated plants at 4 weeks postinfiltration (wpi). The leaves in the images are all attached on the plants. (b) A representative gel image showing the results of viral RNA detection (by RT‐PCR) in the systemic leaves of N. benthamiana plants infiltrated with the pT36CA prototype vectors, either alone or coinfiltrated with p19, at 8 or 12 wpi. 1 and 2 represent the test results for two individual plants. The lanes marked L and T36FL contain a 1 kb DNA ladder and a 1274 bp RT‐PCR product (arrow; obtained using a T36FL‐infected systemic leaf), respectively
FIGURE S3 Analysis of virion formation and/or in planta GFP expression of two chimeric viruses. Virion preparations obtained from the systemic leaves of pHyb2‐ and pHyb7‐inoculated Nicotiana benthamiana plants were analysed by immunocapture transmission electron microscopy. Virion preparations of Hyb2 were also inoculated to Citrus macrophylla plants for the determination of bioactivity. (a) Transmission electron micrographs of virion‐like particles of Hyb2 and Hyb7. (b) Visualization of GFP signals in the peeled bark of an apical branch of a Hyb2 virion preparation‐inoculated C. macrophylla plant at 8 weeks postinfiltration using widefield fluorescence microscopy
FIGURE S4 Schematic procedure for cloning the pT36CA‐ori construct/prototype vector. A two‐part procedure was used to engineer pT36CA‐ori. (a) Part I of the cloning procedure generated the intermediate binary plasmids, pHyb1 and pT36CAori‐full. (b) Part II of the cloning procedure led to the construction of the binary plasmid/prototype vector pT36CA‐ori. Grey and white boxes represent the CTV open reading frames (ORFs) that originated from the C86 (pT36FL) binary plasmid (El‐Mohtar and Dawson, 2014) and T36CA cDNA clones (Chen et al., 2018), respectively. Names of ORFs are as described in Figure 1. Black boxes represent the region containing a ribozyme sequence and nopaline synthase (NOS) terminator. The cauliflower mosaic virus 35S promoter (35S) and a coding region of green fluorescence protein (GFP) are as illustrated. Bars in purple and dark green represent the controller element of the beet yellows virus (BYV) and citrus tristeza virus (CTV) major coat protein, respectively. The restriction enzymes used for cloning and their locations are as shown. The name of each cloned DNA (made up of a combination of letter and numerical identifiers preceded by the letter “p”) or PCR product is indicated at the bottom of, or next to, each illustration
TABLE S1 The oligonucleotide primers used in this study
TEXT S1 Sequence comparison between two T36CA prototypes and their systemic infectivity in HC‐Pro expressing Nicotiana benthamiana plants
TEXT S2 Detailed steps and key intermediates involved in the construction of pT36CA‐ori
ACKNOWLEDGMENTS
We thank Bill Dawson for providing pT36FL and Shou‐wei Ding for helpful discussion. This work was supported, in part, by a grant from the Citrus Research Board of California and a collaborative National Institute of Food and Agriculture SCRI grant (2015‐70016‐2300) to J.C.K.N. The work in K.F.'s laboratory was supported by the National Research Development and Innovation Office, Hungary (K124705). All authors declare no conflict of interest.
Chen AYS, Peng JHC, Polek M, et al. Comparative analysis identifies amino acids critical for citrus tristeza virus (T36CA) encoded proteins involved in suppression of RNA silencing and differential systemic infection in two plant species. Mol Plant Pathol. 2021;22:64–76. 10.1111/mpp.13008
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Agranovsky, A.A. (2016) Closteroviruses: molecular biology, evolution and interactions with cells In: Gaur R.K., Petrov N.M., Patil B.L. and Stoyanova M.I. (Eds.) Plant Viruses: Evolution and Management. Singapore: Springer Singapore, pp. 231–252. [Google Scholar]
- Albiach‐Martí, M.R. , Mawassi, M. , Gowda, S. , Satyanarayana, T. , Hilf, M.E. , Shanker, S. et al. (2000) Sequences of Citrus tristeza virus separated in time and space are essentially identical. Journal of Virology, 74, 6856–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiach‐Martí, M.R. , Robertson, C. , Gowda, S. , Tatineni, S. , Belliure, B. , Garnsey, S.M. et al. (2010) The pathogenicity determinant of Citrus tristeza virus causing the seedling yellows syndrome maps at the 3'‐terminal region of the viral genome. Molecular Plant Pathology, 11, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzhanova, D.V. , Hagiwara, Y. , Peremyslov, V.V. and Dolja, V.V. (2000) Genetic analysis of the cell‐to‐cell movement of beet yellows closterovirus. Virology, 268, 192–200. [DOI] [PubMed] [Google Scholar]
- Alzhanova, D.V. , Napuli, A.J. , Creamer, R. and Dolja, V.V. (2001) Cell‐to‐cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO Journal, 20, 6997–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrós, S. , El‐Mohtar, C. , Ruiz‐Ruiz, S. , Peña, L. , Guerri, J. , Dawson, W.O. et al. (2011) Agroinoculation of Citrus tristeza virus causes systemic Infection and symptoms in the presumed nonhost Nicotiana benthamiana . Molecular Plant‐Microbe Interactions, 24, 1119–1131. [DOI] [PubMed] [Google Scholar]
- Bayne, E.H. , Rakitina, D.V. , Morozov, S.Y. and Baulcombe, D.C. (2005) Cell‐to‐cell movement of Potato potexvirus X is dependent on suppression of RNA silencing. The Plant Journal, 44, 471–482. [DOI] [PubMed] [Google Scholar]
- Burgyán, J. and Havelda, Z. (2011) Viral suppressors of RNA silencing. Trends in Plant Science, 16, 265–272. [DOI] [PubMed] [Google Scholar]
- Chen, A.Y.S. , Watanabe, S. , Yokomi, R. and Ng, J.C.K. (2018) Nucleotide heterogeneity at the terminal ends of the genomes of two California Citrus tristeza virus strains and their complete genome sequence analysis. Virology Journal, 15, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba, T. and Burgyán, J. (2016) Antiviral silencing and suppression of gene silencing in plants In: Wang A. and Zhou X. (Eds.) Current Research Topics in Plant Virology. Cham: Springer International Publishing, pp. 1–33. [Google Scholar]
- Csorba, T. , Kontra, L. and Burgyán, J. (2015) Viral silencing suppressors: tools forged to fine‐tune host–pathogen coexistence. Virology, 479, 85–103. [DOI] [PubMed] [Google Scholar]
- Dolja, V.V. , Kreuze, J.F. and Valkonen, J.P.T. (2006) Comparative and functional genomics of closteroviruses. Virus Research, 117, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Mohtar, C. and Dawson, W.O. (2014) Exploring the limits of vector construction based on Citrus tristeza virus . Virology, 448, 274–283. [DOI] [PubMed] [Google Scholar]
- Garnsey, S.M. and Cambra, M. (1991) Enzyme‐linked immunosorbent assay (ELISA) for citrus pathogens In: Roistacher C.N. (Ed.) Graft‐Transmissible Diseases of Citrus. Handbook for Detection and Diagnosis. Rome: FAO, pp. 193–216. [Google Scholar]
- Gehrig, H.H. , Winter, K. , Cushman, J. , Borland, A. and Taybi, T. (2000) An improved RNA isolation method for succulent plant species rich in polyphenols and polysaccharides. Plant Molecular Biology Reporter, 18, 369–376. [Google Scholar]
- Gursinsky, T. , Pirovano, W. , Gambino, G. , Friedrich, S. , Behrens, S.‐E. and Pantaleo, V. (2015) Homeologs of the Nicotiana benthamiana antiviral ARGONAUTE1 show different susceptibilities to microRNA168‐mediated control. Plant Physiology, 168, 938–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, S.J. (2013) Citrus tristeza virus: evolution of complex and varied genotypic groups. Frontiers in Microbiology, 4, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, S.J. , Cowell, S.J. and Dawson, W.O. (2015a) Finding balance: virus populations reach equilibrium during the infection process. Virology, 485, 205–212. [DOI] [PubMed] [Google Scholar]
- Harper, S.J. , Cowell, S.J. and Dawson, W.O. (2015b) With a little help from my friends: complementation as a survival strategy for viruses in a long‐lived host system. Virology, 478, 123–128. [DOI] [PubMed] [Google Scholar]
- Ivanov, K.I. , Eskelin, K. , Basic, M. , De, S. , Lohmus, A. , Varjosalo, M. et al. (2016) Molecular insights into the function of the viral RNA silencing suppressor HCPro. The Plant Journal, 85, 30–45. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. and Carrington, J.C. (2001) Long‐distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC‐Pro. Virology, 285, 71–81. [DOI] [PubMed] [Google Scholar]
- Kontra, L. , Csorba, T. , Tavazza, M. , Lucioli, A. , Tavazza, R. , Moxon, S. et al. (2016) Distinct effects of p19 RNA silencing suppressor on small RNA mediated pathways in plants. PLoS Pathogens, 12, e1005935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota, K. and Ng, J.C.K. (2016) Lettuce chlorosis virus P23 suppresses RNA silencing and induces local necrosis with increased severity at raised temperatures. Phytopathology, 106, 653–662. [DOI] [PubMed] [Google Scholar]
- Lakatos, L. , Csorba, T. , Pantaleo, V. , Chapman, E.J. , Carrington, J.C. , Liu, Y.‐P. et al. (2006) Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO Journal, 25, 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C. (2010) Virus‐derived small interfering RNAs at the core of plant–virus interactions. Trends in Plant Science, 15, 701–707. [DOI] [PubMed] [Google Scholar]
- Lu, R. , Folimonov, A. , Shintaku, M. , Li, W.X. , Falk, B.W. , Dawson, W.O. et al. (2004) Three distinct suppressors of RNA silencing encoded by a 20‐kb viral RNA genome. Proceedings of the National Academy of Sciences of the United States of America, 101, 15742–15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludman, M. , Burgyán, J. and Fátyol, K. (2017) Crispr/Cas9 mediated inactivation of Argonaute 2 reveals its differential involvement in antiviral responses. Scientific Reports, 7, 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, J.P.B. , Calil, I.P. , Santos, A.A. and Fontes, E.P.B. (2017) Translational control in plant antiviral immunity. Genetics and Molecular Biology, 40, 292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk, C.W. , Molnar, A. and Baulcombe, D.C. (2011) Intercellular and systemic movement of RNA silencing signals. EMBO Journal, 30, 3553–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad, T. , Zhang, F. , Zhang, Y. and Liang, Y. (2019) RNA interference: a natural immune system of plants to counteract biotic stressors. Cells, 8, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odokonyero, D. , Mendoza, M.R. , Alvarado, V.Y. , Zhang, J. , Wang, X. and Scholthof, H.B. (2015) Transgenic down‐regulation of ARGONAUTE2 expression in Nicotiana benthamiana interferes with several layers of antiviral defenses. Virology, 486, 209–218. [DOI] [PubMed] [Google Scholar]
- Peremyslov, V.V. , Hagiwara, Y. and Dolja, V.V. (1999) HSP70 homolog functions in cell‐to‐cell movement of a plant virus. Proceedings of the National Academy of Sciences of the United States of America, 96, 14771–14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, W. and Falk, B.W. (2018) Efficient protein expression and virus‐induced gene silencing in plants using a crinivirus‐derived vector. Viruses, 10, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F. and Morris, T.J. (2005) Suppressors of RNA silencing encoded by plant viruses and their role in viral infections. FEBS Letters, 579, 5958–5964. [DOI] [PubMed] [Google Scholar]
- Robertson, C.J. , Garnsey, S.M. , Satyanarayana, T. , Folimonova, S.Y. and Dawson, W.O. (2005) Efficient infection of citrus plants with different cloned constructs of citrus tristeza virus amplified in Nicotiana benthamiana protoplasts In: Hilf M.E., Duran‐Vila N. and Rocha‐Peña M.A. (Eds.) International Organization of Citrus Virologists Conference Proceedings (1957–2010). Riverside, CA: International Organization of Citrus Virologists, pp. 187–195. [Google Scholar]
- Rubio, L. , Ayllón, M.A. , Kong, P. , Fernández, A. , Polek, M. , Guerri, J. et al. (2001) Genetic variation of Citrus tristeza virus isolates from California and Spain: evidence for mixed infections and recombination. Journal of Virology, 75, 8054–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana, T. , Gowda, S. , Ayllón, M.A. and Dawson, W.O. (2004) Closterovirus bipolar virion: evidence for initiation of assembly by minor coat protein and its restriction to the genomic RNA 5′ region. Proceedings of the National Academy of Sciences of the United States of America, 101, 799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana, T. , Gowda, S. , Mawassi, M. , Albiach‐Martí, M.R. , Ayllón, M.A. , Robertson, C. et al. (2000) Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology, 278, 253–265. [DOI] [PubMed] [Google Scholar]
- Scholthof, H.B. , Alvarado, V.Y. , Vega‐Arreguin, J.C. , Ciomperlik, J. , Odokonyero, D. , Brosseau, C. et al. (2011) Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana . Plant Physiology, 156, 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach, F. , Vaistij, F.E. , Jones, L. and Baulcombe, D.C. (2005) An RNA‐dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiology, 138, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui, S.A. , Sarmiento, C. , Kiisma, M. , Koivumaki, S. , Lemmetty, A. , Truve, E. et al. (2008) Effects of viral silencing suppressors on tobacco ringspot virus infection in two Nicotiana species. Journal of General Virology, 89, 1502–1508. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , Robertson, C.J. , Garnsey, S.M. , Bar‐Joseph, M. , Gowda, S. and Dawson, W.O. (2008) Three genes of Citrus tristeza virus are dispensable for infection and movement throughout some varieties of citrus trees. Virology, 376, 297–307. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , Robertson, C.J. , Garnsey, S.M. and Dawson, W.O. (2011) A plant virus evolved by acquiring multiple nonconserved genes to extend its host range. Proceedings of the National Academy of Sciences of the United States of America, 108, 17366–17371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli, A.A. , Gallo, A. , Rodamilans, B. , José López‐Moya, J. and Antonio García, J. (2018) The HCPro from the Potyviridae family: an enviable multitasking helper component that every virus would like to have. Molecular Plant Pathology, 19, 744–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. and Li, Y. (2018) Dissection of RNAi‐based antiviral immunity in plants. Current Opinion in Virology, 32, 88–99. [DOI] [PubMed] [Google Scholar]
- Yelina, N.E. , Savenkov, E.I. , Solovyev, A.G. , Morozov, S.Y. and Valkonen, J.P.T. (2002) Long‐distance movement, virulence, and RNA silencing suppression controlled by a single protein in hordei‐ and potyviruses: complementary functions between virus families. Journal of Virology, 76, 12981–12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, J.‐Y. , Choi, S.‐K. , Palukaitis, P. and Gray, S.M. (2011) Agrobacterium‐mediated infection of whole plants by yellow dwarf viruses. Virus Research, 160, 428–434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Sequence comparison of the p13 and p20 coding regions between T36CA‐ori and T36CA‐V0. Nucleotide (nt) sequences of the p13 and p20 coding regions in T36CA‐ori and T36CA‐V0 were aligned with those of T36FL using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/). (a) The alignment of nt sequences of p13. (b) The alignment of nt sequences of p20. Nucleotide variants in T36CA‐ori and/or T36CA‐V0 are indicated in red. Nonsynonymous nt variants are highlighted in blue; indicated below them are the resulting change in deduced amino acids (in bold)
FIGURE S2 The infectivity of citrus tristeza virus (T36CA). Two prototype vectors, pT36CA‐ori and pT36CA‐V0, or pT36FL (positive control), were inoculated to transgenic Nicotiana benthamiana plants expressing HC‐Pro by agroinfiltration either alone (no p19) or coinfiltrated with a vector expressing tomato bushy stunt virus p19 and evaluated for bioactivity. (a) GFP fluorescence was observed in the leaves infiltrated with each of the three vectors at 10 days postinfiltration (dpi), and only in the systemic leaves of pT36FL‐infiltrated plants at 4 weeks postinfiltration (wpi). The leaves in the images are all attached on the plants. (b) A representative gel image showing the results of viral RNA detection (by RT‐PCR) in the systemic leaves of N. benthamiana plants infiltrated with the pT36CA prototype vectors, either alone or coinfiltrated with p19, at 8 or 12 wpi. 1 and 2 represent the test results for two individual plants. The lanes marked L and T36FL contain a 1 kb DNA ladder and a 1274 bp RT‐PCR product (arrow; obtained using a T36FL‐infected systemic leaf), respectively
FIGURE S3 Analysis of virion formation and/or in planta GFP expression of two chimeric viruses. Virion preparations obtained from the systemic leaves of pHyb2‐ and pHyb7‐inoculated Nicotiana benthamiana plants were analysed by immunocapture transmission electron microscopy. Virion preparations of Hyb2 were also inoculated to Citrus macrophylla plants for the determination of bioactivity. (a) Transmission electron micrographs of virion‐like particles of Hyb2 and Hyb7. (b) Visualization of GFP signals in the peeled bark of an apical branch of a Hyb2 virion preparation‐inoculated C. macrophylla plant at 8 weeks postinfiltration using widefield fluorescence microscopy
FIGURE S4 Schematic procedure for cloning the pT36CA‐ori construct/prototype vector. A two‐part procedure was used to engineer pT36CA‐ori. (a) Part I of the cloning procedure generated the intermediate binary plasmids, pHyb1 and pT36CAori‐full. (b) Part II of the cloning procedure led to the construction of the binary plasmid/prototype vector pT36CA‐ori. Grey and white boxes represent the CTV open reading frames (ORFs) that originated from the C86 (pT36FL) binary plasmid (El‐Mohtar and Dawson, 2014) and T36CA cDNA clones (Chen et al., 2018), respectively. Names of ORFs are as described in Figure 1. Black boxes represent the region containing a ribozyme sequence and nopaline synthase (NOS) terminator. The cauliflower mosaic virus 35S promoter (35S) and a coding region of green fluorescence protein (GFP) are as illustrated. Bars in purple and dark green represent the controller element of the beet yellows virus (BYV) and citrus tristeza virus (CTV) major coat protein, respectively. The restriction enzymes used for cloning and their locations are as shown. The name of each cloned DNA (made up of a combination of letter and numerical identifiers preceded by the letter “p”) or PCR product is indicated at the bottom of, or next to, each illustration
TABLE S1 The oligonucleotide primers used in this study
TEXT S1 Sequence comparison between two T36CA prototypes and their systemic infectivity in HC‐Pro expressing Nicotiana benthamiana plants
TEXT S2 Detailed steps and key intermediates involved in the construction of pT36CA‐ori
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


