Abstract
Epitopes, in the context of T cell recognition, are short peptides typically derived by antigen processing, and presented on the cell surface bound to MHC molecules (HLA molecules in humans) for TCR scrutiny. The identification of epitopes is a context-dependent process, with consideration given to, for example, the source pathogen and protein, the host organism, and state of the immune reaction (e.g., following natural infection, vaccination, etc.). In the following review, we consider the various approaches used to define T cell epitopes, including both bioinformatic and experimental approaches, and discuss the concepts of immunodominance and immunoprevalence. We also discuss HLA polymorphism and epitope restriction, and the resulting impact on the identification of, and potential population coverage afforded by, epitopes or epitope-based vaccines. Finally, some examples of the practical application of T cell epitope identification are provided, showing how epitopes have been valuable for deriving novel immunological insights in the context of the immune response to various pathogens and allergens.
Keywords: T cell epitopes, MHC, HLA, polymorphism, epitope prediction, vaccines
1. Introduction
In this review, we present our viewpoint on epitope identification. We stress that this is a rather personal and self-centered account. In particular, this review is focused on bacterial and viral microbes and human T cell epitope identification. Our approach hinges on first defining what is an epitope, and the type of data and metadata usually associated with epitopes. In the following sections we discuss epitope prediction, and epitope immunodominance and prevalence. Additional sections discuss HLA1 polymorphism and HLA restriction and their impact on population coverage. After discussing various technical approaches to epitope identification, we close by mentioning specific instances of how epitope identification studies have generated novel insights in microbial diseases.
2. What is a T cell epitope?
An epitope can be defined as the molecular structure recognized by adaptive immune responses, namely by T cell and B cell receptors and soluble antibodies [1]. In this review, we will focus on T cell epitopes, as this is our principal area of expertise. We will further focus on the epitopes recognized by classical alpha-beta TCR receptors, and restricted by conventional HLA molecules. We will not discuss the epitope structures recognized by gamma delta, MAIT and other non-conventional T cells, which have been described and reviewed elsewhere [2–8].
In general, the epitopes recognized by T cells are short peptides derived from processing of larger antigens [9], even though epitopes that are partly or completely of non-peptidic nature have been described [10]. In particular, MHC-II-bound glycan-peptide epitopes, which could be relevant for both infectious disease and cancer, have been described [11, 12]. As a general rule, CD4 “helper” T cells recognize peptides of mostly 13–17 residues in size bound to HLA class II, while CD8 “cytotoxic” T cells recognize peptides of mostly 8–10 residues in size bound to HLA class I [9, 13] (Figure 1A).
Figure 1. Epitope definition.
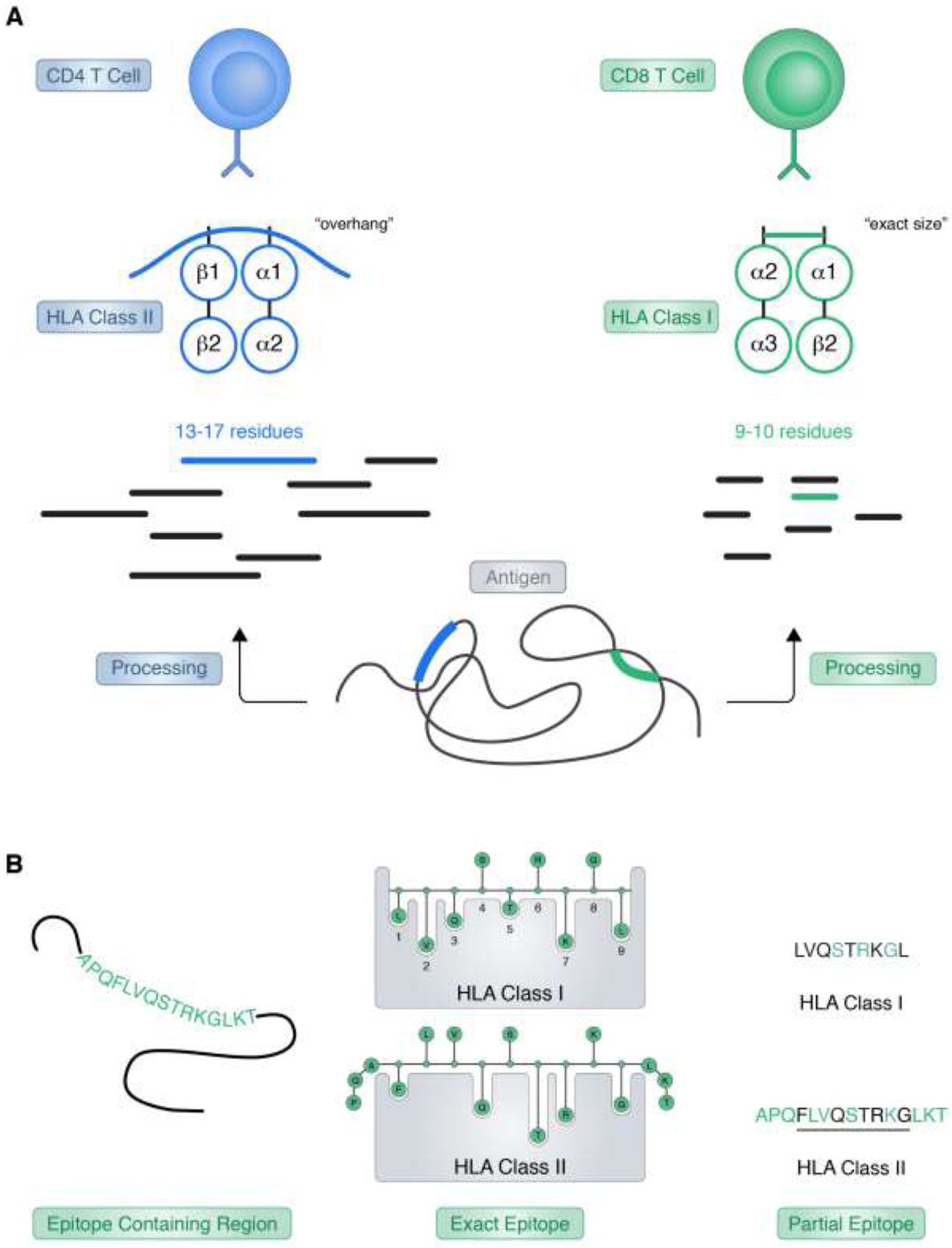
Presentation of peptide ligands by HLA class I and class II molecules is depicted. A) HLA class I and II ligands are generated by proteolytic processing of endogenously expressed proteins (antigen) (class I) or from proteins degraded in endocytic compartments (class II). Longer class II peptides typically overhang the open ends of the HLA class II binding groove, while shorter class I ligands are size constrained due to the closed end of the class I binding groove. With binding, and subsequent presentation on the cell surface, HLA-ligand complexes are available for scrutiny by CD4+ (class II) or CD8+ (class I) T cells. B) Ligands processed from longer protein antigens bind HLA using, in general, a nine-mer core region, where the main energy of binding is provided by interaction of some, but not all, peptide residues with residues forming the main pockets of the HLA binding groove. Definition of partial epitopes reflects that not all residues within an epitope region are necessarily important for HLA binding or T cell recognition, while mutation of other residues may ameliorate or abrogate specific immunity.
Even within the confines of the definition above, it should be pointed out that in practical terms T cell epitopes are defined at different level of granularity. In theory, an unambiguous crystallographic structure of an epitope bound to HLA and its TCR can define all relevant interactions, and unequivocally map the exact structure of the epitope [14]. However, in practice, this exact determination is most often missing, and different experimental epitope definitions are utilized. In some cases, the experimental data points to an epitope “region.” Examples of this situation are the mapping of a CD4 reactivity to a 40–50 residue protein domain, or the identification of a synthetic 15-mer recognized by CD8 T cell responses. In those cases, it is understood that the epitope is contained within the region, but there is no expectation that each and every residue in the region is actually playing a role in HLA binding and/or TCR recognition. In the case of a “partial epitope”, certain peptide residues might be shown to be crucial, for example, because their mutation might abolish or hinder T cell recognition. Here it is clearly understood that those residues are part of the epitope, but there is an expectation that other neighboring residues are actually required for HLA binding and/or TCR recognition (Figure 1B).
Related to these issues, are the concept of defining “optimal” versus “minimal” epitopes. In the case of class I, because of the closed nature of the HLA class I binding groove, an exact size is usually required. While longer sizes may also be recognized, this typically requires their processing in vivo or in vitro to the “right” size, which is remarkably more potent than larger or smaller fragments [15]. Thus, in this case, the optimal size coincides with the minimal size, and it can be unequivocally defined [16]. A rather different situation is observed in the case of HLA class II, where the binding groove is open ended, and an exact size is not required. Natural endogenously bound ligands usually “overhang” by a couple of residues, both at the N- and C- termini, with “ragged ends” families of ligands usually found in ligand elution experiments [17]. Furthermore, beyond the 9-residue core sequence directly involved in class II binding, these “overhang” residues have been shown to influence HLA binding and TCR recognition [18–20]. Because of these structural constraints, an “optimal” CD4 epitope size is not usually unequivocally defined, with several alternative frames equipotent, and also the definition of a “minimal” epitope is not usually biologically relevant (Figure 1A).
3. What metadata is associated with epitope identification?
Epitope identification is a context-dependent process. To simply state that “such and such sequence is an epitope” in not very informative. Whether a given sequence is an epitope, or in other words is recognized by T cells, is highly contingent on several context-dependent variables. To name a few: what is the host, where does the epitope come from (e.g. what is the source organism), how did the immune reaction come about (e.g. natural infection, vaccination, in vitro stimulation), and how did the investigator measure and quantified the observed response. For example, it is intuitive that the fact a given HCV sequence induces cell proliferation in vitro following peptide immunization of C57BL6 mice with adjuvant, does not speak to whether the same peptide induces TNF-alpha production from T cells in PBMC derived from naturally infected humans.
The IEDB resource was created [21, 22] to host epitope data and metadata, as published in the scientific literature, or generated by several NIAID sponsored initiatives, such as the large-scale epitope identification contracts. As of the end of July 2020, it hosts data related to over 50,000 different epitopes, derived from approximately 10,000 different journal articles [23]. Each epitope must be identified by an unequivocal and well-defined molecular structure. For each epitope, a rich contextual set of metadata is also captured (Figure 2).
Figure 2. Metadata associated with epitope identification capture by the IEDB.
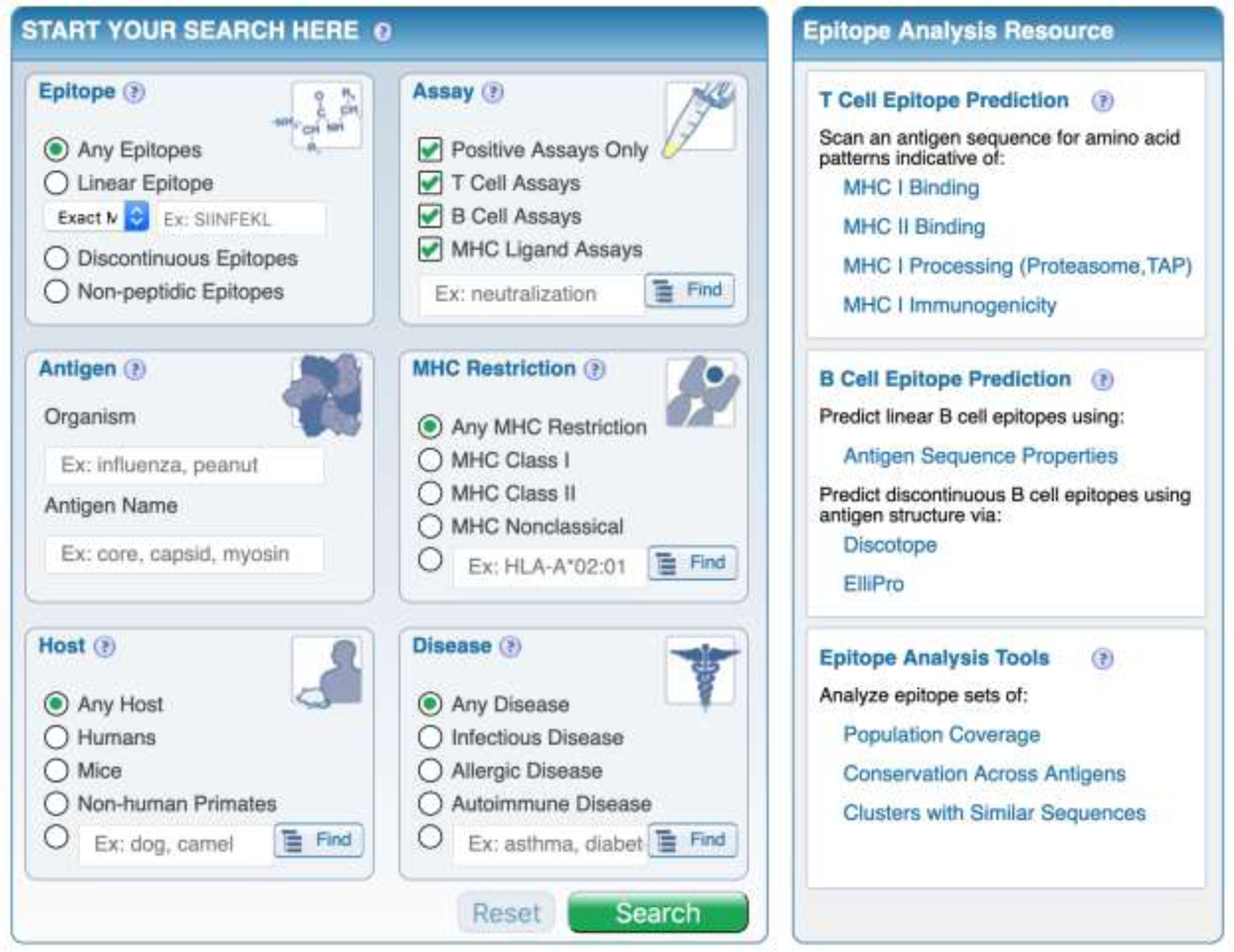
The IEDB homepage (www.iedb.org), and initial search fields are shown. The IEDB is an NIH-NIAID funded publicly available database of T and B cell epitopes curated from the published literature or by direct submission from NIH-NIAID funded large scale epitope discovery contracts. From the homepage, epitopes can be search using selected criteria, and subsequent results can be further filtered with additional criteria, to include specific assays or receptor(s).
In terms of the source of the epitope, key information is captured relating to the organism and strain (following the NCBI taxonomy nomenclature) of provenance, and the specific antigen/protein and its GeneBank accession number. In terms of the host, species information (e.g. mice, humans, NHPs or any other species for which responses are described) and associated features, such the particular mouse strains used in the investigations or, in the case of human data, ethnicity are collected. Age, gender, HLA type of the responding organism, and HLA restriction of the responding T cells are also captured [24]. In particular, the specific features of the responding T cell population are also captured, also including the specific associated TCR sequences, if known [25].
Additional key metadata captured in the IEDB related to any disease process associated with the response (e.g., were the responses associated with donors in an acute or convalescent phase of a viral infection, and so on), and the assay(s) used to detect the response. Examples of assay metadata are whether the response was induced in vivo (e.g. natural infection or exposure, vaccination) or in vitro. Whether in vitro culture or expansion was utilized, and what type of assay was used (proliferation, tetramer staining, ELISPOT and so on) are also noted. All data and metadata hosted in the IEDB are freely available and searchable by selecting and filtering according to the metadata described above [23].
4. Epitope predictions
Several different, yet complimentary approaches are routinely used to identify T cell epitopes (Figure 3). One is based on bioinformatics prediction of peptides likely to bind or to occur as natural ligands of different HLA molecules [26]. Parallel to these approaches are the actual determination of which peptides bind purified HLA in vitro [27], or the experimental determination of peptides occurring as natural HLA ligands [28–30]. All four of these approaches rely on the fact that generation of the peptide by natural processing and subsequent HLA binding are key necessary (but not sufficient) steps for T cell immunogenicity. It is generally true that all epitopes must bind HLA and be generated by processing, but not all of these peptides are immunogenic. In essence, these methodologies seek to apply a powerful filter to reduce the number of candidates to be considered, and are best used in concert with experimental assays and validations, described in more detail in the following section.
Figure 3. Epitope predictions based on HLA interactions.
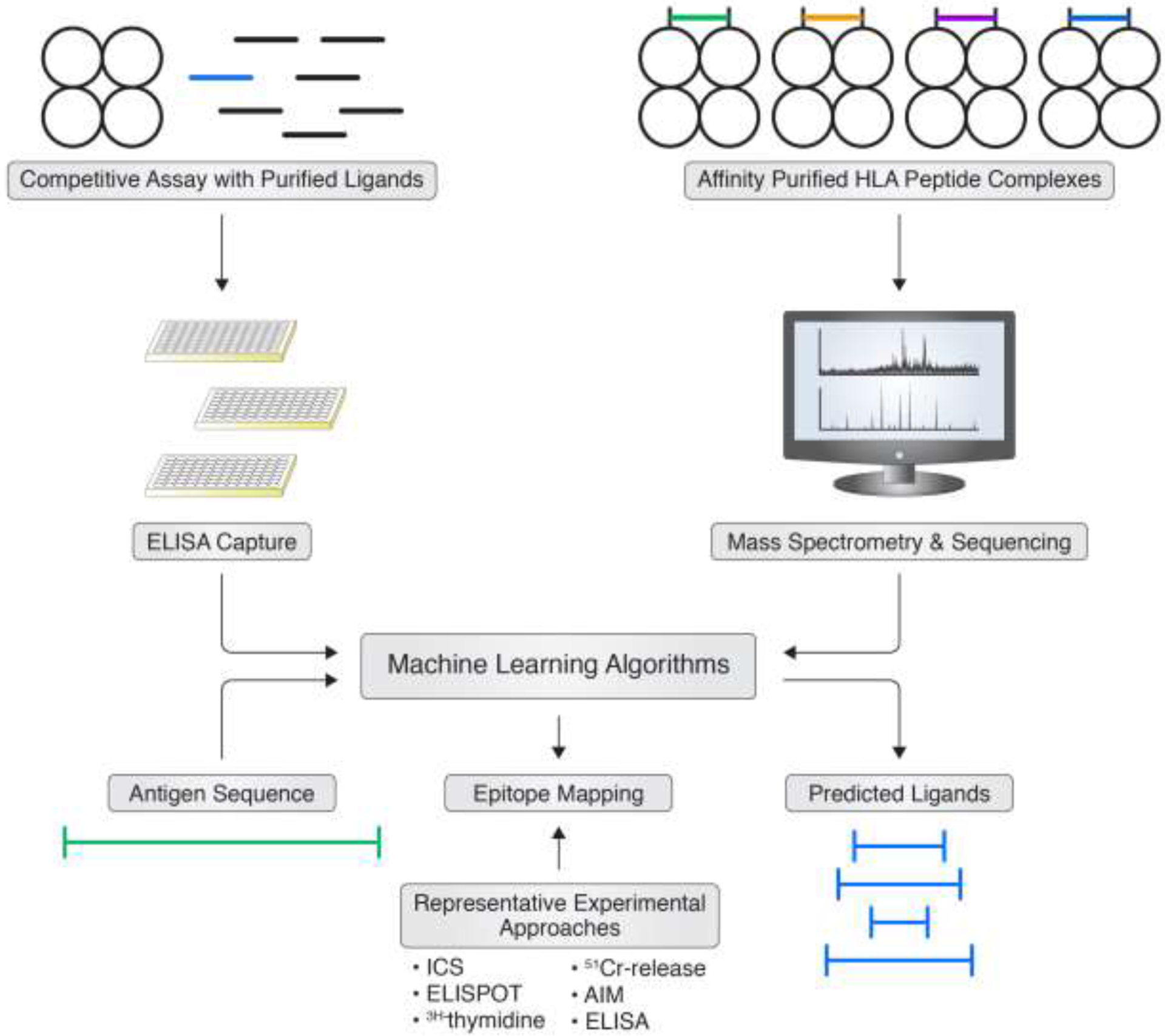
The development of bioinformatic tools to aid in identification of T cell epitopes is based on data generated from, for example, HLA-ligand assays or by mass spectrometry analysis of eluted ligands. This data is then utilized to develop machine learning tools to predict potential HLA binding peptides, and in turn candidate T cell epitopes.
In terms of HLA binding, the most widely used assay is based on a classic receptor-ligand inhibition, utilizing purified HLA molecules and synthetic peptides which, if run in the appropriate stoichiometric conditions, produces IC50 values approximating the true binding affinity (KD) [27]. Alternative assay formats have been proposed which measure dissociation rates, and provide limited increase in predictive power [31, 32]. This methodology was particularly effective in early days, serving to establish the basic parameters of HLA binding as related to T cell recognition [33, 34], including the affinity threshold associated with immunogenicity [35], and has produced a large set of experimental data on the basis of which bioinformatics predictive algorithms were trained and refined [26]. As the performance of predictions has increased over time, the usefulness of measuring binding as a pre-screening assay has decreased, as the cost and labor involved in measuring a large number of affinities is easily offset by the less stringent, but more cost efficient, use of sets of predicted peptides in T cell assays [36].
In terms of analyzing the actual pathogen derived natural ligands, the majority of the data available to date relate to general sets of eluted ligands, mostly of self-origin. These data sets, often referred as “immunopeptidomes” [28–30], have recently outnumbered in terms of data points what is available from measured binding data, and have proven invaluable for training bioinformatics-based predictive algorithms [37, 38]. However, relatively few studies [36, 39, 40] have thus far been able to define actual pathogen derived epitopes, as the majority of the natural ligands are of self-origin and the few pathogen-derived epitopes are relatively difficult to detect [28].
There are many different tools to predict HLA I- and II-restricted T-cell epitopes available online. While the current review is focused on the IEDB analysis resource, a more in-depth analysis of the different algorithms, and a review of their strengths and limitations, has been presented elsewhere [26]. The IEDB analysis resource hosts a large panel of different predictive tools [41]. In terms of bioinformatics predictive algorithms, in general class I predictions are more accurate than their class II counterparts, and methods trained on eluted ligands perform slightly better than the ones trained on binding data [26]. Interestingly, the best performance is obtained when both binding and eluted ligand data are used to train the algorithms [26]. Unbiased systematic benchmarking studies have started to shed an objective light on the matter of the performance of different methodologies [36], showing that predictive methods are remarkably accurate, essentially predicting all epitopes (high true positive rates and low false negative rates), while also showing that not all predicted binders or eluted ligands are recognized by T cell responses (relatively high false positive rates), reflecting that the basic biological fact that HLA binding is a necessary but not sufficient requisite for T cell immunogenicity. An additional methodology that was recently described [42] involves directly using epitope data to train predictive networks to predict epitopes (as opposed to binders or eluted ligands). While conceptually interesting, this method has not yielded substantial improvements over the other methodologies.
5. Immunodominance in T cell immunogenicity
To further discuss epitope identification, it is convenient to introduce some additional terminology. We define as immunodominant (Figure 4A) those epitopes that are recognized within a particular HLA haplotype, and a given protein sequence, with an immune response of relatively greater magnitude than other epitope-specific responses. According to this definition, it is intuitive that high HLA binding and/or efficient processing can contribute to immunodominance [43, 44]. The peptide sequence is determinative for proper binding to specific HLA molecules, and it is equally important to assess whether a particular epitope will be processed from an intact protein. However, the performance of tools that predict processing has been disappointing, in general, as discussed in detail elsewhere [26]. The general current trend is to incorporate the influence of processing by utilizing eluted ligands (peptides that have been generated by processing) in the training of predictive algorithms. Another variable that can contribute to immunodominance is the availability of a repertoire of TCR capable of recognizing the particular epitope/HLA combination [45, 46]. Several groups are interested in systematically studying, and one day predicting, which TCR correspond to particular HLA/epitope combinations [47, 48]. This is a very promising area, but at this point in time is not yet ready for implementation for high throughput epitope identification in large donor cohorts.
Figure 4. Immunodominance and immunoprevalence.
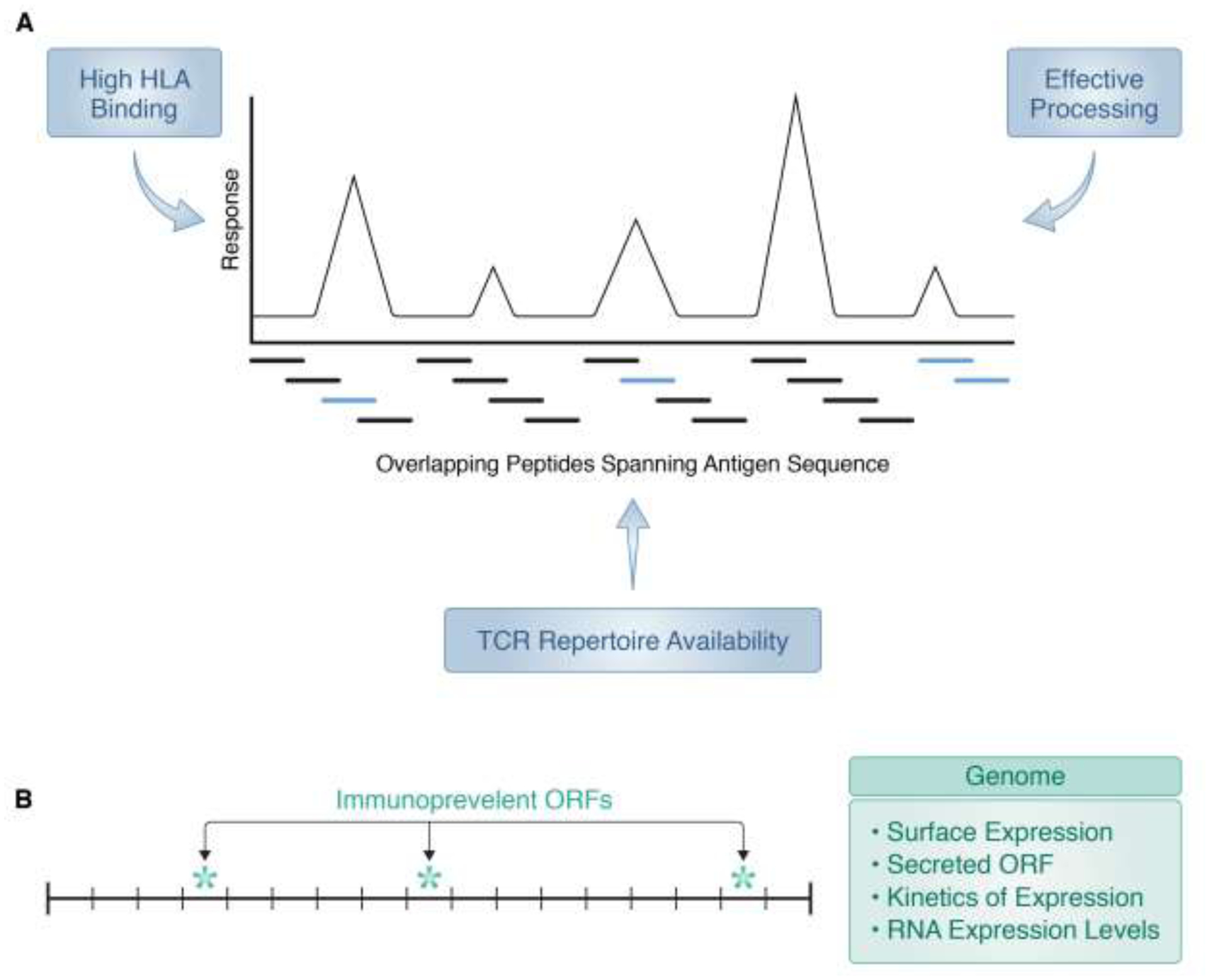
A) Epitopes effectively generated by processing and for which a TCR repertoire is available (light blue bars), and capable of eliciting the (relatively) strongest T cell responses, are termed immunodominant. B) Epitopes, antigens or ORFs that elicit responses with high frequency in an out-bred population are termed immunoprevalent. Typically, only a few epitopes/ORFS/antigens are found to be immunoprevalent, and may be associated with high levels of surface or RNA expression or secretion (class II); kinetics (time) of expression is often associated with dominance of class I or class II responses.
Perhaps the most important variable influencing immunodominance in terms of shaping the TCR repertoire is the relationship of the epitope sequence to other sequences related to the potential epitope. This is because the TCR repertoire is shaped by both positive and negative selection [49], and by constant tuning and expansion based on the actual sequence space encountered by an individual during their lifetime. This influence can be either positive or negative. For example, TCRs recognizing sequences that are self or highly homologous to self might be eliminated or silenced by central [50] or peripheral tolerance [51]. Conversely, cross-reactive TCRs recognizing sequences homologous to other antigens might be preferentially expanded and thus become relatively immunodominant because of repeated exposure to the homologous antigens. Examples include pollen allergen epitopes conserved across different grass species [52, 53], mycobacteria epitopes conserved across multiple bacteria [54–56], Dengue and Zika virus epitopes conserved across different serotypes [57–60], epitopes conserved in multiple herpesvirus and pox species [61–63], influenza [64, 65] and epitopes conserved across multiple coronaviruses [66]. Sequence conservation across the individual microbiome has also been shown to influence immunodominance [67, 68].
6. Immunoprevalence and T cell responsiveness at the antigen level
The previous section reviewed some of the variables that determine, within a given antigen, which epitopes would be recognized as more dominant. Additional variables dictate, when the genome of a pathogen or allergen is considered, which particular antigens would be recognized most prominently. In this context, we also consider the concept of immunoprevalence, (Figure 4B) defined on the basis of how frequently responses to a given epitope or antigen are detected when an out-bread population, such as a human patient cohort, is considered. This is complementary to the concept of immunodominance, defined on the basis of magnitude of responses [69, 70].
At the level of antibody responses, it is routinely observed that certain antigens are recognized with both dominance and prevalence. This is usually directly correlated to protein abundance, expression on the microbe surface, and antigen/epitope surface accessibility [71]. But what determines dominance and prevalence at the level of T cell responses? It is possible different antigens might be recognized as dominant in different individuals, simply because they contain peptide sequences that bind differentially to the specific HLA types expressed by the different individuals. However, for example, in the case of pox viruses it was observed that certain ORFs are immunoprevalent, e.g. recognized in multiple individuals with different HLA types, thus ruling out that prevalent recognition was reflective of HLA type [70]. Further inspection of the data revealed that the targets more prevalently recognized for CD4 responses corresponded to viral products produced abundantly and late in the infection, possibly reflecting uptake and processing of virions. Conversely, targets of CD8 responses tended to map to ORFs that were expressed early in the infection [72], potentially reflecting endogenous generation and presentation before host viral synthesis shut down.
To be able to predict which ORFs might be preferentially recognized is of limited value in the case of viruses with a genome of limited size where all ORFs can be addressed without preselection, but could be of large impact by restricting the potential targets in the case of larger viruses (such as, for example, pox and herpes viruses), bacteria and other pathogens with a complex genome (e. g. plasmodium). Direct determination of mRNA expression levels [73] in combination with HLA binding predictions is a promising approach, that has not been extensively utilized thus far. Gene annotation and expression profiles derived for each target or tools predicting the surface proteome [74–76] or secretome [77] might be of use.
Alternative methods include the experimental determination of surface expressed proteins by proteolysis, and mass spectrometry approaches which have been of value in determining targets of protective antibody immunity in several bacteria [71]. We have utilized an immunoproteomic approach to determine the targets of antibody recognition in complex allergens and shown that this analysis is useful for T cell epitope identification, because of the association between the targets of humoral responses and CD4 T cell immunity [78–80]. This association, however, does not appear to necessarily apply to infectious agents. For example, in the case of Mycobacterium tuberculosis, the targets of antibody and CD4 T cell responses are not correlated [81]. This might reflect that Mycobacterium tuberculosis is an intracellular pathogen and, therefore, recognition of antibody accessible antigens might not be relevant for T cell immunodominance. An additional potential experimental approach involves the elution of peptides bound to a limited set class I or class II HLAs, as a preliminary probe to experimentally determine which ORFs are more represented and therefore most likely to be recognized by CD4 and CD8 responses. This approach has been shown to greatly restrict the number of ORFS to be considered in the search for cancer or autoimmune epitopes [82].
7. HLA polymorphism and epitope recognition
At the genetic level, the HLA is a multi-locus gene complex. Two main loci encode for HLA A and B class I molecules, and additional loci encode for different class II molecules, to include DR (DRB1 and DRB3/4/5), DP and DQ molecules [83]. Thus, each chromosome encodes a total of two main HLA class I and four main HLA class II molecules. Each of the corresponding loci are extremely polymorphic, with several thousand different alleles expressed at different frequencies in different ethnicities [84, 85]. Thus, because of heterozygosity, each individual may express up to four different HLA class I and eight different HLA class II allelic variant molecules (Figure 5).
Figure 5. HLA polymorphism and polygeny.
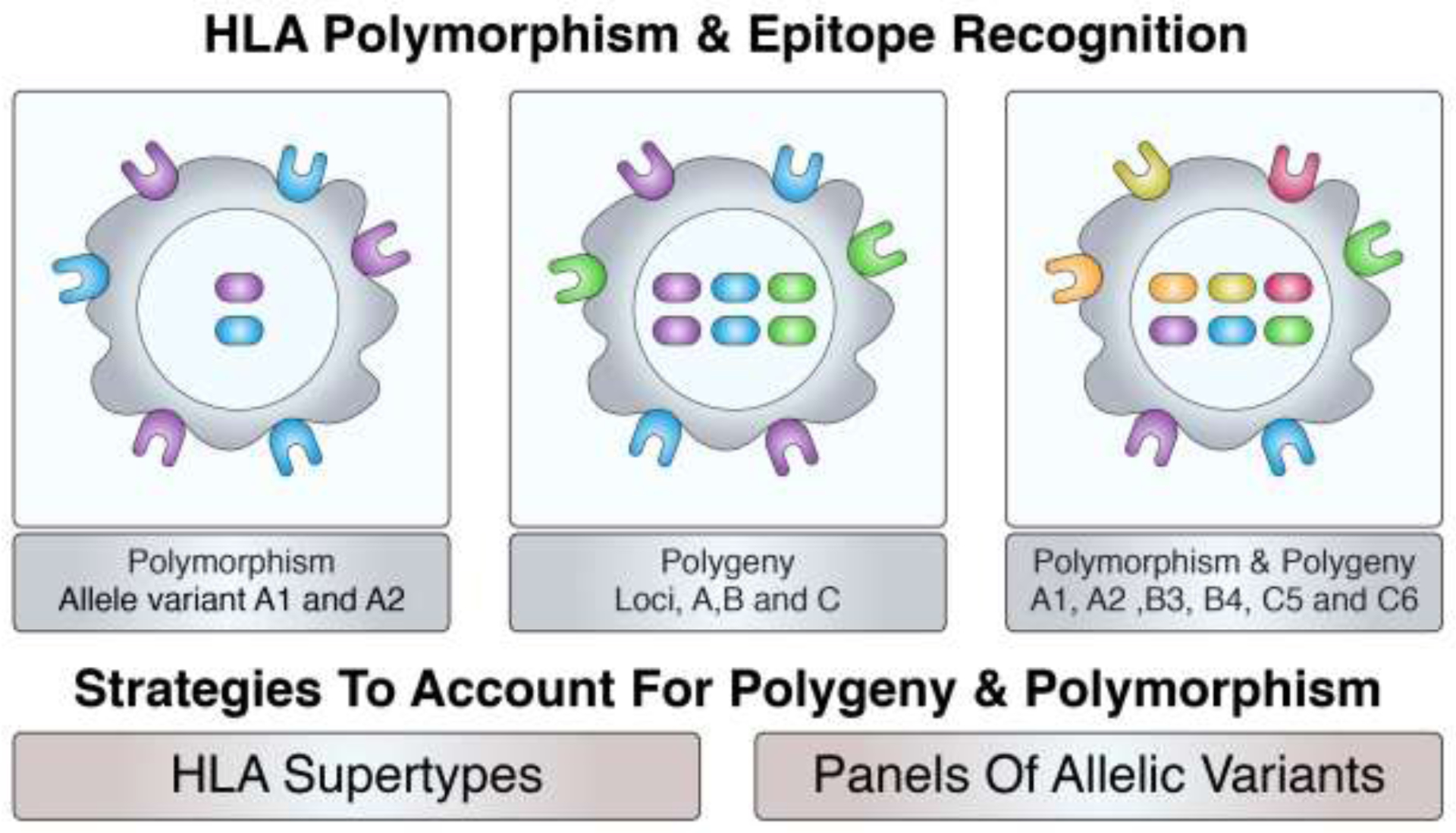
The HLA region is highly polymorphic, with thousands of different allelic variants expressed at each locus. With heterozygosity, each individual will express up to two different alleles at each locus. Further, with class I and II presentation across several loci (polygeny), up to six different class I (2 HLA-A, 2 HLA-B and 2 HLA-C) and eight different class II (2 HLA-DRB1, 2 HLA-DRB3/4/5, 2 DQ and 2 DP) molecules may be expressed by an individual.
We developed targeted approaches to deal with this apparently daunting degree of multi-genicity and genetic polymorphism. Specifically, we have shown that the large majority of different molecules and allelic variants can be classified in relatively few “supertype” specificities [86–99], which are characterized by similar binding specificity (motifs) and, most importantly, largely overlapping peptide binding repertoires. Since several of the HLA allelic variant molecules are relatively more frequent, we have further shown that focusing on approximately 30–40 HLA class I [87, 88, 92, 100, 101] and 20–30 HLA class II molecules [86, 97, 102, 103] allows to cover the vast majority of HLA molecules expressed in different populations, irrespective of ethnicity.
Human T cells recognize epitopes bound to specific HLA molecules. The specific HLA molecule that binds and presents the epitope is called the epitope’s HLA restriction. Determining HLA restriction is an important component of epitope identification studies, since knowledge of HLA restriction is, for example, necessary to produce multimeric staining reagents [104–107], or to project which fraction of given population can respond to a given epitope or set of epitopes (population coverage) [101, 103, 108, 109]. Certain epitopes can bind and be recognized in the context of multiple HLA, and are termed “promiscuous epitopes” [110–115]. It has been shown that promiscuous epitopes also tend to be immunodominant/immunoprevalent [86, 116–119], accounting for a large fraction of responses to a given antigen.
8. HLA restriction of epitope responses
A variety of different approaches are used to determine or infer HLA restriction. Indeed, the IEDB database lists for each scientific study, in the form of “evidence codes,” which methodology is used to assign HLA restriction [120].
The classic approach originally developed to address this question involved the use of antigen presentation assays, utilizing loci or allele specific antibodies to inhibit presentation [121, 122] or using panels of matched/mismatched Antigen Presenting Cells (APC) [102, 117–119, 123–125]. These methodologies are laborious, and suffer from specific drawbacks. Loci/allele specific antibodies are not always available, and the assays require careful titration, as too little antibody will not inhibit, and high doses will kill APCs, or cause non-specific inhibition. Panels of matched/mismatched APCs are also not easy to assemble, especially at the population level, and because of linkage disequilibrium of alleles expressed at different loci. Furthermore, their use often does not allow to determine HLA restriction of promiscuous epitopes.
In recent years, HLA restriction is often inferred based on the presence of specific HLA binding motifs, or on predicted binding affinity. This knowledge is often combined with the knowledge of the HLA expressed in a given donor. For example, in one situation HLA-A*02:01 positive donors are tested for recognition of HLA-A*02:01 binding peptides (either predicted or experimentally verified), and positive responses are assumed to be HLA-A*02:01 restricted. In another experimental setting, a CD8 response was observed in a donor heterozygous for HLA-A*02:01, B*07:02, A*11:03 and B*27:03. Upon further experiments the peptide is mapped to a specific nine-mer predicted to bind B*27:03, but not A*02:01, B*07:02 and A*11:03. Based on these results, the HLA restriction is inferred to be B*27:03. In a further variation, HLA restriction can be inferred based on response frequencies. For example, it might be noted that a given peptide is recognized in 60% of the individuals expressing HLA DRB1*04:01, but only in 5% of individuals that do not express that particular allele. The RATE method [126, 127] automates these types of calculations. Taken together, inference of HLA restriction is effective, but also suffers from several drawbacks, particularly in the case of peptides binding multiple HLAs, individuals expressing HLAs with overlapping binding capacity, or inferences related to HLA loci in strong linkage disequilibrium.
Two additional methodologies are available to experimentally determine restrictions. Multimer staining and use of single HLA transfected cell lines. In the case of multimer staining, a reagent is prepared with a specific epitope /HLA pair, and used to bind the TCR of epitope specific T cells [104, 105, 107, 128, 129]. This is remarkably effective, but does often present a “chicken and egg” situation, in that knowledge (or a reasonable guess) of the HLA restriction is required to prepare the reagent in the first place. This strategy is therefore particularly effective when an educated guess or inference of HLA restriction is available. An alternative methodology is the use of single HLA transfected APCs [102, 130–133]. In these experiments, the peptide is tested in antigen presentation assays utilizing panels of cell lines expressing single HLA molecules that match an allele expressed in the donor from which the T cell response is generated. These assays are laborious and require availability of matching cell lines, but represent to date the most unequivocal methodology to establish HLA restriction.
9. Experimental approaches for actual epitope mapping
A variety of different approaches are used for epitope identification and mapping. The two most popular approaches involve testing overlapping peptides spanning the entire sequence of a given antigen, and the use of predicted epitopes. The relative methods of the two approaches have been compared in murine systems [134] and found to be associated with similar efficiency, even though the sets of epitopes identified by the two methods were not completely overlapping (some epitopes were identified by predictions, but were not identified by the overlapping peptides approach and vice versa). Testing overlapping peptide sets is comprehensive, when compared to predictive methods that are by definition associated with variable levels of accuracy. The drawback is that overlapping peptides require synthesis and testing of a larger number of peptides, which in turn requires availability of larger cell numbers, and some epitopes are missed because of fact that the 15-mer is a suboptimal ligand. Conversely, prediction approaches might miss unusual “non-canonical” motifs and ligand sizes, even though algorithm efficacy is constantly improving. In the case of both predicted or overlapping peptides, it is common practice to test peptide pools that are subsequently deconvoluted to identify the actual epitope recognized.
Regardless of the particular set of peptides tested, different assay and read-out strategies are available to investigators. Often, the cell numbers available to be tested is limiting. An in vitro expansion step is a possible avenue to increase signal by expanding rare T cells [117, 119, 135, 136], but this comes at the expense of a more laborious cost and time intensive set up, and alterations in the phenotype of responding T cells. For these reasons, if practically feasible, a direct ex vivo assay strategy is usually preferred.
Whether an in vitro re-stimulation step or a direct ex vivo assay is used, different final readout options need to be considered, each associated with different degrees of robustness, information content, and costs in terms of effort and resources. Popular assay options include cytokine readouts, such as Enzyme-Linked Immunospot (ELISPOT) [137] and Intracellular Cytokine Staining (ICS) [138]. The ELISPOT is a popular robust assay, but the ICS assay offers the potential advantage to simultaneously perform additional phenotypic determinations, such as determining whether the responding T cells are CD4 or CD8, and whether other common cell markers are expressed. Altogether these assays provide information regarding the functionality of the responding T cells in terms of cytokine responses. Conversely, these assays are by definition only detecting the pre-selected cytokines and are “blind” to additional cytokines.
An alternative assay that has become increasingly popular is the Activation Induced Marker (AIM) assay [139, 140]. This is an agnostic assay in that it detects all activated T cells, and is not focused on a particular cytokine. In particular, this assay is suited to detect T cell subsets that secrete small amounts of cytokines, such as Tfh cells [141–144]. Tetramer staining is also not dependent on any functional activity of the antigen specific T cells, and allows to detect specific T cells in absence of antigen stimulation. However, this methodology is not ideally suited to epitope screening, and more suited for in-depth characterization of epitope specific T cells once a given epitope is identified.
10. Epitope identification and population coverage
Particular attention needs to be devoted to how different approaches achieve and address population coverage. One approach relies on testing a given population/patient group and defining the epitopes recognized. This is an unbiased approach, but suffers the drawback of yielding coverage skewed toward the specific HLA most represented in the test population, raising questions in terms of whether the results will be applicable to other populations with potentially different HLA representation.
The alternative strategy is to define epitopes restricted by specific HLA alleles, thus allowing to project coverage in different populations, and ensuring adequate coverage in different ethnicities if the most common alleles [86, 88] expressed in populations worldwide are selected. A variation along the same lines is the algorithm described by Paul et al. [145], which combined different prototype HLA class II specificities to predict the most dominant promiscuous class II epitopes recognized regardless of ethnicity. Achieving good population coverage is perhaps most challenging in the case of HLA multimeric reagents, because a different reagent is required for each epitope/HLA pair. A quantitative analysis in a Mycobacterium tuberculosis field study [132] revealed that to cover approximately 50% of the total response would require synthesis and validation of several hundred different tetramers.
11. Examples of practical application of epitope identification studies
In this section, we provide a few selected examples of epitope identification studies that led to novel immunological or microbiological insights. A first example is provided by the study of Lindestam Arlehamn et al. [81] that provided the first truly unbiased genomic map of human CD4 T cell responses to Mycobacterium tuberculosis. This study was enabled by the combined use of prediction of promiscuous epitopes and an ELISPOT screen of a large library of over 20,000 different peptides. The results revealed that the CD4 T cell response was generally focused on a few genomic islands associated with Type IV secretion. But, the study also mapped a large number of previously undiscovered antigens and epitopes, highlighting the value of a systematic and unbiased approach.
The study by Lindestam Arlehamn et al. also defined hundreds of different epitopes, and subsequent work showed that these epitopes could be arranged in a “megapool” which could in turn be utilized as a reagent to comprehensively quantitate antigens specific responses [132, 146]. The epitopes defined in those studies were further utilized to map relationships with Mycobacterium tuberculosis recognition to non-tubercoloid-complex Mycobacteria [56], and map the evolution of responses as a function of response to therapy and microbiome similarity [68]. Similar genome-wide analyses are underway for Bordetella pertussis [147], another bacterium of high significance for human health. The megapool approach has also been validated for a variety of different targets, including, amongst others, tetanus toxoid [148], Zika virus [57], and various allergens [78, 149, 150]. In the case of Dengue virus, CD4 and CD8 megapools have been produced and utilized to quantitate responses in endemic areas and in response to vaccination [58, 151, 152].
Megapools have also played a key role in extracting and isolating antigen specific T cells for a variety of different indications, and establishing specific immune signatures [153–162]. The purpose of these studies is to identify specific correlates of immune protection versus immunopathology. Epitope identification studies have played a key role in defining the extent to which responses against antigens from different flaviviruses are cross-reactive with each other, which is of relevance for interpreting results from Dengue virus vaccines that utilize other flaviviruses as a delivery backbone [163]. Similarly, in recent studies, the megapool and epitope identification approaches outlined here have played a key role in characterizing T cell responses in SARS-CoV-2 exposed and unexposed subjects alike [66, 164, 165].
12. Conclusions
Identification of T cell epitopes is a crucial step in establishing a capacity to understand the immune response to pathogens and allergens. Various bioinformatic approaches have been developed over the years that have afforded significant advances in the epitope identification process. In addition, various experimental approaches using defined epitopes, particularly with an understanding of the impact of HLA polymorphism, immunodominance and immunoprevalence, have provided important insights into disease mechanisms and correlates of immunity.
Acknowledgements
For very helpful discussions, we would like to thank Shane Crotty, Daniela Weiskopf, Cecilia Lindestam Arlehamn, Alba Grifoni, and Ricardo Da Silva Antunes. We also thank Christina Corbaci for generating the illustrations. This work was supported by funding provided by the National Institutes of Health, National Institutes for Allergy and Infectious Diseases contracts 75N93019C00001, HHSN272201400045C and 75N9301900065 and grant U19 AI135731.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations utilized: Major histocompatibility complex (MHC); Human leukocyte antigen (HLA); T cell receptor (TCR); Peripheral blood mononuclear cell (PBMC); Immune Epitope Database and Analysis Resource (IEDB); Intracellular Cytokine Staining (ICS); Mucosal-associated invariant T cells (MAIT); Enzyme-Linked Immunospot (ELISPOT); Activation Induced Marker (AIM); Antigen presenting cell (APC); Open reading frame (ORF); 50% inhibitory capacity (IC50).
Disclosure
Declarations of interest: none.
References
- [1].Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, Nemazee D, Ponomarenko JV, Sathiamurthy M, Schoenberger SP, Stewart S, Surko P, Way S, Wilson S, Sette A, The design and implementation of the immune epitope database and analysis resource, Immunogenetics 57(5) (2005) 326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shiromizu CM, Jancic CC, gammadelta T Lymphocytes: An Effector Cell in Autoimmunity and Infection, Frontiers in immunology 9 (2018) 2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Karamooz E, Harriff MJ, Lewinsohn DM, MR1-dependent antigen presentation, Semin Cell Dev Biol 84 (2018) 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Terabe M, Berzofsky JA, Tissue-Specific Roles of NKT Cells in Tumor Immunity, Frontiers in immunology 9 (2018) 1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lepore M, Mori L, De Libero G, The Conventional Nature of Non-MHC-Restricted T Cells, Frontiers in immunology 9 (2018) 1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hinks TSC, Zhang XW, MAIT Cell Activation and Functions, Frontiers in immunology 11 (2020) 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Alfonso C, Karlsson L, Nonclassical MHC class II molecules, Annual review of immunology 18 (2000) 113–42. [DOI] [PubMed] [Google Scholar]
- [8].Braud VM, Allan DS, McMichael AJ, Functions of nonclassical MHC and non-MHC-encoded class I molecules, Current opinion in immunology 11(1) (1999) 100–8. [DOI] [PubMed] [Google Scholar]
- [9].Blum JS, Wearsch PA, Cresswell P, Pathways of antigen processing, Annual review of immunology 31 (2013) 443–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ishioka GY, Lamont AG, Thomson D, Bulbow N, Gaeta FC, Sette A, Grey HM, Major histocompatibility complex class II association and induction of T cell responses by carbohydrates and glycopeptides, Springer Semin Immunopathol 15(2–3) (1993) 293–302. [DOI] [PubMed] [Google Scholar]
- [11].Avci FY, Li X, Tsuji M, Kasper DL, A mechanism for glycoconjugate vaccine activation of the adaptive immune system and its implications for vaccine design, Nat Med 17(12) (2011) 1602–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sun L, Paschall AV, Middleton DR, Ishihara M, Ozdilek A, Wantuch PL, Aceil J, Duke JA, LaBranche CC, Tiemeyer M, Avci FY, Glycopeptide epitope facilitates HIV-1 envelope specific humoral immune responses by eliciting T cell help, Nat Commun 11(1) (2020) 2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pamer E, Cresswell P, Mechanisms of MHC class I--restricted antigen processing, Annu Rev Immunol 16 (1998) 323–58. [DOI] [PubMed] [Google Scholar]
- [14].Rudolph MG, Stanfield RL, Wilson IA, How TCRs bind MHCs, peptides, and coreceptors, Annual review of immunology 24 (2006) 419–66. [DOI] [PubMed] [Google Scholar]
- [15].Rotzschke O, Falk K, Stevanovic S, Jung G, Walden P, Rammensee HG, Exact prediction of a natural T cell epitope, Eur J Immunol 21(11) (1991) 2891–4. [DOI] [PubMed] [Google Scholar]
- [16].Bertoletti A, Chisari FV, Penna A, Guilhot S, Galati L, Missale G, Fowler P, Schlicht HJ, Vitiello A, Chesnut RC, et al. , Definition of a minimal optimal cytotoxic T-cell epitope within the hepatitis B virus nucleocapsid protein, J Virol 67(4) (1993) 2376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hunt DF, Michel H, Dickinson TA, Shabanowitz J, Cox AL, Sakaguchi K, Appella E, Grey HM, Sette A, Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad, Science 256(5065) (1992) 1817–20. [DOI] [PubMed] [Google Scholar]
- [18].Lamont AG, Powell MF, Colon SM, Miles C, Grey HM, Sette A, The use of peptide analogs with improved stability and MHC binding capacity to inhibit antigen presentation in vitro and in vivo, J Immunol 144(7) (1990) 2493–8. [PubMed] [Google Scholar]
- [19].Madden DR, The three-dimensional structure of peptide-MHC complexes, Annu Rev Immunol 13 (1995) 587–622. [DOI] [PubMed] [Google Scholar]
- [20].Carson RT, Vignali KM, Woodland DL, Vignali DA, T cell receptor recognition of MHC class II-bound peptide flanking residues enhances immunogenicity and results in altered TCR V region usage, Immunity 7(3) (1997) 387–99. [DOI] [PubMed] [Google Scholar]
- [21].Martini S, Nielsen M, Peters B, Sette A, The Immune Epitope Database and Analysis Resource Program 2003–2018: reflections and outlook, Immunogenetics 72(1–2) (2020) 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Salimi N, Fleri W, Peters B, Sette A, The immune epitope database: a historical retrospective of the first decade, Immunology 137(2) (2012) 117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, Peters B, The Immune Epitope Database (IEDB): 2018 update, Nucleic Acids Res 47(D1) (2019) D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vita R, Overton JA, Sette A, Peters B, Better living through ontologies at the Immune Epitope Database, Database (Oxford) 2017(1) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mahajan S, Vita R, Shackelford D, Lane J, Schulten V, Zarebski L, Jespersen MC, Marcatili P, Nielsen M, Sette A, Peters B, Epitope Specific Antibodies and T Cell Receptors in the Immune Epitope Database, Frontiers in immunology 9 (2018) 2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Peters B, Nielsen M, Sette A, T Cell Epitope Predictions, Annu Rev Immunol 38 (2020) 123–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sidney J, Southwood S, Moore C, Oseroff C, Pinilla C, Grey HM, Sette A, Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture, Curr Protoc Immunol Chapter 18 (2013) Unit 18 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Paul S, Grifoni A, Peters B, Sette A, Major Histocompatibility Complex Binding, Eluted Ligands, and Immunogenicity: Benchmark Testing and Predictions, Frontiers in immunology 10 (2019) 3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gfeller D, Bassani-Sternberg M, Predicting Antigen Presentation-What Could We Learn From a Million Peptides?, Frontiers in immunology 9 (2018) 1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Freudenmann LK, Marcu A, Stevanovic S, Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry, Immunology 154(3) (2018) 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Harndahl M, Rasmussen M, Roder G, Dalgaard Pedersen I, Sorensen M, Nielsen M, Buus S, Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity, European journal of immunology 42(6) (2012) 1405–16. [DOI] [PubMed] [Google Scholar]
- [32].Harndahl M, Rasmussen M, Roder G, Buus S, Real-time, high-throughput measurements of peptide-MHC-I dissociation using a scintillation proximity assay, J Immunol Methods 374(1–2) (2011) 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sette A, Buus S, Colon S, Smith JA, Miles C, Grey HM, Structural characteristics of an antigen required for its interaction with Ia and recognition by T cells, Nature 328(6129) (1987) 395–9. [DOI] [PubMed] [Google Scholar]
- [34].Buus S, Sette A, Colon SM, Miles C, Grey HM, The relation between major histocompatibility complex (MHC) restriction and the capacity of Ia to bind immunogenic peptides, Science 235(4794) (1987) 1353–8. [DOI] [PubMed] [Google Scholar]
- [35].Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast WM, Melief CJ, Oseroff C, Yuan L, Ruppert J, Sidney J, del Guercio MF, Southwood S, Kubo RT, Chesnut RW, Grey HM, Chisari FV, The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes, J Immunol 153(12) (1994) 5586–92. [PubMed] [Google Scholar]
- [36].Paul S, Croft NP, Purcell AW, Tscharke DC, Sette A, Nielsen M, Peters B, Benchmarking predictions of MHC class I restricted T cell epitopes in a comprehensively studied model system, PLoS Comput Biol 16(5) (2020) e1007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].O’Donnell TJ, Rubinsteyn A, Laserson U, MHCflurry 2.0: Improved Pan-Allele Prediction of MHC Class I-Presented Peptides by Incorporating Antigen Processing, Cell Syst 11(1) (2020) 42–48 e7. [DOI] [PubMed] [Google Scholar]
- [38].Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M, NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data, Nucleic Acids Res 48(W1) (2020) W449–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wu T, Guan J, Handel A, Tscharke DC, Sidney J, Sette A, Wakim LM, Sng XYX, Thomas PG, Croft NP, Purcell AW, La Gruta NL, Quantification of epitope abundance reveals the effect of direct and cross-presentation on influenza CTL responses, Nat Commun 10(1) (2019) 2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Croft NP, Smith SA, Pickering J, Sidney J, Peters B, Faridi P, Witney MJ, Sebastian P, Flesch IEA, Heading SL, Sette A, La Gruta NL, Purcell AW, Tscharke DC, Most viral peptides displayed by class I MHC on infected cells are immunogenic, Proc Natl Acad Sci U S A 116(8) (2019) 3112–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dhanda SK, Mahajan S, Paul S, Yan Z, Kim H, Jespersen MC, Jurtz V, Andreatta M, Greenbaum JA, Marcatili P, Sette A, Nielsen M, Peters B, IEDB-AR: immune epitope database-analysis resource in 2019, Nucleic acids research 47(W1) (2019) W502–W506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dhanda SK, Karosiene E, Edwards L, Grifoni A, Paul S, Andreatta M, Weiskopf D, Sidney J, Nielsen M, Peters B, Sette A, Predicting HLA CD4 Immunogenicity in Human Populations, Frontiers in immunology 9 (2018) 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yewdell JW, Bennink JR, Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses, Annu Rev Immunol 17 (1999) 51–88. [DOI] [PubMed] [Google Scholar]
- [44].Yewdell JW, Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses, Immunity 25(4) (2006) 533–43. [DOI] [PubMed] [Google Scholar]
- [45].Schaeffer EB, Sette A, Johnson DL, Bekoff MC, Smith JA, Grey HM, Buus S, Relative contribution of “determinant selection” and “holes in the T-cell repertoire” to T-cell responses, Proc Natl Acad Sci U S A 86(12) (1989) 4649–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui HH, Frahm N, Brander C, Peters B, Grey H, Sette A, A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection, J Immunol 178(12) (2007) 7890–901. [DOI] [PubMed] [Google Scholar]
- [47].Huang H, Wang C, Rubelt F, Scriba TJ, Davis MM, Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening, Nat Biotechnol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, Haas N, Arlehamn CSL, Sette A, Boyd SD, Scriba TJ, Martinez OM, Davis MM, Identifying specificity groups in the T cell receptor repertoire, Nature 547(7661) (2017) 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Klein L, Kyewski B, Allen PM, Hogquist KA, Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see), Nature reviews. Immunology 14(6) (2014) 377–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Klein L, Robey EA, Hsieh CS, Central CD4(+) T cell tolerance: deletion versus regulatory T cell differentiation, Nature reviews. Immunology 19(1) (2019) 7–18. [DOI] [PubMed] [Google Scholar]
- [51].Paterson AM, Sharpe AH, Taming tissue-specific T cells: CTLA-4 reins in self-reactive T cells, Nat Immunol 11(2) (2010) 109–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pham J, Oseroff C, Hinz D, Sidney J, Paul S, Greenbaum J, Vita R, Phillips E, Mallal S, Peters B, Sette A, Sequence conservation predicts T cell reactivity against ragweed allergens, Clin Exp Allergy 46(9) (2016) 1194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Westernberg L, Schulten V, Greenbaum JA, Natali S, Tripple V, McKinney DM, Frazier A, Hofer H, Wallner M, Sallusto F, Sette A, Peters B, T-cell epitope conservation across allergen species is a major determinant of immunogenicity, J Allergy Clin Immunol 138(2) (2016) 571–578 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Scriba TJ, Carpenter C, Pro SC, Sidney J, Musvosvi M, Rozot V, Seumois G, Rosales SL, Vijayanand P, Goletti D, Makgotlho E, Hanekom W, Hatherill M, Peters B, Sette A, Arlehamn CSL, Differential Recognition of Mycobacterium tuberculosis-Specific Epitopes as a Function of Tuberculosis Disease History, Am J Respir Crit Care Med 196(6) (2017) 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Shah JA, Lindestam Arlehamn CS, Horne DJ, Sette A, Hawn TR, Nontuberculous Mycobacteria and Heterologous Immunity to Tuberculosis, J Infect Dis 220(7) (2019) 1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lindestam Arlehamn CS, Paul S, Mele F, Huang C, Greenbaum JA, Vita R, Sidney J, Peters B, Sallusto F, Sette A, Immunological consequences of intragenus conservation of Mycobacterium tuberculosis T-cell epitopes, Proc Natl Acad Sci U S A 112(2) (2015) E147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Grifoni A, Pham J, Sidney J, O’Rourke PH, Paul S, Peters B, Martini SR, de Silva AD, Ricciardi MJ, Magnani DM, Silveira CGT, Maestri A, Costa PR, de-Oliveira-Pinto LM, de Azeredo EL, Damasco PV, Phillips E, Mallal S, de Silva AM, Collins M, Durbin A, Diehl SA, Cerpas C, Balmaseda A, Kuan G, Coloma J, Harris E, Crowe JE Jr., Stone M, Norris PJ, Busch M, Vivanco-Cid H, Cox J, Graham BS, Ledgerwood JE, Turtle L, Solomon T, Kallas EG, Watkins DI, Weiskopf D, Sette A, Prior Dengue Virus Exposure Shapes T Cell Immunity to Zika Virus in Humans, J Virol 91(24) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Weiskopf D, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, de Silva AD, Lindow JC, Diehl SA, Whitehead S, Durbin A, Kirkpatrick B, Sette A, The human CD8+ T cell responses induced by a live attenuated tetravalent dengue vaccine are directed against highly conserved epitopes, J Virol 89(1) (2015) 120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Weiskopf D, Angelo MA, Sidney J, Peters B, Shresta S, Sette A, Immunodominance changes as a function of the infecting dengue virus serotype and primary versus secondary infection, J Virol 88(19) (2014) 11383–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Grifoni A, Voic H, Dhanda SK, Kidd CK, Brien JD, Buus S, Stryhn A, Durbin AP, Whitehead S, Diehl SA, De Silva AD, Balmaseda A, Harris E, Weiskopf D, Sette A, T Cell Responses Induced by Attenuated Flavivirus Vaccination Are Specific and Show Limited Cross-Reactivity with Other Flavivirus Species, J Virol 94(10) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chiu C, McCausland M, Sidney J, Duh FM, Rouphael N, Mehta A, Mulligan M, Carrington M, Wieland A, Sullivan NL, Weinberg A, Levin MJ, Pulendran B, Peters B, Sette A, Ahmed R, Broadly reactive human CD8 T cells that recognize an epitope conserved between VZV, HSV and EBV, PLoS Pathog 10(3) (2014) e1004008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Siciliano NA, Hersperger AR, Lacuanan AM, Xu RH, Sidney J, Sette A, Sigal LJ, Eisenlohr LC, Impact of distinct poxvirus infections on the specificities and functionalities of CD4+ T cell responses, J Virol 88(17) (2014) 10078–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Walsh SR, Gillis J, Peters B, Mothe BR, Sidney J, Sette A, Johnson RP, Diverse recognition of conserved orthopoxvirus CD8+ T cell epitopes in vaccinated rhesus macaques, Vaccine 27(36) (2009) 4990–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Greenbaum JA, Kotturi MF, Kim Y, Oseroff C, Vaughan K, Salimi N, Vita R, Ponomarenko J, Scheuermann RH, Sette A, Peters B, Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population, Proc Natl Acad Sci U S A 106(48) (2009) 20365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Skevaki C, Hudemann C, Matrosovich M, Mobs C, Paul S, Wachtendorf A, Alashkar Alhamwe B, Potaczek DP, Hagner S, Gemsa D, Garn H, Sette A, Renz H, Influenza-derived peptides cross-react with allergens and provide asthma protection, J Allergy Clin Immunol 142(3) (2018) 804–814. [DOI] [PubMed] [Google Scholar]
- [66].Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, Burger ZC, Rawlings SA, Smith DM, Phillips E, Mallal S, Lammers M, Rubiro P, Quiambao L, Sutherland A, Yu ED, da Silva Antunes R, Greenbaum J, Frazier A, Markmann AJ, Premkumar L, de Silva A, Peters B, Crotty S, Sette A, Weiskopf D, Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans, Science (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bresciani A, Paul S, Schommer N, Dillon MB, Bancroft T, Greenbaum J, Sette A, Nielsen M, Peters B, T-cell recognition is shaped by epitope sequence conservation in the host proteome and microbiome, Immunology 148(1) (2016) 34–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Carrasco Pro S, Lindestam Arlehamn CS, Dhanda SK, Carpenter C, Lindvall M, Faruqi AA, Santee CA, Renz H, Sidney J, Peters B, Sette A, Microbiota epitope similarity either dampens or enhances the immunogenicity of disease-associated antigenic epitopes, PLoS One 13(5) (2018) e0196551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Laheurte C, Galaine J, Beziaud L, Dosset M, Kerzerho J, Jacquemard C, Gaugler B, Ferrand C, Dormoy A, Aubin F, Jacoulet P, Westeel V, Borg C, Tartour E, Godet Y, Maillere B, Adotevi O, Immunoprevalence and magnitude of HLA-DP4 versus HLA-DR-restricted spontaneous CD4(+) Th1 responses against telomerase in cancer patients, Oncoimmunology 5(5) (2016) e1137416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Oseroff C, Peters B, Pasquetto V, Moutaftsi M, Sidney J, Panchanathan V, Tscharke DC, Maillere B, Grey H, Sette A, Dissociation between epitope hierarchy and immunoprevalence in CD8 responses to vaccinia virus western reserve, J Immunol 180(11) (2008) 7193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Moxon R, Reche PA, Rappuoli R, Editorial: Reverse Vaccinology, Frontiers in immunology 10 (2019) 2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Moutaftsi M, Tscharke DC, Vaughan K, Koelle DM, Stern L, Calvo-Calle M, Ennis F, Terajima M, Sutter G, Crotty S, Drexler I, Franchini G, Yewdell JW, Head SR, Blum J, Peters B, Sette A, Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens, Future Microbiol 5(2) (2010) 221–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A, Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes, Proc Natl Acad Sci U S A 105(6) (2008) 2140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Gene ontology: tool for the unification of biology. The Gene Ontology Consortium, Nature genetics 25(1) (2000) 25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bausch-Fluck D, Hofmann A, Bock T, Frei AP, Cerciello F, Jacobs A, Moest H, Omasits U, Gundry RL, Yoon C, Schiess R, Schmidt A, Mirkowska P, Hartlova A, Van Eyk JE, Bourquin JP, Aebersold R, Boheler KR, Zandstra P, Wollscheid B, A mass spectrometric-derived cell surface protein atlas, PLoS One 10(3) (2015) e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Bausch-Fluck D, Goldmann U, Muller S, van Oostrum M, Muller M, Schubert OT, Wollscheid B, The in silico human surfaceome, Proc Natl Acad Sci U S A 115(46) (2018) E10988–E10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Vathipadiekal V, Wang V, Wei W, Waldron L, Drapkin R, Gillette M, Skates S, Birrer M, Creation of a Human Secretome: A Novel Composite Library of Human Secreted Proteins: Validation Using Ovarian Cancer Gene Expression Data and a Virtual Secretome Array, Clinical cancer research : an official journal of the American Association for Cancer Research 21(21) (2015) 4960–9. [DOI] [PubMed] [Google Scholar]
- [78].Schulten V, Westernberg L, Birrueta G, Sidney J, Paul S, Busse P, Peters B, Sette A, Allergen and Epitope Targets of Mouse-Specific T Cell Responses in Allergy and Asthma, Frontiers in immunology 9 (2018) 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Schulten V, Greenbaum JA, Hauser M, McKinney DM, Sidney J, Kolla R, Lindestam Arlehamn CS, Oseroff C, Alam R, Broide DH, Ferreira F, Grey HM, Sette A, Peters B, Previously undescribed grass pollen antigens are the major inducers of T helper 2 cytokine-producing T cells in allergic individuals, Proc Natl Acad Sci U S A 110(9) (2013) 3459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Oseroff C, Christensen LH, Westernberg L, Pham J, Lane J, Paul S, Greenbaum J, Stranzl T, Lund G, Hoof I, Holm J, Wurtzen PA, Meno KH, Frazier A, Schulten V, Andersen PS, Peters B, Sette A, Immunoproteomic analysis of house dust mite antigens reveals distinct classes of dominant T cell antigens according to function and serological reactivity, Clin Exp Allergy 47(4) (2017) 577–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, Kim Y, Sidney J, James EA, Taplitz R, McKinney DM, Kwok WW, Grey H, Sallusto F, Peters B, Sette A, Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset, PLoS pathogens 9(1) (2013) e1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Pearson H, Daouda T, Granados DP, Durette C, Bonneil E, Courcelles M, Rodenbrock A, Laverdure JP, Cote C, Mader S, Lemieux S, Thibault P, Perreault C, MHC class I-associated peptides derive from selective regions of the human genome, The Journal of clinical investigation 126(12) (2016) 4690–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Parham P, Immunological Reviews: Genomic organisation of the MHC: structure, origin and function, Immunological Reviews, Munksgaard, Copenhagen, 1999. [Google Scholar]
- [84].Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE, IPD-IMGT/HLA Database, Nucleic acids research 48(D1) (2020) D948–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Gonzalez-Galarza FF, McCabe A, Santos E, Jones J, Takeshita L, Ortega-Rivera ND, Cid-Pavon GMD, Ramsbottom K, Ghattaoraya G, Alfirevic A, Middleton D, Jones AR, Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools, Nucleic acids research 48(D1) (2020) D783–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A, Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes, Immunogenetics 63(6) (2011) 325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sette A, Sidney J, HLA supertypes and supermotifs: a functional perspective on HLA polymorphism, Current opinion in immunology 10(4) (1998) 478–82. [DOI] [PubMed] [Google Scholar]
- [88].Sette A, Sidney J, Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism, Immunogenetics 50(3–4) (1999) 201–12. [DOI] [PubMed] [Google Scholar]
- [89].Sidney J, del Guercio MF, Southwood S, Engelhard VH, Appella E, Rammensee HG, Falk K, Rotzschke O, Takiguchi M, Kubo RT, et al. , Several HLA alleles share overlapping peptide specificities, J Immunol 154(1) (1995) 247–59. [PubMed] [Google Scholar]
- [90].Sidney J, Grey HM, Kubo RT, Sette A, Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs, Immunol Today 17(6) (1996) 261–6. [DOI] [PubMed] [Google Scholar]
- [91].Sidney J, Grey HM, Southwood S, Celis E, Wentworth PA, del Guercio MF, Kubo RT, Chesnut RW, Sette A, Definition of an HLA-A3-like supermotif demonstrates the overlapping peptide-binding repertoires of common HLA molecules, Hum Immunol 45(2) (1996) 79–93. [DOI] [PubMed] [Google Scholar]
- [92].Sidney J, Peters B, Frahm N, Brander C, Sette A, HLA class I supertypes: a revised and updated classification, BMC Immunol 9 (2008) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sidney J, Southwood S, del Guercio MF, Grey HM, Chesnut RW, Kubo RT, Sette A, Specificity and degeneracy in peptide binding to HLA-B7-like class I molecules, J Immunol 157(8) (1996) 3480–90. [PubMed] [Google Scholar]
- [94].Sidney J, Southwood S, Mann DL, Fernandez-Vina MA, Newman MJ, Sette A, Majority of peptides binding HLA-A*0201 with high affinity crossreact with other A2-supertype molecules, Hum Immunol 62(11) (2001) 1200–16. [DOI] [PubMed] [Google Scholar]
- [95].Sidney J, Southwood S, Pasquetto V, Sette A, Simultaneous prediction of binding capacity for multiple molecules of the HLA B44 supertype, J Immunol 171(11) (2003) 5964–74. [DOI] [PubMed] [Google Scholar]
- [96].Sidney J, Southwood S, Sette A, Classification of A1- and A24-supertype molecules by analysis of their MHC-peptide binding repertoires, Immunogenetics 57(6) (2005) 393–408. [DOI] [PubMed] [Google Scholar]
- [97].Southwood S, Sidney J, Kondo A, del Guercio M, Appella E, Hoffman S, Kubo R, Chesnut R, Grey H, Sette A, Several common HLA-DR types share largely overlapping peptide binding repertoires, J Immunol 160 (1998) 3363–3373. [PubMed] [Google Scholar]
- [98].Lund O, Nielsen M, Kesmir C, Petersen AG, Lundegaard C, Worning P, Sylvester-Hvid C, Lamberth K, Roder G, Justesen S, Buus S, Brunak S, Definition of supertypes for HLA molecules using clustering of specificity matrices, Immunogenetics 55(12) (2004) 797–810. [DOI] [PubMed] [Google Scholar]
- [99].Nielsen M, Lund O, Buus S, Lundegaard C, MHC class II epitope predictive algorithms, Immunology 130(3) (2010) 319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, Broadwater A, Kolla RV, De Silva AD, de Silva AM, Mattia KA, Doranz BJ, Grey HM, Shresta S, Peters B, Sette A, Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells, Proc Natl Acad Sci U S A 110(22) (2013) E2046–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Reche PA, Reinherz EL, PEPVAC: a web server for multi-epitope vaccine development based on the prediction of supertypic MHC ligands, Nucleic Acids Res 33(Web Server issue) (2005) W138–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].McKinney DM, Southwood S, Hinz D, Oseroff C, Arlehamn CS, Schulten V, Taplitz R, Broide D, Hanekom WA, Scriba TJ, Wood R, Alam R, Peters B, Sidney J, Sette A, A strategy to determine HLA class II restriction broadly covering the DR, DP, and DQ allelic variants most commonly expressed in the general population, Immunogenetics 65(5) (2013) 357–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Oyarzun P, Ellis JJ, Gonzalez-Galarza FF, Jones AR, Middleton D, Boden M, Kobe B, A bioinformatics tool for epitope-based vaccine design that accounts for human ethnic diversity: application to emerging infectious diseases, Vaccine 33(10) (2015) 1267–73. [DOI] [PubMed] [Google Scholar]
- [104].Nepom GT, MHC class II tetramers, J Immunol 188(6) (2012) 2477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Nepom GT, Buckner JH, Novak EJ, Reichstetter S, Reijonen H, Gebe J, Wang R, Swanson E, Kwok WW, HLA class II tetramers: tools for direct analysis of antigen-specific CD4+ T cells, Arthritis and rheumatism 46(1) (2002) 5–12. [DOI] [PubMed] [Google Scholar]
- [106].Cecconi V, Moro M, Del Mare S, Dellabona P, Casorati G, Use of MHC class II tetramers to investigate CD4+ T cell responses: problems and solutions, Cytometry A 73(11) (2008) 1010–8. [DOI] [PubMed] [Google Scholar]
- [107].Christophersen A, Peptide-MHC class I and class II tetramers: From flow to mass cytometry, HLA 95(3) (2020) 169–178. [DOI] [PubMed] [Google Scholar]
- [108].Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A, Predicting population coverage of T-cell epitope-based diagnostics and vaccines, BMC bioinformatics 7 (2006) 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Liu G, Carter B, Bricken T, Jain S, Viard M, Carrington M, Gifford DK, Computationally Optimized SARS-CoV-2 MHC Class I and II Vaccine Formulations Predicted to Target Human Haplotype Distributions, Cell Syst (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A, Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells, Eur J Immunol 19(12) (1989) 2237–42. [DOI] [PubMed] [Google Scholar]
- [111].Sinigaglia F, Guttinger M, Kilgus J, Doran DM, Matile H, Etlinger H, Trzeciak A, Gillessen D, Pink JR, A malaria T-cell epitope recognized in association with most mouse and human MHC class II molecules, Nature 336(6201) (1988) 778–80. [DOI] [PubMed] [Google Scholar]
- [112].Busch R, Strang G, Howland K, Rothbard JB, Degenerate binding of immunogenic peptides to HLA-DR proteins on B cell surfaces, Int Immunol 2(5) (1990) 443–51. [DOI] [PubMed] [Google Scholar]
- [113].Roche PA, Cresswell P, High-affinity binding of an influenza hemagglutinin-derived peptide to purified HLA-DR, J Immunol 144(5) (1990) 1849–56. [PubMed] [Google Scholar]
- [114].O’Sullivan D, Sidney J, Del Guercio MF, Colon SM, Sette A, Truncation analysis of several DR binding epitopes, J Immunol 146(4) (1991) 1240–6. [PubMed] [Google Scholar]
- [115].O’Sullivan D, Sidney J, Appella E, Walker L, Phillips L, Colon SM, Miles C, Chesnut RW, Sette A, Characterization of the specificity of peptide binding to four DR haplotypes, J Immunol 145(6) (1990) 1799–808. [PubMed] [Google Scholar]
- [116].Alexander J, Sidney J, Southwood S, Ruppert J, Oseroff C, Maewal A, Snoke K, Serra HM, Kubo RT, Sette A, et al. , Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides, Immunity 1(9) (1994) 751–61. [DOI] [PubMed] [Google Scholar]
- [117].Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, Wasserman SI, Weiskopf D, McKinney DM, Chung JL, Petersen A, Grey H, Peters B, Sette A, Molecular determinants of T cell epitope recognition to the common Timothy grass allergen, J Immunol 185(2) (2010) 943–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Oseroff C, Sidney J, Tripple V, Grey H, Wood R, Broide DH, Greenbaum J, Kolla R, Peters B, Pomes A, Sette A, Analysis of T cell responses to the major allergens from German cockroach: epitope specificity and relationship to IgE production, J Immunol 189(2) (2012) 679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Oseroff C, Sidney J, Vita R, Tripple V, McKinney DM, Southwood S, Brodie TM, Sallusto F, Grey H, Alam R, Broide D, Greenbaum JA, Kolla R, Peters B, Sette A, T cell responses to known allergen proteins are differently polarized and account for a variable fraction of total response to allergen extracts, J Immunol 189(4) (2012) 1800–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Vita R, Peters B, Sette A, The curation guidelines of the immune epitope database and analysis resource, Cytometry A 73(11) (2008) 1066–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Brodsky FM, Parham P, Barnstable CJ, Crumpton MJ, Bodmer WF, Monoclonal antibodies for analysis of the HLA system, Immunol Rev 47 (1979) 3–61. [DOI] [PubMed] [Google Scholar]
- [122].McMichael AJ, HLA restriction of human cytotoxic T cells, Springer Semin Immunopathol 3(1) (1980) 3–22. [DOI] [PubMed] [Google Scholar]
- [123].Wang RF, Molecular cloning and characterization of MHC class I- and II-restricted tumor antigens recognized by T cells, Curr Protoc Immunol Chapter 20 (2009) Unit 20 10. [DOI] [PubMed] [Google Scholar]
- [124].Wilson CC, Palmer B, Southwood S, Sidney J, Higashimoto Y, Appella E, Chesnut R, Sette A, Livingston BD, Identification and antigenicity of broadly cross-reactive and conserved human immunodeficiency virus type 1-derived helper T-lymphocyte epitopes, J Virol 75(9) (2001) 4195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Lamb JR, Eckels DD, Lake P, Johnson AH, Hartzman RJ, Woody JN, Antigen-specific human T lymphocyte clones: induction, antigen specificity, and MHC restriction of influenza virus-immune clones, J Immunol 128(1) (1982) 233–8. [PubMed] [Google Scholar]
- [126].Paul S, Arlehamn CSL, Schulten V, Westernberg L, Sidney J, Peters B, Sette A, Experimental validation of the RATE tool for inferring HLA restrictions of T cell epitopes, BMC Immunol 18(Suppl 1) (2017) 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Paul S, Dillon MB, Lindestam Arlehamn CS, Huang H, Davis MM, McKinney DM, Scriba TJ, Sidney J, Peters B, Sette A, A population response analysis approach to assign class II HLA-epitope restrictions, J Immunol 194(12) (2015) 6164–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Altman JD, Davis MM, MHC-peptide tetramers to visualize antigen-specific T cells, Curr Protoc Immunol Chapter 17 (2003) Unit 17 3. [DOI] [PubMed] [Google Scholar]
- [129].Bakker AH, Schumacher TN, MHC multimer technology: current status and future prospects, Curr Opin Immunol 17(4) (2005) 428–33. [DOI] [PubMed] [Google Scholar]
- [130].Ihantola EL, Ilmonen H, Kailaanmaki A, Rytkonen-Nissinen M, Azam A, Maillere B, Lindestam Arlehamn CS, Sette A, Motwani K, Seay HR, Brusko TM, Knip M, Veijola R, Toppari J, Ilonen J, Kinnunen T, Characterization of Proinsulin T Cell Epitopes Restricted by Type 1 Diabetes-Associated HLA Class II Molecules, J Immunol 204(9) (2020) 2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Surenaud M, Montes M, Lindestam Arlehamn CS, Sette A, Banchereau J, Palucka K, Lelievre JD, Lacabaratz C, Levy Y, Anti-HIV potency of T-cell responses elicited by dendritic cell therapeutic vaccination, PLoS pathogens 15(9) (2019) e1008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Lindestam Arlehamn CS, McKinney DM, Carpenter C, Paul S, Rozot V, Makgotlho E, Gregg Y, van Rooyen M, Ernst JD, Hatherill M, Hanekom WA, Peters B, Scriba TJ, Sette A, A Quantitative Analysis of Complexity of Human Pathogen-Specific CD4 T Cell Responses in Healthy M. tuberculosis Infected South Africans, PLoS Pathog 12(7) (2016) e1005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Campbell VL, Nguyen L, Snoey E, McClurkan CL, Laing KJ, Dong L, Sette A, Lindestam Arlehamn CS, Altmann DM, Boyton RJ, Roby JA, Gale M Jr., Stone M, Busch MP, Norris PJ, Koelle DM, Proteome-Wide Zika Virus CD4 T Cell Epitope and HLA Restriction Determination, Immunohorizons 4(8) (2020) 444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Kotturi MF, Peters B, Buendia-Laysa F Jr., Sidney J, Oseroff C, Botten J, Grey H, Buchmeier MJ, Sette A, The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus, J Virol 81(10) (2007) 4928–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Geiger R, Duhen T, Lanzavecchia A, Sallusto F, Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells, J Exp Med 206(7) (2009) 1525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Koelle DM, Expression cloning for the discovery of viral antigens and epitopes recognized by T cells, Methods 29(3) (2003) 213–26. [DOI] [PubMed] [Google Scholar]
- [137].Czerkinsky CC, Nilsson LA, Nygren H, Ouchterlony O, Tarkowski A, A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells, J Immunol Methods 65(1–2) (1983) 109–21. [DOI] [PubMed] [Google Scholar]
- [138].Pala P, Hussell T, Openshaw PJ, Flow cytometric measurement of intracellular cytokines, J Immunol Methods 243(1–2) (2000) 107–24. [DOI] [PubMed] [Google Scholar]
- [139].Havenar-Daughton C, Reiss SM, Carnathan DG, Wu JE, Kendric K, Torrents A de la Pena, S.P. Kasturi, J.M. Dan, M. Bothwell, R.W. Sanders, B. Pulendran, G. Silvestri, S. Crotty, Cytokine-Independent Detection of Antigen-Specific Germinal Center T Follicular Helper Cells in Immunized Nonhuman Primates Using a Live Cell Activation-Induced Marker Technique, J Immunol 197(3) (2016) 994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Reiss S, Baxter AE, Cirelli KM, Dan JM, Morou A, Daigneault A, Brassard N, Silvestri G, Routy JP, Havenar-Daughton C, Crotty S, Kaufmann DE, Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells, PLoS One 12(10) (2017) e0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Dan JM, Lindestam Arlehamn CS, Weiskopf D, da Silva Antunes R, Havenar-Daughton C, Reiss SM, Brigger M, Bothwell M, Sette A, Crotty S, A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4+ T Cells in Blood, J Immunol 197(3) (2016) 983–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Dan JM, Havenar-Daughton C, Kendric K, Al-Kolla R, Kaushik K, Rosales SL, Anderson EL, LaRock CN, Vijayanand P, Seumois G, Layfield D, Cutress RI, Ottensmeier CH, Lindestam Arlehamn CS, Sette A, Nizet V, Bothwell M, Brigger M, Crotty S, Recurrent group A Streptococcus tonsillitis is an immunosusceptibility disease involving antibody deficiency and aberrant TFH cells, Science translational medicine 11(478) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Herati RS, Muselman A, Vella L, Bengsch B, Parkhouse K, Del Alcazar D, Kotzin J, Doyle SA, Tebas P, Hensley SE, Su LF, Schmader KE, Wherry EJ, Successive annual influenza vaccination induces a recurrent oligoclonotypic memory response in circulating T follicular helper cells, Sci Immunol 2(8) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Morou A, Brunet-Ratnasingham E, Dube M, Charlebois R, Mercier E, Darko S, Brassard N, Nganou-Makamdop K, Arumugam S, Gendron-Lepage G, Yang L, Niessl J, Baxter AE, Billingsley JM, Rajakumar PA, Lefebvre F, Johnson RP, Tremblay C, Routy JP, Wyatt RT, Finzi A, Douek DC, Kaufmann DE, Altered differentiation is central to HIV-specific CD4(+) T cell dysfunction in progressive disease, Nat Immunol 20(8) (2019) 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Paul S, Lindestam Arlehamn CS, Scriba TJ, Dillon MB, Oseroff C, Hinz D, McKinney DM, Carrasco Pro S, Sidney J, Peters B, Sette A, Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes, J Immunol Methods 422 (2015) 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Carrasco Pro S, Sidney J, Paul S, Lindestam Arlehamn C, Weiskopf D, Peters B, Sette A, Automatic Generation of Validated Specific Epitope Sets, Journal of immunology research 2015 (2015) 763461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].da Silva Antunes R, Quiambao LG, Sutherland A, Soldevila F, Dhanda SK, Armstrong SK, Brickman TJ, Merkel T, Peters B, Sette A, Development and Validation of a Bordetella pertussis Whole-Genome Screening Strategy, Journal of immunology research 2020 (2020) 8202067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].da Silva Antunes R, Paul S, Sidney J, Weiskopf D, Dan JM, Phillips E, Mallal S, Crotty S, Sette A, Lindestam Arlehamn CS, Definition of Human Epitopes Recognized in Tetanus Toxoid and Development of an Assay Strategy to Detect Ex Vivo Tetanus CD4+ T Cell Responses, PLoS One 12(1) (2017) e0169086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Hinz D, Oseroff C, Pham J, Sidney J, Peters B, Sette A, Definition of a pool of epitopes that recapitulates the T cell reactivity against major house dust mite allergens, Clin Exp Allergy 45(10) (2015) 1601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Hinz D, Seumois G, Gholami AM, Greenbaum JA, Lane J, White B, Broide DH, Schulten V, Sidney J, Bakhru P, Oseroff C, Wambre E, James EA, Kwok WW, Peters B, Vijayanand P, Sette A, Lack of allergy to timothy grass pollen is not a passive phenomenon but associated with the allergen-specific modulation of immune reactivity, Clin Exp Allergy 46(5) (2016) 705–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Grifoni A, Angelo MA, Lopez B, O’Rourke PH, Sidney J, Cerpas C, Balmaseda A, Silveira CGT, Maestri A, Costa PR, Durbin AP, Diehl SA, Phillips E, Mallal S, De Silva AD, Nchinda G, Nkenfou C, Collins MH, de Silva AM, Lim MQ, Macary PA, Tatullo F, Solomon T, Satchidanandam V, Desai A, Ravi V, Coloma J, Turtle L, Rivino L, Kallas EG, Peters B, Harris E, Sette A, Weiskopf D, Global Assessment of Dengue Virus-Specific CD4(+) T Cell Responses in Dengue-Endemic Areas, Frontiers in immunology 8 (2017) 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Weiskopf D, Cerpas C, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, Sanches FP, Silvera CG, Costa PR, Kallas EG, Gresh L, de Silva AD, Balmaseda A, Harris E, Sette A, Human CD8+ T-Cell Responses Against the 4 Dengue Virus Serotypes Are Associated With Distinct Patterns of Protein Targets, J Infect Dis 212(11) (2015) 1743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Arlehamn CL, Seumois G, Gerasimova A, Huang C, Fu Z, Yue X, Sette A, Vijayanand P, Peters B, Transcriptional profile of tuberculosis antigen-specific T cells reveals novel multifunctional features, J Immunol 193(6) (2014) 2931–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lindestam Arlehamn CS, Sette A, Definition of CD4 Immunosignatures Associated with MTB, Frontiers in immunology 5 (2014) 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Burel JG, Lindestam Arlehamn CS, Khan N, Seumois G, Greenbaum JA, Taplitz R, Gilman RH, Saito M, Vijayanand P, Sette A, Peters B, Transcriptomic Analysis of CD4(+) T Cells Reveals Novel Immune Signatures of Latent Tuberculosis, J Immunol 200(9) (2018) 3283–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Burel JG, Babor M, Pomaznoy M, Lindestam Arlehamn CS, Khan N, Sette A, Peters B, Host Transcriptomics as a Tool to Identify Diagnostic and Mechanistic Immune Signatures of Tuberculosis, Frontiers in immunology 10 (2019) 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Grifoni A, Costa-Ramos P, Pham J, Tian Y, Rosales SL, Seumois G, Sidney J, de Silva AD, Premkumar L, Collins MH, Stone M, Norris PJ, Romero CME, Durbin A, Ricciardi MJ, Ledgerwood JE, de Silva AM, Busch M, Peters B, Vijayanand P, Harris E, Falconar AK, Kallas E, Weiskopf D, Sette A, Cutting Edge: Transcriptional Profiling Reveals Multifunctional and Cytotoxic Antiviral Responses of Zika Virus-Specific CD8(+) T Cells, J Immunol 201(12) (2018) 3487–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Tian Y, Babor M, Lane J, Schulten V, Patil VS, Seumois G, Rosales SL, Fu Z, Picarda G, Burel J, Zapardiel-Gonzalo J, Tennekoon RN, De Silva AD, Premawansa S, Premawansa G, Wijewickrama A, Greenbaum JA, Vijayanand P, Weiskopf D, Sette A, Peters B, Unique phenotypes and clonal expansions of human CD4 effector memory T cells re-expressing CD45RA, Nat Commun 8(1) (2017) 1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Patil VS, Madrigal A, Schmiedel BJ, Clarke J, O’Rourke P, de Silva AD, Harris E, Peters B, Seumois G, Weiskopf D, Sette A, Vijayanand P, Precursors of human CD4(+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis, Sci Immunol 3(19) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Tian Y, Babor M, Lane J, Seumois G, Liang S, Goonawardhana NDS, De Silva AD, Phillips EJ, Mallal SA, da Silva Antunes R, Grifoni A, Vijayanand P, Weiskopf D, Peters B, Sette A, Dengue-specific CD8+ T cell subsets display specialized transcriptomic and TCR profiles, J Clin Invest 129(4) (2019) 1727–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Grifoni A, Tian Y, Sette A, Weiskopf D, Transcriptomic immune profiles of human flavivirus-specific T-cell responses, Immunology 160(1) (2020) 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Tian Y, Seumois G, De-Oliveira-Pinto LM, Mateus J, Herrera-de la Mata S, Kim C, Hinz D, Goonawardhana NDS, de Silva AD, Premawansa S, Premawansa G, Wijewickrama A, Balmaseda A, Grifoni A, Vijayanand P, Harris E, Peters B, Sette A, Weiskopf D, Molecular Signatures of Dengue Virus-Specific IL-10/IFN-gamma Co-producing CD4 T Cells and Their Association with Dengue Disease, Cell Rep 29(13) (2019) 4482–4495 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Grifoni A, Voic H, Dhanda SK, Kidd CK, Brien JD, Buus S, Stryhn A, Durbin AP, Whitehead S, Diehl SA, De Silva AD, Balmaseda A, Harris E, Weiskopf D, Sette A, T cell responses induced by attenuated flavivirus vaccination are specific and show limited cross-reactivity with other flavivirus species, J Virol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A, Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals, Cell 181(7) (2020) 1489–1501 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, van den Akker JPC, Molenkamp R, Koopmans MPG, van Gorp ECM, Haagmans BL, de Swart RL, Sette A, de Vries RD, Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome, Sci Immunol 5(48) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]


