Abstract
Approximately 15%-20% of patients infected with SARS-CoV-2 coronavirus (COVID-19) progress beyond mild and self-limited disease to require supplemental oxygen for severe pneumonia; 5% of COVID-19-infected patients further develop acute respiratory distress syndrome (ARDS) and multiorgan failure. Despite mortality rates surpassing 40%, key insights into COVID-19-induced ARDS pathology have not been fully elucidated and multiple unmet needs remain. This review focuses on the unmet need for effective therapies that target unchecked innate immunity-driven inflammation which drives unchecked vascular permeability, multiorgan dysfunction and ARDS mortality. Additional unmet needs including the lack of insights into factors predicting pathogenic hyperinflammatory viral host responses, limited approaches to address the vast disease heterogeneity in ARDS, and the absence of clinically-useful ARDS biomarkers. We review unmet needs persisting in COVID-19-induced ARDS in the context of the potential role for damage-associated molecular pattern proteins in lung and systemic hyperinflammatory host responses to SARS-CoV-2 infection that ultimately drive multiorgan dysfunction and ARDS mortality. Insights into promising stratification-enhancing, biomarker-based strategies in COVID-19 and non-COVID ARDS may enable the design of successful clinical trials of promising therapies.
Abbreviations: ACE2, angiotensin converting enzyme 2; ANG-2, angiopoietin-2; ARDS, acute respiratory distress syndrome; COVID-19, coronavirus disease 19 infection; CRP, C-reactive protein; DAMPs, damage-associated molecular pattern proteins; eNAMPT, extracellular nicotinamide phosphoribosyl-transferase; IFNγ, interferon gamma; IL-1RA, interleukin 1 receptor antagonist; IL-6, interleukin 6; IP-10, interferon gamma-induced protein 10; IRF7, interferon regulatory factor 7; MCP1, monocyte chemoattractant protein 1; MIF, macrophage migration inhibition factor; HMGB1, the high mobility group box 1 protein; NO, Nitric oxide; PAMPs, pathogen-associated molecular pattern proteins; RIPK1, Receptor-interacting serine/threonine-protein kinase; ROS, reactive oxygen species; SARS-CoV-2, Severe Acute Respiratory Syndrome-related Coronavirus 2; SMI, small molecule inhibitor; TLRs, Toll-like family of receptors; TNFα, tumor necrosis factor alpha; VILI, ventilator-induced lung injury
Overview: Serious Unmet Needs in the COVID-19 Pandemic Landscape
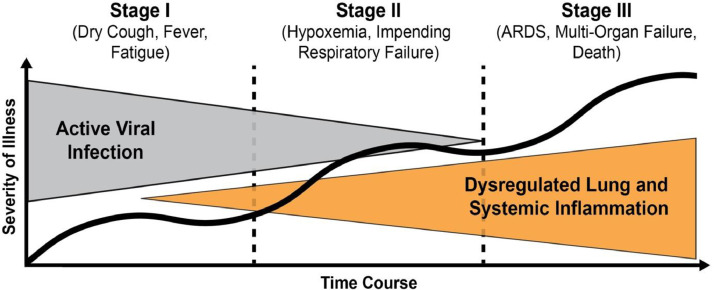
Within the 10 months following notification by the World Health Organization's (WHO) China Country Office of a pneumonia of unknown cause detected in the city of Wuhan (Hubei province, China) (January 2020), the COVID-19 pandemic, now known to be caused by Severe Acute Respiratory Syndrome-related Coronavirus 2 (SARS-CoV-2), has fundamentally altered global health and economies and changed our country, community and families.1 To date (November 2020), the pandemic has resulted in more than 55 million infected individuals worldwide and ∼1.3 million deaths.2 Whereas the majority (80%) of patients with confirmed SARS-CoV-2 infection exhibit mildly symptomatic, self-limited coronavirus disease (COVID-19), approximately 15% of infected subjects progress to severe pneumonia requiring supplemental oxygen and 5% further progress to develop acute respiratory distress syndrome (ARDS), and/or multiorgan failure (MOF)3 (Fig 1 ) which drives mortality rates that surpass 40%.
Fig 1.
Stages of illness severity following SARS-CoV-2 infection. Over 80% of COVID-19-infected patients resolve a self-limited disease in Stage I. Approximately 15% of infected subjects, however, progress to severe pneumonia requiring supplemental oxygen (Stage II) with 5% further developing acute respiratory distress syndrome (ARDS), and/or multiorgan failure (Stage III). Stages II and III are characterized by dysregulated lung and systemic inflammation which are essential contributors to COVID-19 disease severity. Adapted from Siddiqi et al.4
The unprecedented nature of the SARS-CoV-2/COVID-19 pandemic has highlighted multiple unmet medical needs including the glaring absence of effective FDA–approved targeted pharmacotherapies.5 This has created urgency within basic science, translational, and clinical research communities, particularly as immunity-derived protection against the novel coronaviruses such as SARS (severe acute respiratory syndrome, 2002-2004), and MERS (Middle East respiratory syndrome, 2012-2015) proved to be inefficient and unpredictable.6 , 7 The availability of effective SARS-CoV-2 vaccines and anti-SARS-CoV-2 drugs are imminent and of obvious utility, however, neither therapeutic strategy addresses the pathobiology of COVID-19-ARDS, that is, the unremitting activation of innate immunity inflammatory pathways with unchecked lung and systemic inflammation. The activation of evolutionarily-conserved inflammatory cascades by SARS-CoV-2, as well as significant amplification of these pathways by exposure to excessive levels of ventilator-induced mechanical stress,8 results in massive increases in circulating levels of inflammatory cytokines producing vascular leak, edema of multiple critical organ (lung, kidneys, heart, brain, liver, gastrointestinal tract), and dysregulated activation of the coagulation cascade, ultimately leading to multiorgan dysfunction and death.9, 10, 11
Studies of past coronavirus outbreaks, involving SARS-CoV-112 , 13 and MERS-CoV,14 have increased our understanding of the inflammation-induced pathobiology in SARS-CoV-2 revealing such commonalities as the cytokine-release syndrome. Other immune responses to SARS-CoV-2, however, remain distinctive15 , 16 including the unique expanded tropism of SARS-CoV-2 for vascular endothelial cells, likely relevant to the hypercoagulability observed in severe COVID-19.16 , 17 In addition, SARS-CoV-2 infectivity peaks before the onset of symptoms, a stark contrast to SARS-CoV-1,3 , 18 reflecting current understanding of longer SARS-CoV-2 incubation periods and higher rates of transmission compared to other coronaviruses.5 Potential mechanisms for the significant differences in the host immune response to SARS-CoV-2 infection include viral escape from innate sensing, hyperinflammatory responsiveness referable to inflammatory myeloid subpopulations, and lymphopenia marked by T cell and Natural Killer cell dysfunction among other alterations such as hypercoagulability.4 , 19 , 20 Clearly, additional studies are needed to more fully define immune responses to specific coronavirus infections, including SARS-CoV-2, that dictate disease severity.
In this report, we summarize potential mechanisms of COVID-19 pathobiology regarding increases in vascular leakage, hypercoagulation initiation, and the promotion of lung and systemic inflammation. In addition, we examine the underpinnings of the multiple unmet needs in COVID-19 ARDS which includes the absence of effective therapies that target unchecked activation of lung and systemic inflammatory cascades by damage-associated molecular pattern proteins which ultimately drives ARDS multiorgan dysfunction and mortality. Acknowledging the difficulty in identifying specific ARDS subphenotypes, due to heterogeneity in host responses to SARS-CoV-2 infection, we also highlight the unmet need for biomarkers predictive of a pathologic hyperinflammatory viral host response vs a physiologic host response.21, 22, 23 Insights into promising stratification-enhancing, biomarker-based strategies in COVID-19 and non-COVID ARDS may enable the design of successful clinical trials of promising therapies.
SARS-CoV-2 Pathobiology: Viral Invasion and Clinical Presentation
An important first step to addressing the multiple and critical unmet needs in severe COVID-19 infection is to understand the pathogen-activated innate immunity pathways designed to contain the infection but which produce unremitting inflammation in a subset of infected patients. Both SARS-CoV and SARS-CoV-2 consist of 4 main structural glycoproteins; the spike (S) protein responsible for viral binding and entry into host cell, and membrane (M), envelope (E), and nucleocapsid (N) proteins which play an important role in viral particle assembly and release.24 Human angiotensin converting enzyme 2 (ACE2) has been identified as an entry receptor for SARS-CoV25 and SARS-CoV-2.24 Membrane fusion after host cell binding is facilitated by S-protein cleavage by host cell proteases, specifically, the transmembrane serine protease 2.26 The dependence on ACE2 for viral entry contributes to the tropism for nasal ciliated epithelium and alveolar type II pneumocytes.5 Increased infectivity and novel clinical features of SARs-CoV-2 have been associated with the tropism for other ACE2-expressing cells such as endothelial cells which is conferred by acquisition of a furin cleavage site on the S-protein.24
SARS-CoV-2 Pathobiology: Vascular Leakage in Severe COVID-19 ARDS
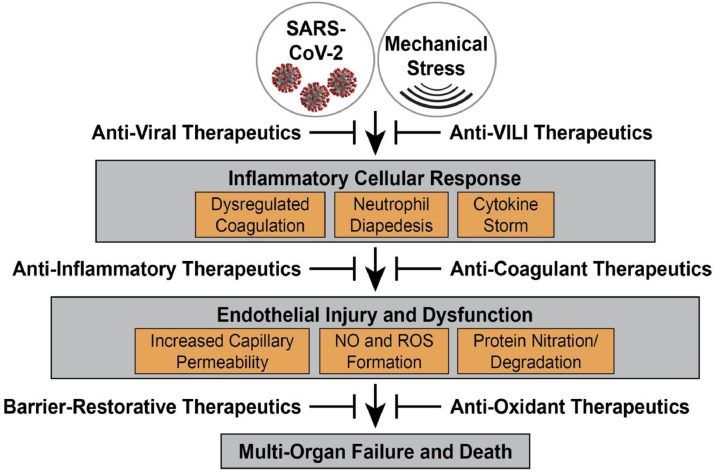
Key homeostatic functions of the vascular endothelium include presenting a non- thrombotic surface to circulating components and serving as a semi-permeable barrier to the extravasation of blood proteins, fluid, and infiltrating leukocytes.27 The hallmark of COVID-19- and non-COVID-19 ARDS is the loss of endothelial cell barrier integrity resulting in vascular leakage and increased tissue edema in multiple vital organs, most prominently the lung (Fig 2 ). The pathobiological mechanisms that potentially cause increased vascular permeability in severe SARS-Co-V-2 infection may include a direct viral infection of endothelial cells,17 activation of the kallikrein-bradykinin pathway, and the activation of permeability-inducing signaling pathways by adherent platelets, activated neutrophils28 and circulating cytokines and vasoactive molecules.20 The increased mechanical stress produced by mechanical ventilation as well as impaired endothelial cell ACE2 activity may also contribute to increases in vascular permeability.26 Multiple studies, including our own, have underscored the critical role of the endothelial cell cytoskeleton in vascular barrier regulation and repair that is central to the pathobiology of ARDS and ventilator-induced lung injury (VILI).27 , 29 Inflammatory cell mediator-induced activation of vascular barrier-disruptive signaling pathways, in combination with increases in reactive oxygen species (ROS), result in enhanced endothelial cells contractility, loosening of interendothelial junctions, formation of paracellular gaps, and development of profound vascular leakage and organ edema (Fig 2).27 In the lung, endothelial cell cytoskeletal target proteins are clearly involved in: (1) Unchecked inflammation-induced vascular permeability and injury; (2) vascular responses to excessive mechanical stress in VILI, and (3) regulation of leukocyte transmigration to the alveolar space. In addition, several cytoskeletal target genes harbor variants which contribute to the genetic basis for observed ARDS health disparities in Americans of African descent.30, 31, 32, 33, 34 Despite the identification of unchecked vascular permeability as a critical feature of progression to ARDS mortality, to date, there are no FDA-approved therapies that directly address the severe vascular leak that is critical to COVID- and non-COVID ARDS pathobiology and outcomes.
Fig 2.
SARS-CoV-2 infection- and mechanical ventilation (VILI)-induced inflammation and vascular injury/dysfunction. Critical sequence of coronaviral- and mechanical stress-mediated activation of evolutionarily-conserved inflammatory cascades resulting in dysregulated cytokine release, hypercoagulation, and leukocyte-endothelial cell interactions and diapedesis. In combination with excessive levels of reactive oxygen species (ROS) generated in target endothelial cells, these combined effects result in vascular injury and triggering of the cytoskeletal contractile apparatus to increase permeability leading to organ edema, multiorgan failure and COVID-19 ARDS mortality.
SARS-CoV-2 Pathobiology: Dysregulated Coagulation and Hypercoagulable Responses
After the initial phase of SARS-CoV-2 infection, 20% of patients develop severe or critical illness based on the severity of lung inflammation, hypoxemia, respiratory failure including ARDS, septic shock and/or multiorgan dysfunction (Fig 1).19 , 20 The progressive lung damage is a combination of a profound proinflammatory cytokine storm as well as coagulation abnormalities with a high incidence of thrombotic events observed in patients with severe COVID-19-ARDS (Fig 2).35 As noted above, although the nasal ciliated epithelium and alveolar type II pneumocytes are the initial point of invasion by SARS-CoV-2, activated endothelial cells play a critical role in the pathogenesis of COVID-19 associated ARDS and multiorgan dysfunction.20 Unique features of SARS-CoV-2 include the ability of the virus to directly affect endothelial cells, causing endothelialitis17 and dysregulated coagulation pathways.15 , 16 Granulocyte-containing microthrombi are a common feature of severe COVID-19 disease,35 a phenomenon described as immuno-thrombosis. This highly coagulative and inflammatory state is the result of a complex interplay between activated platelets, primed neutrophils and activated vascular endothelial cells.36 The formation of platelet neutrophil complexes at the surface of endothelial cells, leads to recruitment of highly cytotoxic neutrophils and inflammation-activated platelets that translocate through the pulmonary microvasculature into the alveolar and pulmonary interstitial spaces contributing to endothelial cell injury (Fig 2).37 Sequestration of platelet/neutrophil complexes in the pulmonary vasculature produces microthrombi and microemboli in the alveolocapillary circulation. This highly inflammatory and procoagulant state also leads to hyperactivation of the coagulation cascade and a relative exhaustion of the fibrinolytic system.36 , 38 The regulatory mechanisms of these hyperinflammatory and hypercoagulable responses in COVID-19 are still under investigation, however, it merits stressing that COVID-19-related coagulation disorders (arterial thrombosis, pulmonary embolism, etc.) are closely related to innate immune activation and inflammation, with a strong influence of Toll-like receptors.39 Currently, the role of anticoagulant therapies in COVID disease is speculative and under investigation although patients with elevated D Dimer or confirmed clots appear to benefit from systemic anticoagulation. Despite the critical importance for risk of thrombotic complications and the contribution to COVID-19 morbidity and mortality, there are no FDA-approved therapies that directly address endothelial cell dysregulation associated with increased coagulation/thrombosis in COVID and non-COVID ARDS.
SARS-CoV-2 Pathobiology: DAMP-Regulated Inflammatory Cascade Amplification
SARS-CoV-2 is a cytopathic virus which induces cellular death and local release of various damage-associated molecular pattern proteins (DAMPs) and pathogen–associated molecular pattern proteins (PAMPs).40 Pattern-recognition receptors such as Toll-like family of receptors (TLRs) recognize PAMPs in the extracellular space, triggering expression and activation of proinflammatory transcription factors such as NF-κB.41 , 42 These interactions also trigger the activation of interferon-regulatory factors that mediate antiviral responses.41 , 42 Given their master regulatory role in innate immunity, DAMPs are an attractive therapeutic target in inflammatory injuries such as COVID and non-COVID ARDS. For example, the high mobility group box 1 protein (HMGB1), is a recognized DAMP which is upstream of IL-6 release43, 44, 45 that like other DAMPs activates the inflammasome via recognition by the nucleotide-binding domain, leucine-rich containing (NLR) protein family. This triggers inflammasome activation resulting in secretion of cytokines such as IL-1β (after conversion of proIL-1β to IL-1β)41 , 42 and likely participates in the magnitude of inflammatory responses occurring in the lungs of COVID-19 patients. Type I interferon responses also lead to the cytokine storm characterized by marked elevations in IL-6, IL-8, monocyte chemoattractant protein 1 (MCP1), type II interferon (IFNγ), and interferon gamma-induced protein 10 (IP-10). In the lung, these factors result in subsequent pulmonary recruitment of immune cells such as dendritic cells and macrophages.20 Cytokines and chemokines are further released from virally-infected macrophages and dendritic cells with subsequent activation of late-phase immune-cell recruitment of antigen-specific T cells to destroy virally-infected alveolar cells.46 Antibody-mediated viral neutralization potentially contributes to successful viral clearance although the kinetics of different immunoglobulins is continuing to be characterized.47 , 48 The hyperinflammatory phenotype observed in a portion of COVID subjects is in contrast to the majority of COVID-19-infected patients who exhibit a more regulated physiologic immune response to infection remaining as Stage I subjects who are asymptomatic or experience only mild to moderate clinical illness with adequate oxygen saturation (Fig 1).3 Interferon-related proteins are a complex array of proteins that influence immune function and may play a role in determining individual responses to COVID-19 infection. Consistent with the notion that interferons can contribute to inflammatory injury, mutations in interferon regulatory factor 7 (IRF7), a member of the interferon regulatory factor family of transcription factors, was associated with life-threatening COVID-19 disease.49 However, the complexity of interferon responses is exemplified by studies showing interferon beta to exhibit antiviral effects against coronaviruses, including SARS-CoV and MERS-CoV. Autoantibodies against type I IFN were more common among patients with more severe life-threatening COVID-1950 and inhaled interferon beta-1a has been proposed as a potential treatment against COVID-19. The efficacy of such an approach is unclear, however, as a Phase 3 ARDS clinical trial (INTEREST, Faron Pharma), evaluating Traumakine, an IV-delivered recombinant human interferon beta-1a, was uniformly unsuccessful.51 Clearly, the genetic and nongenetic predictors of disparate phenotypes of the dysregulated inflammatory response in COVID-19 patients require further investigation. Strategies to identify the “at risk” hyperinflammatory subgroup are critical in guiding innovative clinical trial designs of novel anti-inflammatory therapeutics.
Anti-Inflammatory Therapeutic Strategies in COVID-19
A key feature of patients who progress from Stage I COVID-19 disease to increasingly severe Stage II and Stage III disease is blood biomarker evidence of the maladaptive DAMP-mediated loss of innate immunity regulation with excessive levels of circulating proinflammatory cytokines and chemokines (IL-6, IL-2, IL-7, IL-10, G-CSF, CXCL-10, MCP1, IFNγ, MIP1α, TNFα).5 Traditional biomarkers of acute systemic inflammation include C-reactive protein (CRP) and ferritin which positively correlate with severity of COVID-19 disease,52 , 53 but are of limited utility as they lack specificity and do not inform therapeutic decisions. In addition to dysregulated cytokine release, excessive secretion of proteases and generation of reactive oxygen species (ROS) by infiltrating immune cells contribute to the hyperinflammation characteristic of severe COVID-19 disease (Fig 2). The role of circulating levels of these proteins as diagnostic or prognostic biomarkers has been extensively investigated in non-COVID-19 ARDS21 , 54 , 55 but to date have not yet been utilized in clinical decision-making.55 In COVID-19 ARDS, the utility of plasma biomarkers as important diagnostic or prognostic biomarkers and as targets for therapeutics remains under investigation.
Discussion of relevant or promising ARDS biomarkers is germane to addressing the previously emphasized major unmet need for successful ARDS therapies. The prominent role of DAMP-mediated hyperinflammation in COVID-19 ARDS initially led to rapid repurposing of anti-inflammatory drugs for COVID-19 ARDS that were previously shown to be effective for cancer and autoimmune disorders indications. Unfortunately, multiple clinical trials with anti-inflammatory drugs have failed leading to speculation that therapeutic targeting of the “cytokine storm” that is emblematic of non-COVID ARDS may be of limited utility in COVID-19-ARDS.56 The primary basis for this line of reasoning has evolved from 2 observations which center on the interleukin-6 (IL-6) pathway, a well-recognized inflammatory pathway in ARDS pathobiology.57 , 58 First, median circulating IL-6 levels in 5 COVID-19 cohorts were observed to be lower when compared to cohorts of non-COVID-19 ARDS.56 Secondly, several high-profile efforts targeting IL-6 and IL-6 receptor antagonism (Tocilizumab-Roche/Sanofi, Sarilumab-Regeneron/Sanofi), in COVID ARDS failed to improve COVID-19 ARDS mortality in Phase 2/3 clinical trials of COVID-19 patients with severe disease59 (“Cleanup on IL-6”). The accumulated failure of targeting the interleukin-6 (IL-6) pathway may reflect several factors that are of specific relevance to IL-6, including the downstream nature of this cytokine in the inflammatory cascade, as well as the complex role of IL-6 in innate immunity, that is, IL-6 exhibits both context-dependent pro-and anti-inflammatory properties.57 , 58 , 60 IL-6 targeting may in fact increase risk to other infections, a particular risk in regions where drug-resistant bacteria are common.59
Despite the lack of success with anti-IL-6 strategies, there are currently more than 300 active trials of therapeutic agents by the global academic and biopharma communities which target SARS-CoV-2 and the complications of COVID-19-ARDS including anti-inflammatory therapies (Table 1 ). For example, studies evaluating the efficacy of GM-CSF, a pleiotropic growth factor and proinflammatory cytokine, were recently halted and the sponsor is seeking FDA approval, but details have not been published. To date only 3 therapies are approved for adult COVID-19 patients, the antiviral treatments Remdesivir (USA, Japan, Australia) and Favilavir (China, Italy, Russia) and the steroid preparation Dexamethasone (United Kingdom, Japan). The UK RECOVERY trial found dexamethasone to reduce COVID-19 fatalities when administered to severely hypoxemic patients or those requiring mechanical ventilation.61 Dexamethasone has not been shown to benefit patients with mild COVID-19 (Stage I, Fig 1) who do not require oxygen support and may carry the risk of reducing antivirus defenses. Details on various antivirals, antibodies, cell-based therapies, devices, RNA-based treatments, repurposed compounds, convalescent serum62 and other therapeutic modalities63, 64, 65 are constantly curated and cataloged.
Table 1.
Summary of anti-inflammatory therapeutics for COVID-19 pneumonia/ARDS*
| Medication Class/Mechanism | Trade name (generic name) | Sponsor/Trial phase | Status |
|---|---|---|---|
| Glucocorticoid | Dexamethasone | University of Oxford/Phase 2/3 | Approved UK |
| IL-6 receptor mAb | Actemra (Tocilizumab) Kevzara (Sarilumab) |
Roche/Phase 2/3 Sanofi; Regeneron/Phase 2/3 |
COVACTA EMPACTA CURIMUNO NCT04327388 |
| IL-6 mAb | Siltuximab Anakinra |
Judit Pich Martinez/Phase 2 Fundacion Miguel Servet/Phase 2/3 |
NCT04329650 NCT04443881 |
| TNF mAb | Humira (Adalimumab) Remicade (Infliximab) Remsima (Infliximab) |
University of Oxford/Phase 2 Janssen/Phase 2/3 |
CATALYST trial NCT04593940 |
| Celltrion/Phase 2 | NCT04425538 | ||
| CSF2/GM-CSF mAb | Lenzilumab | Humanigen; Catalent/Phase 3 |
NCT04351152 |
| Bradykinin Antagonist | Takhzyro (Lanadelumab) |
Takeda/Phase 1 |
NCT04460105 |
| IL-1β mAb | Ilaris (Canakinumab) |
Novartis/Phase 3 |
NCT04362813 NCT04365153 |
| CCR5 co-receptor mAb | Pro 140 (Leronlimab) |
CytoDyn/Phase 2 |
NCT04343651 NCT04347239 |
| GM-CSF Receptor mAb | Mavrilimumab | Kiniksa Pharmaceuticals/Phase 2/3 |
NCT04399980 NCT04447469 |
| GM-CSF mAb | Gimsilumab | Roivant Sciences/Phase 2/3 | NCT04351243 |
| Otilimab TJM2 Lenzilumab |
MorphoSys; GSK/Phase 2 I-MAB Biopharma Humanigen Inc./Catalent Biologics/Phase 3 |
NCT04376684 NCT04341116 NCT04351152 |
|
| Interferon gamma mAb | Gamifant (Emapalumab) |
Swedish Orphan Biovitrum/Phase 2/3 | NCT04324021 |
| Angiopoietin 2 (ANG2) mAb | LY3127804 | Eli Lilly/Phase 2 | NCT04342897 |
| C5 complement inhibitor mAb | Ultomiris (Ravulizumab) |
Alexion/Phase 3 | NCT04369469 |
| Vasoactive intestinal peptide (VIP) antagonist | RLF-100 (Aviptadil) |
NeuroRx; Relief Therapeutics/Phase2/3 |
NCT04360096 NCT04311697 NCT04453839 |
| Tyrosine Kinase inhibitor | STI-5656 (Abivertinib) |
Sorrento Therapeutics/Phase 2 |
NCT04440007 NCT04528667 |
| Recombinant Fusion Protein – binds DAMPS | SACCOVID (CD24Fc) |
Oncoimmune/Phase 3 | NCT04317040 |
| VIP receptor agonist | PB1046 | PhaseBio/Phase 2 | NCT04433546 |
| Anti-gout agent | Colchicine | NHLBI/Phase 3 | NCT04322682 |
| Calpain SMI | BLD-2660 | Blade Therapeutics/Phase 2 | NCT04334460 |
| Recombinant human plasma | Rhu-pGSN (Gelsolin) |
BioAegis Therapeutics/Phase 2 | NCT04358406 |
| Dihydroorotate dehydrogenase (DHODH) inhibitor | PTC299 | PTC/Phase2/3 | NCT04439071 |
| Angiotensin-(1-7) peptide agonist | TXA127 | Constant Therapeutics/Phase 2 | NCT04401423 |
| RIPK1 Inhibitor | DNL758 (SAR44122) |
Danofi; Denali Therapeutics/Phase 1b | NCT04469621 |
| p38α/β MAPK) inhibitor | Losmapimod | Fulcrum Therapeutics/Phase 3 | NCT04511819 |
| Bruton's tyrosine kinase inhibitor | Calquence (Acalabrutinib) |
AstraZeneca/Phase 2 |
NCT04380688 NCT04346199 |
| Auto adipose-derived MSC | AdMSCs | Celtrex Therapeutics/Phase 2 | NCT04428801 |
Therapies directed at blocking viral entry or neutralizing the virus are not included. Abbreviation: DAMPs, damage-associated molecular pattern proteins; RIPK1, receptor-interacting serine/threonine-protein kinase; SMI, small molecule inhibitor.
Biomarkers of Systemic Inflammation Associated with COVID-19 Severity
Although a hyperinflammatory response to SARS-CoV-2 is a major cause for morbidity and mortality in COVID-19, there are currently no biomarkers that are predictive of pathogenic inflammatory responses, reliably predict did ease severity/mortality, or that are able to identify targetable immune pathways. General biomarkers of inflammation such as C-reactive protein, ferritin, and lactic dehydrogenase (LDH), and dysregulated coagulation (D Dimer) are persistently elevated in patients with severe SOVID-19 disease. Additionally, cytokines such as IL-1β, IL-6, IL-8, IL-10 and TNFα are significantly elevated in COVID-19 patients when compared to controls.66 , 67 The levels of IL-1β, IL-6, IL-8, and TNFα in patients with severe COVID-19 are comparable to those of patients receiving Chimeric antigen receptor T-cell (CAR) therapy.66 These biomarkers reflect general systemic inflammation and currently COVID-19 specific biomarkers are not known.
Extracellularly-Secreted Nicotinamide Phosphoribosyl-Transferase (eNAMPT) is a DAMP and Novel COVID-19 ARDS Target
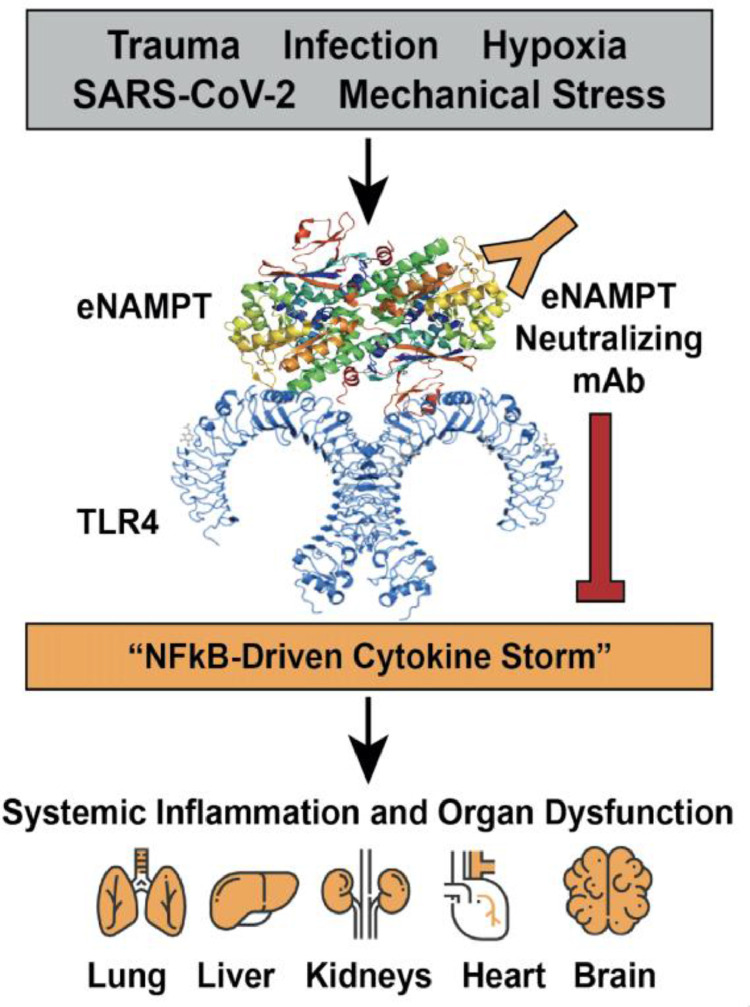
The absence of successful ARDS clinical trials that target inflammatory pathway components poses a vexing problem that potentially implicates several issues including: (1) poor target selection, that is, targeting cytokines downstream in the inflammatory cascade, and (2) delayed administration of the anti-inflammatory therapeutic ,that is, at a point where influencing the DAMP-mediated inflammatory cascade activation is minimal.68 , 69 We have previously utilized genomic–intensive approaches70, 71, 72 and cellular and preclinical studies10 , 73 , 74 of bacterial pneumonia and excessive mechanical stress/ventilator-induced lung injury (VILI) to identify eNAMPT as a novel DAMP and essential participant in ARDS/VILI pathobiology. Circulating eNAMPT functions as a master regulator of evolutionarily-conserved inflammatory cascades via ligation of Toll–like receptor 4 (TLR4)73 eliciting profound NFkB-driven inflammatory processes involved in ARDS/VILI pathobiology, including the loss of lung vascular barrier integrity (Fig 3 ). 10 , 73 , 74 We and others have demonstrated key NAMPT SNPs present at minor allelic frequencies >5% in both non-Hispanic whites and in Blacks, confer increased risk of developing sepsis/trauma-induced ARDS/VILI and confer increased ARDS severity and mortality (reduced ventilator-free days, increased ARDS mortality).72 , 75, 76, 77 Importantly, leveraging preclinical murine, rat and porcine studies of ARDS/VILI, we demonstrated that eNAMPT is a highly druggable target10 , 73 with a humanized eNAMPT-neutralizing mAb proven to be efficacious in preclinical ARDS/ VILI models (Fig 3).74 , 78
Fig 3.
eNAMPT is a novel DAMP in COVID-19 infection and in the development of ARDS. In response to a variety of injurious ARDS-relevant stimuli, including trauma, hypoxia, mechanical stress (generated by mechanical ventilation) and SARS-CoV2 infection, the NAMPT gene is activated, primarily in epithelial cells, leukocytes and vascular endothelial cells, to generate and secrete eNAMPT into the blood.10,73 Circulating eNAMPT functions as a damage-associated molecular pattern protein or DAMP via ligation of pathogen-recognition receptor, TLR4, eliciting NFkB-mediated gene expression and activation of systemic inflammatory cascades.73 The elaborated cytokines, that is, the “cytokine storm,” produce systemic inflammation with increases in vascular permeability, organ edema and multiorgan failure, the main contributor to ARDS mortality. ARDS, acute respiratory distress syndrome; DAMP, damage-associated molecular pattern protein; eNAMPT, extracellularly-secreted nicotinamide phosphoribosyl-transferase.
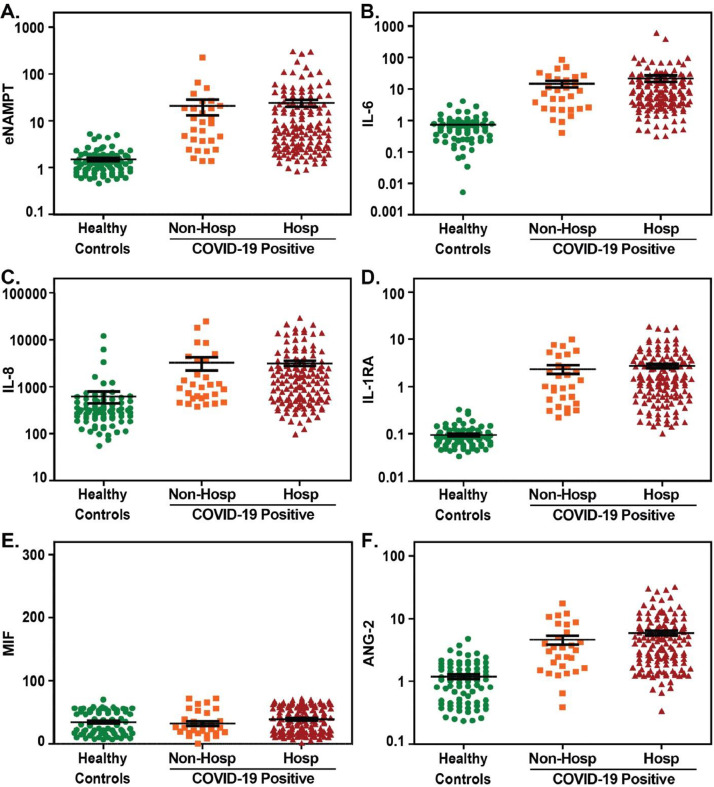
In addition to ARDS- associated NAMPT SNPs and genotypes, eNAMPT is tightly linked to human ARDS by eNAMPT levels that are elevated in in plasma and BAL in ARDS subjects and serve as a biomarker for human ARDS severity.55 , 72 , 79 , 80 Blood levels of secreted eNAMPT are linked to NAMPT transcriptional regulation which is highly induced by ARDS-relevant stimuli (hypoxia, trauma, infection, ventilator stress) (Fig 3).77 , 81, 82, 83 Our preliminary evaluation of plasma eNAMPT levels in hospitalized and non-hospitalized COVID-19-positive patients (Fig 4 ) show significantly elevated plasma eNAMPT levels in COVID-19-positive subjects compared to healthy controls, with higher eNAMPT levels in hospitalized COVID-19-positive subjects compared to non- hospitalized subjects. The magnitude of plasma eNAMPT elevation in COVID-19 subjects with ARDS was similar to our prior reports in non-COVID ARDS due to sepsis and trauma.55 Although the sample size is small, with the exception of MIF, significantly elevations in additional ARDS-associated biomarkers (IL-6, IL-8, IL-1RA, ANG-2) were also noted in the same COVID-19-positive subjects (compared to controls) (Fig 4). With the availability of an eNAMPT-neutralizing humanized mAb effective in dampening VILI and inflammation,74 , 78 there appears substantial and compelling foundational basis for eNAMPT as a viable therapeutic target in ARDS/VILI to dampen inflammatory cascade amplification and improve ARDS survival.
Fig 4.
Plasma biomarkers levels in COVID-19 positive subjects. Plasma was obtained from healthy controls (n = 78), COVID-19 positive subjects (n = 168) including subjects that did not require hospitalization (n = 29) and subjects that required hospitalization (n = 139). Measurements of plasma biomarkers were performed using U-PLEX and R-PLEX MesoScale Discovery platform as we have previously reported74 and included assessment of eNAMPT (A), IL-6 (B), IL-8 (C), IL1-RA (D), macrophage migration inhibition factor or MIF (E), and angiopoietin-2 or ANG-2 (F). With the exception of MIF, multiple comparisons of the median values using analysis of variance for non-parametric Mann-Whitney and Kruskal-Wallis tests, revealed significant difference in each marker in the 3 groups compared to controls (P value <0.0001). All analyses performed with Stata and Graphpad Prism software. All subject recruitment was IRB-approved.
Summary: Unanswered Questions and Future Directions
Multiple challenging issues including the heterogeneity of the ARDS phenotype (COVID-19 and non-COVID-19), the complex interactions involving mediators of dysregulated inflammation, and the absence of clinically-useful biomarkers, all contribute to the lack of progress in addressing unmet needs in COVID and non-COVID ARDS. While the diagnostic and prognostic value of biomarkers of dysregulated inflammation in COVID-19 ARDS may be enhanced by combining multiple candidates, these issues remain a critical challenge to the conduct of successful therapeutic clinical trials by ARDS clinical trial networks in the United States. and world-wide. We have summarized the current understanding of key pathobiologic features of dysregulated lung and systemic inflammation associated with SARS-Co-V-2 infection and the landscape for anti-inflammatory pharmacotherapies. We highlight SARS-CoV-2-induced local and systemic release of various DAMPs and PAMPs to produce unremitting activation of the inflammatory cascade, increased vascular leakage, dysregulated coagulation, and multiorgan dysfunction contributing to COVID-19 ARDS mortality. There is an urgent need for national and international cohort studies to obtain detailed clinical data and biologic samples from hospitalized and nonhospitalized COVID-19-infected individuals to identify immune signatures/molecular biomarkers associated with clinical disease course. Given the well-recognized racial and ethnic disparities in ARDS mortality in the United States with racial and ethnic minorities, especially Blacks, Hispanics and Native Americans at increased risk of death from ARDS30 , 32 , 84 , 85 that has been dramatically highlighted in the current COVID-19 pandemic,86 the recruitment of ARDS cohorts with substantial diversity is mandated. Such diverse cohorts would improve our current understanding of ARDS biomarkers, allow for discovery of novel biomarkers, improve our ability to stratify patients for enrollment into clinical trials, and allow the prioritization of clinical interventions. Studies conducted with diverse “at risk” cohorts for ARDS mortality should focus on identification of high-risk ARDS subjects potentially incorporating an anti-inflammatory platform which combines the availability of predictive biomarkers, point of care ARDS genotype testing, and a highly efficacious biologic or small molecule therapy. These strategies may provide the best opportunity to deliver personalized medicine in the current COVID-19 pandemic landscape and to directly address the unmet need for strategies to improve ARDS outcomes.
Acknowledgments
We would like to acknowledge the contributions of the following: Dr. David T. Harris, Director, University of Arizona Health Sciences Biorepository, and Dr. Mrinalini Kala, Director, Department of Medicine Biospecimen Core, College of Medicine Phoenix, for preprocessing and storage of COVID-19 samples that were used in the biomarker analysis. We also wish to thank Anna Valencia MPH and Dr. David Horn for invaluable assistance in recruitment of COVID-19 patients. In addition, we thank Taylor Gregory, Vivian Reyes-Hernon, Mathew K. Hufford, and Heather D. Lynn for sample processing and performance of the MSD assays on COVID-19 and control samples reported herein.
Declaration of Competing Interest: Dr. Garcia is Founder and CEO of Aqualung Therapeutics Corp. (Tucson, AZ).
Funding: This work was supported by K08 HL141623 (CB); P01 HL126609 (JGNG); R41 HL147769 (JGNG); R42 HL145930 (JGNG).
References
- 1.Garg S, Kim L, Whitaker M, et al. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019 - COVID-NET, 14 States, March 1-30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:458–464. doi: 10.15585/mmwr.mm6915e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sokouti M, Sadeghi R, Pashazadeh S, et al. Comparative Global epidemiological investigation of SARS-CoV-2 and SARS-CoV diseases using meta-MUMS tool through incidence, mortality, and recovery rates. Arch Med Res. 2020;51:458–463. doi: 10.1016/j.arcmed.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 4.Siddiqi HK, Mehra MR. COVID-19 illness in native and immunosuppressed states: A clinical–therapeutic staging proposal. J Heart Lung Transplant. 2020;39:405–407. doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohn MK, Hall A, Sepiashvili L, Jung B, Steele S, Adeli K. Pathophysiology of COVID-19: mechanisms underlying disease severity and progression. Physiology. 2020;35:288–301. doi: 10.1152/physiol.00019.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li CK-f, Xu X. Host immune responses to SARS coronavirus in humans. Mol Biol SARS-Coronavirus. 2009:259–278. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7123234/pdf/978-3-642-03683-5_Chapter_16.pdf [Google Scholar]
- 7.Kumar S, Nyodu R, Maurya VK, Saxena SK. Host Immune Response and Immunobiology of Human SARS-CoV-2 Infection. Coronavirus Dis 2019 (COVID-19) 2020:43–53. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7189399/pdf/978-981-15-4814-7_Chapter_5.pdf [Google Scholar]
- 8.Wu J, Yan Z, Schwartz DE, Yu J, Malik AB, Hu G. Activation of NLRP3 inflammasome in alveolar macrophages contributes to mechanical stretch-induced lung inflammation and injury. J Immunol. 2013;190:3590–3599. doi: 10.4049/jimmunol.1200860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acute Respiratory Distress Syndrome N. Brower RG, Matthay MA, et al. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N Engl J Med. 2000;342:1301–1308. doi: 10.1056/NEJM200005043421801. [DOI] [PubMed] [Google Scholar]
- 10.Hong SB, Huang Y, Moreno-Vinasco L, et al. Essential role of pre-B-cell colony enhancing factor in ventilator-induced lung injury. Am J Respir Crit Care Med. 2008;178:605–617. doi: 10.1164/rccm.200712-1822OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katira BH. Ventilator-induced lung injury: classic and novel concepts. Respir Care. 2019;64:629–637. doi: 10.4187/respcare.07055. [DOI] [PubMed] [Google Scholar]
- 12.Ding Y, Wang H, Shen H, et al. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J Pathol. 2003;200:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu J, Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am J Pathol. 2007;170:1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Brand JM, Smits SL, Haagmans BL. Pathogenesis of Middle East respiratory syndrome coronavirus. J Pathol. 2015;235:175–184. doi: 10.1002/path.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klok FA, Kruip MJHA, van der Meer NJM, et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magro C, Mulvey JJ, Berlin D, et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh KA, Jordan K, Clyne B, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres Acosta MA, Singer BD. Pathogenesis of COVID-19-induced ARDS: implications for an ageing population. Eur Respir J. 2020;56 doi: 10.1183/13993003.02049-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teuwen L-A, Geldhof V, Pasut A, Carmeliet P. COVID-19: the vasculature unleashed. Nat Rev Immunol. 2020;20:389–391. doi: 10.1038/s41577-020-0343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bime C, Camp SM, Casanova N, et al. The acute respiratory distress syndrome biomarker pipeline: crippling gaps between discovery and clinical utility. Transl Res. 2020;226:105–115. doi: 10.1016/j.trsl.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spadaro S, Park M, Turrini C, et al. Biomarkers for acute respiratory distress syndrome and prospects for personalised medicine. J Inflamm. 2019;16:1. doi: 10.1186/s12950-018-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blondonnet R, Constantin JM, Sapin V, Jabaudon M. A pathophysiologic approach to biomarkers in acute respiratory distress syndrome. Dis Markers. 2016;2016 doi: 10.1155/2016/3501373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudek SM, Garcia JG. Cytoskeletal regulation of pulmonary vascular permeability. J Appl Physiol. 2001;91:1487–1500. doi: 10.1152/jappl.2001.91.4.1487. [DOI] [PubMed] [Google Scholar]
- 28.Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nat Rev Immunol. 2007;7:803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- 29.Mirzapoiazova T, Moitra J, Moreno-Vinasco L, et al. Non-muscle myosin light chain kinase isoform is a viable molecular target in acute inflammatory lung injury. Am J Respir Cell Mol Biol. 2011;44:40–52. doi: 10.1165/rcmb.2009-0197OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bime C, Poongkunran C, Borgstrom M, et al. Racial differences in mortality from severe acute respiratory failure in the United States: 2008-2012. Ann Am Thorac Soc. 2016;13:2184–2189. doi: 10.1513/AnnalsATS.201605-359OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christie JD, Ma SF, Aplenc R, et al. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med. 2008;36:2794–2800. doi: 10.1097/ccm.0b013e318186b843. [DOI] [PubMed] [Google Scholar]
- 32.Erickson SE, Shlipak MG, Martin GS, et al. Racial and ethnic disparities in mortality from acute lung injury. Crit Care Med. 2009;37:1–6. doi: 10.1097/CCM.0b013e31819292ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao L, Grant A, Halder I, et al. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol. 2006;34:487–495. doi: 10.1165/rcmb.2005-0404OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moss M, Mannino DM. Race and gender differences in acute respiratory distress syndrome deaths in the United States: an analysis of multiple-cause mortality data (1979–1996)*. Crit Care Med. 2002;30:1679–1685. doi: 10.1097/00003246-200208000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Nicolai L, Leunig A, Brambs S, et al. Immunothrombotic dysregulation in COVID-19 pneumonia is associated with respiratory failure and coagulopathy. Circulation. 2020;142:1176–1189. doi: 10.1161/CIRCULATIONAHA.120.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris G, Bortolasci CC, Puri BK, et al. Preventing the development of severe COVID-19 by modifying immunothrombosis. Life Sci. 2020 doi: 10.1016/j.lfs.2020.118617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito T. PAMPs and DAMPs as triggers for DIC. J Intensive Care. 2014;2:67. doi: 10.1186/s40560-014-0065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frantzeskaki F, Armaganidis A, Orfanos SE. Immunothrombosis in acute respiratory distress syndrome: cross talks between inflammation and coagulation. Respiration. 2017;93:212–225. doi: 10.1159/000453002. [DOI] [PubMed] [Google Scholar]
- 39.Biswas I, Khan GA. Coagulation disorders in COVID-19: role of toll-like receptors. J Inflamm Res. 2020;13:823–828. doi: 10.2147/JIR.S271768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnappauf O, Chae JJ, Kastner DL, Aksentijevich I. The pyrin inflammasome in health and disease. Front Immunol. 2019;10:1745. doi: 10.3389/fimmu.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soy M, Keser G, Atagündüz P, Tabak F, Atagündüz I, Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. 2020;39:2085–2094. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andersson U, Ottestad W, Tracey KJ. Extracellular HMGB1: a therapeutic target in severe pulmonary inflammation including COVID-19? Mol Med. 2020;26:42. doi: 10.1186/s10020-020-00172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersson U, Wang H, Palmblad K, et al. High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J Exp Med. 2000;192:565–570. doi: 10.1084/jem.192.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerr N, de Rivero Vaccari JP, Dietrich WD, Keane RW. Neural-respiratory inflammasome axis in traumatic brain injury. Exp Neurol. 2020;323 doi: 10.1016/j.expneurol.2019.113080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zohar T, Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat Rev Immunol. 2020;20:392–394. doi: 10.1038/s41577-020-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, Bastard P, Liu Z, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bastard P, Rosen LB, Zhang Q, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ranieri VM, Pettila V, Karvonen MK, et al. Effect of intravenous interferon beta-1a on death and days free from mechanical ventilation among patients with moderate to severe acute respiratory distress syndrome: a randomized clinical trial. JAMA. 2020;323:725–733. doi: 10.1001/jama.2019.22525. [DOI] [PubMed] [Google Scholar]
- 52.Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020;58:1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 53.Gandini O, Criniti A, Ballesio L, et al. Serum Ferritin is an independent risk factor for acute respiratory distress syndrome in COVID-19. J Infect. 2020;81:979–997. doi: 10.1016/j.jinf.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bime C, Casanova N, Oita RC, et al. Development of a biomarker mortality risk model in acute respiratory distress syndrome. Crit Care. 2019;23:410. doi: 10.1186/s13054-019-2697-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinha P, Matthay MA, Calfee CS. Is a “Cytokine Storm” Relevant to COVID-19? JAMA Intern Med. 2020;180:1152–1154. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 57.Flores C, Ma SF, Maresso K, Wade MS, Villar J, Garcia. JG. IL6 gene-wide haplotype is associated with susceptibility to acute lung injury. Transl Res. 2008;152:11–17. doi: 10.1016/j.trsl.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 58.Goldman JL, Sammani S, Kempf C, et al. Pleiotropic effects of interleukin-6 in a "two-hit" murine model of acute respiratory distress syndrome. Pulm Circ. 2014;4:280–288. doi: 10.1086/675991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stone JH, Frigault MJ, Serling-Boyd NJ, et al. Efficacy of tocilizumab in patients hospitalized with Covid-19. N Engl J Med. 2020;383:2333–2344. doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta. 2011;1813:878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 61.RECOVERY Collaborative Group. Horby P, Lim WS, et al. Dexamethasone in hospitalized patients with Covid-19 - preliminary report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2021436. https://www.nejm.org/doi/full/10.1056/NEJMoa2021436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agarwal A, Mukherjee A, Kumar G, et al. Convalescent plasma in the management of moderate covid-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial) BMJ. 2020;371:m3939. doi: 10.1136/bmj.m3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Society RAP. COVID-19 therapeutics tracker. 2020:https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-therapeutics-tracker.
- 64.Institute M COVID-19 treatment and vaccine tracker. 2020:https://covid-19tracker.milkeninstitute.org.
- 65.Bio. BIO COVID-19 therapeutic development tracker. 2020:https://www.bio.org/policy/human-health/vaccines-biodefense/coronavirus/pipeline-tracker.
- 66.Del Valle DM, Kim-Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keddie S, Ziff O, Chou MKL, et al. Laboratory biomarkers associated with COVID-19 severity and management. Clin Immunol. 2020;221 doi: 10.1016/j.clim.2020.108614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fuller BM, Mohr NM, Hotchkiss RS, Kollef MH. Reducing the burden of acute respiratory distress syndrome: the case for early intervention and the potential role of the emergency department. Shock. 2014;41:378–387. doi: 10.1097/SHK.0000000000000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malaviya R, Laskin JD, Laskin DL. Anti-TNFalpha therapy in inflammatory lung diseases. Pharmacol Ther. 2017;180:90–98. doi: 10.1016/j.pharmthera.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grigoryev D, Ma S-F, Irizarry R, Ye S, Quackenbush J, Garcia J. Orthologous gene-expression profiling in multi-species models: search for candidate genes. Genome Biology. 2004;5:R34. doi: 10.1186/gb-2004-5-5-r34. R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simon BA, Easley RB, Grigoryev DN, et al. Microarray analysis of regional cellular responses to local mechanical stress in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2006;291:L851–L861. doi: 10.1152/ajplung.00463.2005. [DOI] [PubMed] [Google Scholar]
- 72.Ye SQ, Simon BA, Maloney JP, et al. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med. 2005;171:361–370. doi: 10.1164/rccm.200404-563OC. [DOI] [PubMed] [Google Scholar]
- 73.Camp SM, Ceco E, Evenoski CL, et al. Unique toll-like receptor 4 activation by NAMPT/PBEF induces NFkappaB signaling and inflammatory lung injury. Sci Rep. 2015;5:13135. doi: 10.1038/srep13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quijada H, Bermudez T, Kempf CL, et al. Endothelial eNAMPT amplifies preclinical acute lung injury: efficacy of an eNAMPT-neutralising mAb. Eur Respir J. 2020 doi: 10.1183/13993003.02536-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bajwa EK, Yu CL, Gong MN, Thompson BT, Christiani DC. Pre-B-cell colony-enhancing factor gene polymorphisms and risk of acute respiratory distress syndrome. Crit Care Med. 2007;35:1290–1295. doi: 10.1097/01.CCM.0000260243.22758.4F. [DOI] [PubMed] [Google Scholar]
- 76.O'Mahony DS, Glavan BJ, Holden TD, et al. Inflammation and immune-related candidate gene associations with acute lung injury susceptibility and severity: a validation study. PLoS One. 2012;7:e51104. doi: 10.1371/journal.pone.0051104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun X, Elangovan VR, Mapes B, et al. The NAMPT promoter is regulated by mechanical stress, signal transducer and activator of transcription 5, and acute respiratory distress syndrome-associated genetic variants. Am J Respir Cell Mol Biol. 2014;51:660–667. doi: 10.1165/rcmb.2014-0117OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bermudez T, Sammani S, Song JH, et al. An eNAMPT-neutralizing mAb reduces rat and porcine lung injury severity in preclinical ARDS/VILI. Am J Resp Crit Care Med. 2020 submitted. [Google Scholar]
- 79.Lee K, Huh JW, Lim CM, Koh Y, Hong SB. Clinical role of serum pre-B cell colony-enhancing factor in ventilated patients with sepsis and acute respiratory distress syndrome. Scand J Infect Dis. 2013;45:760–765. doi: 10.3109/00365548.2013.797600. [DOI] [PubMed] [Google Scholar]
- 80.Lee KA, Gong MN. Pre-B-cell colony-enhancing factor and its clinical correlates with acute lung injury and sepsis. Chest. 2011;140:382–390. doi: 10.1378/chest.10-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adyshev DM, Elangovan VR, Moldobaeva N, Mapes B, Sun X, Garcia JG. Mechanical stress induces pre-B-cell colony-enhancing factor/NAMPT expression via epigenetic regulation by miR-374a and miR-568 in human lung endothelium. Am J Respir Cell Mol Biol. 2014;50:409–418. doi: 10.1165/rcmb.2013-0292OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elangovan VR, Camp SM, Kelly GT, et al. Endotoxin- and mechanical stress-induced epigenetic changes in the regulation of the nicotinamide phosphoribosyltransferase promoter. Pulm Circ. 2016;6:539–544. doi: 10.1086/688761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun X, Sun BL, Babicheva A, et al. Direct extracellular NAMPT involvement in pulmonary hypertension and vascular remodeling. Transcriptional regulation by SOX and HIF-2alpha. Am J Respir Cell Mol Biol. 2020;63:92–103. doi: 10.1165/rcmb.2019-0164OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Erickson S, Martin G, Davis J, Matthay M, Eisner M. Recent trends in acute lung injury mortality: 1996-2005. Crit Care Med. 2009;37:1574–1579. doi: 10.1097/CCM.0b013e31819fefdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garcia JG, Sznajder JI. Healthcare disparities in patients with acute respiratory distress syndrome. Toward equity. Am J Respir Crit Care Med. 2013;188:631–632. doi: 10.1164/rccm.201307-1394ED. [DOI] [PubMed] [Google Scholar]
- 86.Thakur N, Lovinsky-Desir S, Bime C, Wisnivesky JP, Celedón JC. The structural and social determinants of the racial/ethnic disparities in the U.S. COVID-19 Pandemic. What's our role? Am J Respir Crit Care Med. 2020;202:943–949. doi: 10.1164/rccm.202005-1523PP. [DOI] [PMC free article] [PubMed] [Google Scholar]