Abstract
The current health crisis of corona virus disease 2019 (COVID-19) highlights the urgent need for vaccine systems that can generate potent and protective immune responses. Protein vaccines are safe, but conventional approaches for protein-based vaccines often fail to elicit potent and long-lasting immune responses. Nanoparticle vaccines designed to co-deliver protein antigens and adjuvants can promote their delivery to antigen-presenting cells and improve immunogenicity. However, it remains challenging to develop vaccine nanoparticles that can preserve and present conformational epitopes of protein antigens for induction of neutralizing antibody responses. Here, we have designed a new lipid-based nanoparticle vaccine platform (NVP) that presents viral proteins (HIV-1 and SARS-CoV-2 antigens) in a conformational manner for induction of antigen-specific antibody responses. We show that NVP was readily taken up by dendritic cells (DCs) and promoted DC maturation and antigen presentation. NVP loaded with BG505.SOSIP.664 (SOSIP) or SARS-CoV-2 receptor-binding domain (RBD) was readily recognized by neutralizing antibodies, indicating the conformational display of antigens on the surfaces of NVP. Rabbits immunized with SOSIP-NVP elicited strong neutralizing antibody responses against HIV-1. Furthermore, mice immunized with RBD-NVP induced robust and long-lasting antibody responses against RBD from SARS-CoV-2. These results suggest that NVP is a promising platform technology for vaccination against infectious pathogens.
Keywords: COVID-19, SARS-CoV-2, HIV, Vaccine, Nanoparticle
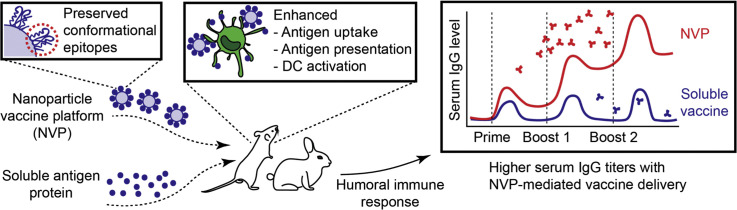
Graphical abstract
A nanoparticle vaccine platform that preserves conformational epitopes of protein antigens elicits robust humoral immune responses against HIV-1 and SARS-CoV-2.
List of abbreviations
| ACE2 | angiotensin-converting enzyme-2 |
| AF647 | alexa Fluor 647 |
| AIDS | acquired immunodeficiency syndrome |
| BTP | bis-tris propane |
| COVID-19 | corona virus disease 2019 |
| DC | dendritic cell |
| DLS | dynamic light scattering |
| DOPC | 1,2-dioleoyl-sn-glycero-3-phosphocholine |
| DOPE-NHS | N-(Succinimidyloxy-glutaryl)-L-α-phosphatidylethanolamine, dioleoyl |
| DQ-OVA | DQ-ovalbumin |
| HIV-1 | human immunodeficiency virus |
| MPLA | monophosphoryl lipid A |
| NVP | nanoparticle vaccine platform |
| PEI | polyethyleneimine |
| RBD | receptor binding domain |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus-2 |
| SDS-PAGE | sodium dodecyl sulphate–polyacrylamide gel electrophoresis |
| SOSIP | trimeric HIV-1 glycoprotein 140 stabilized by disulfide bond (SOS) and Ile/Pro substitution (IP) |
1. Introduction
As shown during the current COVID-19 pandemic, reliable and efficient vaccine delivery systems are urgently needed for vaccine development against COVID-19 as well as other emerging pathogens [1,2]. Traditional vaccines based on the live attenuated virus and inactivated virus vaccines are potent activators of the immune system, but they are limited by potential viral reversion and long development and regulatory timeline. On the other hand, protein vaccines with favorable safety profiles have been widely used for prophylactic vaccination against various pathogens, such as hepatitis B and influenza viruses [3]. Yet, protein subunit vaccines often fail to elicit potent and long-lasting immune responses.
These challenges may be addressed by co-administering subunit protein vaccines with potent adjuvants [4], especially in nanoparticle formulations that allow for their co-delivery to antigen-presenting cells for strong immune activation [5]. There are various nanoparticle vaccine platforms under development, including polymers [6,7], gold [8,9], silica [10,11], and others [12,13]. In particular, lipid-based nanoparticles are generally considered to have excellent biocompatibility and safety, and they have been used as a vaccine carrier to deliver mRNA [14], DNA [[15], [16], [17]], and peptides [18]. However, for protein antigens, it remains challenging to preserve their conformational epitopes and achieve robust neutralizing antibody responses using nano-vaccines. In particular, conformational display of antigens in vaccine formulations is crucial as immunogens should present epitopes to which the immune cells recognize, interact, and generate immune responses. Here, we sought to address these challenges by designing a new lipid-based nanoparticle vaccine platform (NVP) that can load and present viral proteins (HIV-1 and SARS-CoV-2 antigens) in a conformational manner for induction of antigen-specific antibody responses.
In particular, previous studies on acquired immunodeficiency syndrome (AIDS) have revealed the presence of broadly neutralizing antibodies in a subset of AIDS patients [19,20]. As the human immunodeficiency virus-1 (HIV-1) envelope glycoprotein (Env gp) has been identified to induce broadly neutralizing antibodies, various HIV-1 Env gp immunogens have been developed. Among them, BG505.SOSIP.664 (SOSIP) has emerged as a promising immunogen for inducing neutralizing antibodies against HIV-1 [[21], [22], [23]]. SOSIP is derived from the BG505 HIV-1 clade A virus, which was isolated from a 6-week-old infant who later developed broadly neutralizing antibodies [23,24]. The native form of the glycoprotein gp160 is cleaved into gp120 and gp41 subunits during HIV-1 entry into host cells. To utilize gp160 for vaccination purpose, the membrane-associated and cytoplasmic domains were truncated and stabilized by insertion of a disulfide bond (referred to as “SOS”) and an Ile/Pro (“IP”) substitution at residue 559 (I559P), resulting in an immunogen termed as SOSIP [[21], [22], [23]]. SOSIP self-assembles into a soluble HIV-1 Env trimer; therefore, nano-vaccine formulation with SOSIP should maintain the structural integrity and neutralizing epitopes of SOSIP.
During the initial COVID-19 outbreak, the structural similarities between SARS-CoV and SARS-CoV-2 were discovered [25]. It was subsequently revealed that spike glycoprotein (S protein) of SARS-CoV-2 was responsible for viral infection via interaction with angiotensin-converting enzyme 2 (ACE2) receptors on human cell membranes and that the antibodies generated against the S protein effectively neutralize viral entry to human cells [26,27]. The receptor-binding domain (RBD) is the functional domain within the S protein that first engages with ACE2, is considered a prime target for COVID-19 vaccine development, and can be produced as a recombinant antigen to generate directed antibody responses [28,29].
It should be noted that both SOSIP and RBD possess tertiary molecular structures through various bonds, including disulfide bond [30,31], that are prone to denaturation if placed under harsh condition, e.g., extreme pH, temperature, and physical stress. While we have previously reported lipid-based vaccine nanoparticles that employ thiol-maleimide crosslinking reaction to form nanoparticles [[32], [33], [34], [35]], they are not ideal for immunogens held together by disulfide bonds, such as in SOSIP. Therefore, we sought to design a new nano-formulation for loading HIV-1 SOSIP and SARS-CoV-2 RBD while preserving epitopes for inducing antibody responses. We show that pre-formed lipid vesicles incubated with protein antigens, followed by complexation and stabilization of lipid layers with branched polyethyleneimine (PEI), forms a nanoparticle vaccine platform (NVP) capable of loading viral antigens in a conformational manner. NVP co-loaded with antigens and monophosphoryl lipid A (MPLA, a potent Toll-like receptor-4 agonist) was readily taken up by dendritic cells (DCs) and promoted DC maturation and antigen presentation. NVP carrying SOSIP or RBD was recognized and surface-bound by neutralizing antibodies, indicating the conformational display of antigens on the surfaces of NVP. Rabbits immunized with SOSIP-NVP elicited neutralizing antibody responses against HIV-1. Moreover, mice immunized with RBD-NVP induced robust and durable antibody responses against RBD from SARS-CoV-2. These results suggest that NVP is a promising platform technology for vaccination against infectious pathogens.
2. Materials & methods
2.1. Reagents
SOSIP proteins were kindly provided by Drs. John Moore and Rogier Sanders from Weill Medical College, Cornell University, New York. SARS-CoV-2 RBD was produced in HEK293T cells from a clone kindly provided by Prof. Florian Kramer (Icahn School of Medicine at Mt. Sinai). Antigen (SOSIP)-specific primary monoclonal antibodies b6 and PGV04 were kindly provided by the International AIDS Vaccine Initiative. Rabbit Anti-Mouse IgG H&L-HRP was purchased from Abcam. Goat Anti-Mouse IgG1-HRP and Goat Anti-Mouse IgG2a-HRP were purchased from Southern Biotech. Following antibodies were used for antigen display assay: human IgG1 kappa isotype (EMD Millipore), PE-conjugated anti-human IgG (Fcγ) secondary antibody (ebioscience), Alexa Fluor 488-labeled anti-human IgG1 Fc secondary antibody (Invitrogen), Anti-SARS-CoV-2 RBD Neutralizing Antibody, Human IgG1 (SAD-S35) (Acrobiosystems).
2.2. Synthesis and characterization of nanoparticle vaccine platform (NVP)
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) (Avanti Polar Lipids), n-(succinimidyloxy-glutaryl)-L-α-phosphatidylethanolamine, dioleoyl (DOPE-NHS) (NOF America Corporation) and monophosphoryl lipid A (MPLA) (Avanti Polar Lipids), all dissolved in chloroform, were mixed in 50:50:0.5 M ratio in a glass vial. Lipids were dried under nitrogen gas, followed by a further complete drying step by putting the vial inside a desiccator. The dried lipid film was rehydrated with 100 μl of bis-tris propane (BTP) buffer (pH 7.0) by vortexing for 10 s every minute for 7 min. The resulting multilamellar vesicles were probe tip-sonicated at 40% amplitude for 5 min while placed in ice. The resulting unilamellar vesicles (ULV) were mixed with 100 μl of either SOSIP (25 μg) - or RBD (20 μg)-containing BTP buffer and incubated in 37 °C for 30 min under constant shaking. Branched PEI of 1800 Da (number-based average) (Sigma Aldrich) was added at 0.35:1 primary amine:NHS ratio (NHS groups present in liposome), and then the mixture was further incubated at 37 °C for 30 min. The resulting antigen-loaded NVP suspension was centrifuged at 14,000g for 5 min, then were washed with PBS twice, followed by final resuspension with 200 μl of PBS. The particles were transferred to deionized water for size and surface charge measurement using the Zetasizer Nano (Malvern, UK). The loading efficiencies of proteins in NVP were measured by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), followed by Coomassie Blue staining (for SOSIP and RBD). Gels were imaged with a gel doc machine, Fluorchem M Imaging System (Protein Simple).
2.3. In vitro DC uptake assays
Mouse BMDCs were isolated from bone marrow obtained from hind femurs of C57BL/6 mice. Cells were cultured in media supplemented with GM-CSF for 10 days in 37 °C, 5% CO2. Mature BMDCs were seeded in 2 × 105 per well of a 96-well tissue culture plate (flow cytometry) or 1 × 105 per well of an 8-well chambered cover glass (confocal microscopy), incubated for at least 6 h for cell adhesion, and then treated with DQ-OVA (Invitrogen)-containing vaccine formulations for 1–24 h. For flow cytometry analysis, cells were recovered after trypsin treatment for 10 min. Retrieved cells were incubated with anti-CD16/32 blocking antibody for 10 min in room temperature (RT), followed by incubation with anti-CD11c, anti-CD80, and anti-SIINFEKL/MHC-I antibodies for 20 min in RT and a fixable viability dye (eFluor 450, eBioscience) for 10 min in RT. Cells were then fixed with 4% formaldehyde in PBS for 10 min, washed and resuspended in PBS containing 1% bovine serum albumin (BSA) and analyzed with a flow cytometer (Bio-Rad ZE5). For confocal microscopy, BMDCs were treated with DQ-OVA formulations for 4 h, followed by three times of washing with PBS. Cells were then stained with 0.1 μM Lysotracker (ThermoFisher L7528) and 1 μg/ml Hoechst (ThermoFisher H3570) in 37 °C for 30 min. After washing with PBS, cells were fixed with 4% formaldehyde in PBS for 15 min, followed by washing with PBS. Cells were then analyzed with Nikon A1Rsi confocal microscope.
2.4. Antigen display on NVP
For assessing antigen display on NVP, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine (DiD) (Invitrogen, 0.2 mol%) was added in the lipid composition of NVP. Fluorescence signal from DiD was used to normalize the amount of NVP for comparison between different formulations due to variance in the recovery of formulations. For SOSIP-NVP, SOSIP-specific antibodies, b6 and PGV04, were incubated with NVP, followed by washing in PBS and addition of PE-conjugated anti-human IgG (Fcγ) secondary antibody (ebioscience). For RBD-NVP, monoclonal neutralizing antibody (SAD-S35, Acrobiosystems) against SARS-CoV-2 was treated (1:100 dilution), followed by washing in PBS (x3) and addition of Alexa Fluor 488-labeled anti-human IgG1 Fc secondary antibody (1:50 dilution) (Invitrogen). Antibody incubations were performed at room temperature for 30 min with constant shaking. Resulting samples were measured with a fluorometer (Biotek Synergy Neo microplate reader) at excitation/emission wavelengths of 485 nm/528 nm and 630 nm/680 nm or by “NanoFACS” as we recently reported [33].
2.5. In vivo vaccination study
Animals were cared for following federal, state, and local guidelines. All experiments performed on animals were in accordance with and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Michigan, Ann Arbor. New Zealand White rabbits (6–8 weeks old females from Jackson Laboratory) were vaccinated subcutaneously at four sites on both caudal thighs (2 sites per side) with either soluble mixture SOSIP and MPLA or NVP co-loaded with SOSIP and MPLA. Doses used for primary and boost injections were 30 μg SOSIP +50 μg MPLA and 12.4 μg SOSIP +20.6 MPLA, respectively. Primary vaccination was injected on day 0, followed by boost vaccinations on days 28 and 84. 2–3 ml of blood was sampled from each rabbit via marginal ear vein on days 28, 56, 105 and 169, which was placed in room temperature undisturbed for 1 h to induce coagulation, followed by centrifugation at 2000g for 12 min at 4 °C to obtain serum. Rabbit immune sera were analyzed for neutralizing activities against tier 1A (MW965.26) and autologous tier 2 (BG505/T332N) viral entry using the TZM-bl cell assay, which measures transactivation of a luciferase reporter gene by an infecting virus [36,37]. BALB/c mice (6–8 weeks old females from Jackson Laboratory) were vaccinated subcutaneously at the tail-base on days 0, 14, and 28 with either soluble mixture RBD and MPLA or NVP co-loaded with RBD and MPLA. PBS was included as a negative control. The dose used for each injection was 0.5 μg RBD and 1 μg MPLA. Blood was sampled from each mouse via submandibular vein on days 14, 28 and 42. Samples were collected in a gel-containing tube (Microvette 500 SER-GEL, Sarstedt Inc.), followed by centrifugation at 10,000g for 5 min to obtain serum. Mouse immune sera were analyzed for RBD-specific IgG, IgG1, and IgG2a antibody titers using ELISA. Briefly, RBD protein was coated on 96-well ELISA plates (0.1 μg/well) and serially diluted sera samples were added. After one hr of incubation and multiple washings, horse radish peroxidase (HRP)-labeled secondary antibodies were added and further incubated for 1 h in room temperature. Secondary antibodies used included rabbit anti-mouse IgG H&L-HRP (Abcam), goat anti-mouse IgG1-HRP (Southern Biotech) or goat anti-mouse IgG2a-HRP (Southern Biotech). Next, TMB substrate solution was added, and the reaction was stopped by the addition of NaF. The absorbance was measured at a 620 nm wavelength using a plate reader (Synergy Neo, BioTek). To measure antibody titers, titration curves were drawn based on the absorbance and the dilution factor, from which half maximal effective concentration (EC50) values were calculated using a software Gen5 (BioTek).
2.6. Lymph node trafficking study
BALB/c mice were subcutaneously injected at the tail-base with PBS, soluble antigen + MPLA, or antigen-MPLA-NVP. As a model antigen for investigating lymph node trafficking, Alexa Fluor 647-labeled ovalbumin (AF647OVA) (Invitrogen) was used. A dose consisting of 10 μg of AF647OVA and 11 μg of MPLA was injected to each mouse. After 4, 8, 24 and 48 h of injection, serum samples were collected for ELISA to measure proinflammatory cytokine levels. After 48 h, draining lymph nodes were collected for fluorescence imaging to quantify the amount of antigen. Lymph nodes were placed in the imaging machine (IVIS spectrum, PerkinElmer) and imaged using 640 nm and 680 nm ex/em filters. Lymph nodes were then processed into single cells for flow cytometric analysis. Briefly, lymph nodes were homogenized with a mini-tissue homogenizer, then passed through a 40 μm strainer to collect single cells. After washing the cells twice with PBS (1% BSA), anti-CD16/32 Ab was added for 10 min at 4 °C, followed by the addition of anti-mouse CD80 Ab (FITC) (BD Biosciences), anti-mouse CD86 Ab (PECy7) (eBioscience), and anti-mouse CD11c Ab (PE) (BioLegend) Ab for 30 min at 4 °C. Then the cells were washed twice with PBS, stained for live/dead (EBioscience, Fixable viability dye eFluor 450), and fixed with 4% formaldehyde. Resulting cells were washed twice with PBS (1% BSA) and analyzed by flow cytometry.
2.7. Blue native PAGE
For Blue Native PAGE, samples were run on NativePAGE™ Novex® Bis-Tris gel system (Life Technologies). Briefly, samples were diluted in Native PAGE sample buffer, bath sonicated to disrupt aggregates, and were incubated with N-Dodecyl β-D-maltoside (DDM, Invitrogen) at a 1.11% working concentration for 30 min on ice. Immediately before loading onto gels (3%–12% Bis-Tris), Coomassie G-250 was added to the samples following manufacturer's instructions. Gels were run at room temperature using dark blue cathode buffer for approximately 100 min. Gels were destained according to manufacturer's instructions, and protein migration was assessed by silver staining (Thermo Fisher).
2.8. Statistical analysis
The results are expressed as means ± SEM. Prism 7.0e (GraphPad Software) software was used for statistical analyses. Statistical comparisons were performed using either unpaired student's t-test or one-way or two-way ANOVA, followed by Tukey's HSD multiple comparison test. Statistical significances are indicated as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
3. Results and discussion
3.1. Synthesis and characterization of NVP
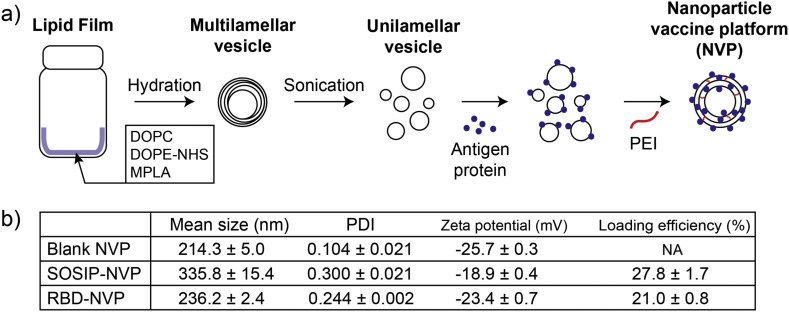
We synthesized a nanoparticle vaccine platform (NVP) by adding viral antigens to pre-formed unilamellar lipid vesicles, followed by incubation with branched PEI that allows for complexation and stabilization of vesicles into NVP via interaction between NHS-functionalized phospholipids and primary amine groups in PEI. Briefly, multilamellar vesicles were prepared by hydrating a lipid film composed of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), n-(Succinimidyloxy-glutaryl)-L-α-phosphatidylethanolamine, dioleoyl (DOPE-NHS) and monophosphoryl lipid A (MPLA, a Toll-like receptor-4 agonist) (50:50:0.5 mol ratio). Multilamellar vesicles were tip sonicated to form into unilamellar vesicles, which then were mixed and surface-loaded with antigen proteins. Branched PEI (1800 Da) was subsequently added in a primary amine:NHS ratio of 0.35:1 to induce complexation of lipid vesicles and nanoparticle formation by the reaction between the primary amines on PEI and the NHS groups on the unilamellar vesicles (Fig. 1a). The resulting NVP exhibited a hydrodynamic size of 200–400 nm and a polydispersity index of 0.1–0.3 depending on the antigen added, with slightly negative surface charge as shown by dynamic light scattering (DLS) measurement (Fig. 1b).
Fig. 1.
Preparation and characterization of NVP. (a) Schematic drawing of NVP preparation process. (b) Mean hydrodynamic size, PDI, and surface charge of NVPs as measured by DLS. Protein loading efficiency was calculated based SDS-PAGE analysis.
3.2. NVP improves antigen uptake by DCs and enhances DC activation and antigen presentation
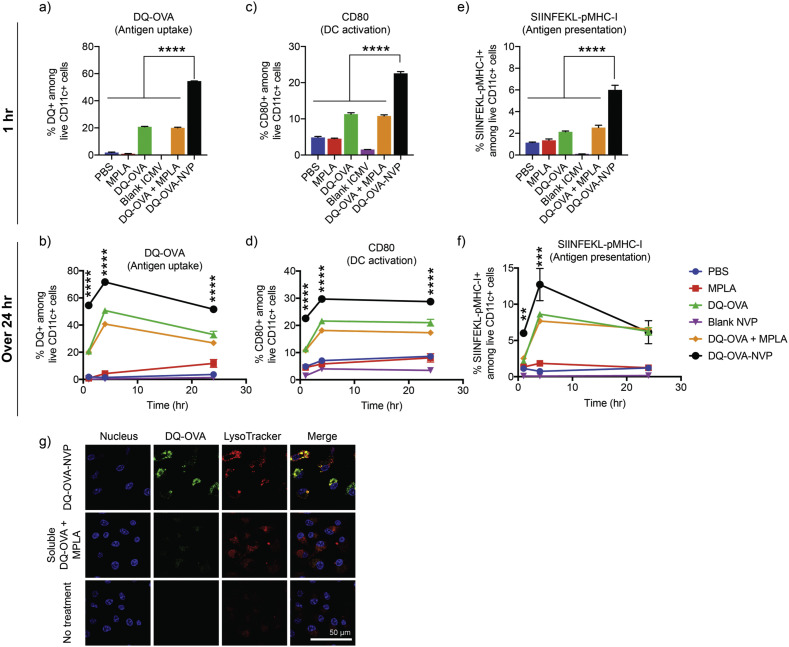
As a model antigen, DQ-labeled ovalbumin (DQ-OVA) was loaded to NVP to examine antigen delivery to and activation of DCs in vitro (Fig. 2a-f) DQ is a self-quenched dye that emits fluorescence upon degradation after cellular entry [38]. PAGE analysis showed efficient loading (~60%) of DQ-OVA to NVP (Fig. S1). BMDCs treated for 1 h with NVP carrying DQ-OVA (DQ-OVA-NVP) exhibited a 2.72-fold increase in the DQ signal, compared with those treated with soluble DQ-OVA and MPLA mixture, as confirmed by flow cytometry (Fig. 2a). A similar trend was observed over 24 h (Fig. 2b). As shown by confocal microscopy, BMDCs treated with DQ-OVA-NVP for 4 h showed significantly higher DQ signal within the cytosol, with a high level of co-localization with lysosomes (Fig. 2g). In addition, DQ-OVA-NVP induced robust DC maturation, as shown by the up-regulation of CD80 co-stimulatory marker on DCs within 1 h of incubation (Fig. 2c) and throughout 24 h time window (Fig. 2d). Moreover, we examined antigen presentation on DCs by flow cytometry assay after staining DCs with a monoclonal antibody specific to an immunodominant OVA epitope (SIINFEKL) loaded in major histocompatibility complex-I (MHC-I) molecule. NVP-mediated DQ-OVA delivery led to significantly greater antigen presentation on DCs (Fig. 2e-f). Taken together, these results show that NVP significantly increases DC uptake of vaccines, leading to improved DC activation and antigen presentation.
Fig. 2.
NVP enhances antigen uptake, activation and antigen processing by dendritic cells in vitro. (a-g) BMDCs were treated with DQ-OVA and MPLA using the indicated formulations, and DQ-OVA signal was quantified by (a-b) flow cytometry after 1, 4, and 24 h of incubation. BMDCs were also assessed for (c-d) CD80 expression and (e-f) antigen presentation of SIINFEKL on MHC-I. (g) After 4 h of incubation, BMDCs were stained with LysoTracker and Hoechst, followed by confocal microscopy. Data are presented as mean ± SEM. **p < 0.01, ****p < 0.0001, analyzed by one-way ANOVA (a,c,e) or two-way ANOVA (b,d,f), followed by Tukey's HSD multiple comparison post hoc test. Asterisks in (b,d,f) indicate statistical comparison between DQ-OVA-NVP and DQ-OVA.
3.3. NVP effectively delivers antigen to DCs in lymph nodes
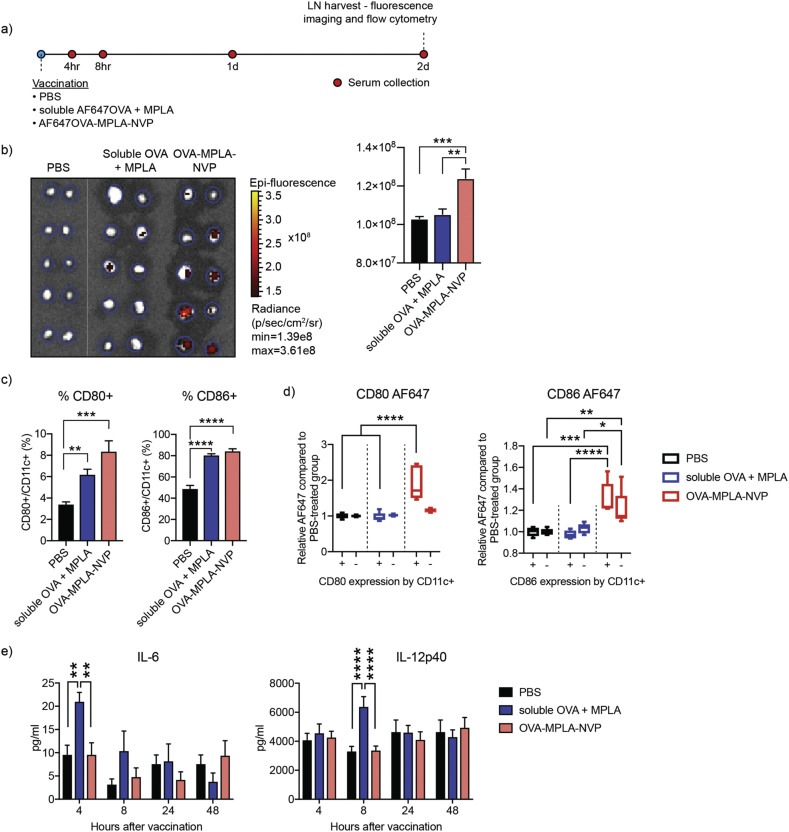
Next, we investigated NVP-mediated lymph node trafficking of antigen using a model antigen, Alexa Fluor 647 (AF647)-labeled ovalbumin (AF647OVA). Mice were given tail-base subcutaneous injection of PBS, soluble AF647OVA + MPLA, or AF647OVA-loaded NVP (OVA-MPLA-NVP), followed by flow cytometry or ELISA analyses (Fig. 3a). After 48 h of vaccination, draining inguinal lymph nodes were visualized by IVIS fluorescence imaging. Mice administered with OVA-MPLA-NVP had significantly stronger AF647 signal in lymph nodes, compared with those treated with the soluble formulation (Fig. 3b). Flow cytometric analysis showed that CD11c + DCs in lymph nodes from the OVA-MPLA-NVP group exhibited signs of maturation, as shown by CD80 and CD86 staining (Fig. 3c). Interestingly, among CD80+ and CD86+ DCs, the OVA-MPLA-NVP group had significantly higher mean fluorescence intensity of AF647OVA, compared with the soluble vaccine group (Fig. 3d), showing robust DC-targeted delivery of antigen by NVP. Lastly, the serum levels of proinflammatory cytokines, IL-6 and IL-12p40, were measured using ELISA. Serum concentrations of IL-6 and IL-12 were elevated at 4 h and 8 h post injection, respectively, for the soluble vaccine group (Fig. 3e), whereas there was no spike of either cytokines in the NVP group. Taken together, these results showed that NVP provides an efficient and safe platform for antigen delivery to antigen-presenting cells in lymph nodes.
Fig. 3.
NVP improves antigen delivery to antigen-presenting cells in lymph nodes. (a) Mice were administered subcutaneously with PBS, soluble AF647OVA + MPLA, or AF647OVA-MPLA-NVP, followed by tissue analysis on the indicated time points. (b) Inguinal lymph nodes were excised and imaged for AF617OVA signal at 48 h after vaccination. (c) CD11c + DCs in inguinal lymph nodes were analyzed for CD80 and CD86 maturation markers. (d) CD11c + DCs with or without upregulation of CD80 and CD86 were analyzed for AF647OVA. (e) At the indicated time points after vaccination, serum concentrations of IL-6 and IL-12p40 were analyzed by ELISA. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, analyzed by (b,c) one-way ANOVA or (d,e) two-way ANOVA, followed by Tukey's HSD multiple comparison test.
3.4. Loading recombinant HIV envelope glycoprotein (SOSIP) into NVP
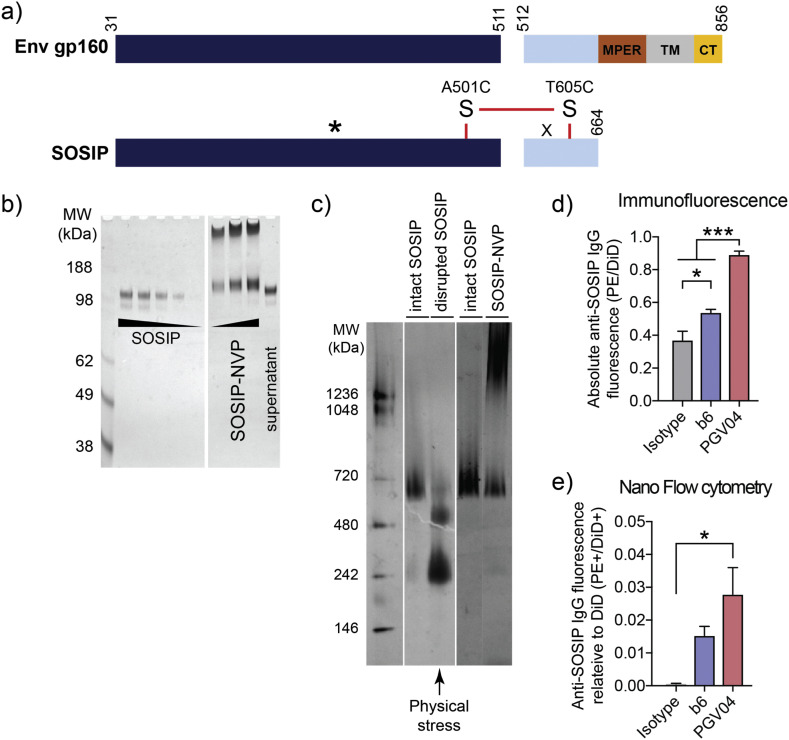
BG505.SOSIP.664 (SOSIP) is a recombinant HIV-1 envelope glycoprotein derived from the BG505 clade A virus. SOSIP is held together by a disulfide bond and self-assembles into a soluble HIV-1 Env trimer (Fig. 4a). SOSIP is a promising immunogen for HIV-1 vaccine development, as shown by prior pre-clinical studies reporting SOSIP-mediated induction of neutralizing antibodies against HIV-1 [21,22,39]. Here, we prepared NVP carrying SOSIP antigen and examined its efficacy to induce neutralizing antibody response against HIV-1. Using the procedure described above, we loaded SOSIP into NVP (SOSIP-NVP), which exhibited a hydrodynamic size of ~330 nm, as determined by DLS analysis (Fig. 1b). PAGE-based quantification indicated a ~ 25% loading efficiency of SOSIP in NVP (Fig. 4b). Notably, it is crucial to maintain the conformational epitopes and trimeric structure of HIV-1 Env for the induction of broadly neutralizing antibody responses [40]. Therefore, we examined whether SOSIP-NVP preserves the structure and epitopes of SOSIP during the vaccine formulation. Our non-reducing PAGE analysis performed on SOSIP retrieved from SOSIP-NVP indicated that SOSIP-NVP maintained the disulfide bond in SOSIP without disruption during the loading process (Fig. 4b). SOSIP also appeared in the high molecular weight area in the PAGE gel, which may have been due to complexation with PEI and incomplete retrieval process from SOSIP-NVP. In addition, the preservation of quaternary structure of SOSIP trimer after NVP loading was examined by blue-native PAGE. Interestingly, application of a significant physical stress (e.g. tip sonication) while SOSIP trimer is present in solution induced dissociation of the trimer into monomer and dimer, demonstrating the delicate binding force between the subunits (Fig. 4c, 3rd lane). Thus, we modified the SOSIP-NVP preparation by adding SOSIP to the reaction mixture after any physical stresses were taken place, which resulted in the preservation of intact quaternary structure after SOSIP-NVP formulation (Fig. 4c, 5th lane). Nevertheless, these results indicated that SOSIP was effectively loaded into NVP.
Fig. 4.
BG505.SOSIP.664 (SOSIP) protein and its display on NVP surface after loading. a) Genetic modifications from Env gp160 resulted in SOSIP, having truncation at residue 664, added glycan (indicated by *; T332N) and a disulfide bond, and other point mutations. MPER: membrane proximal region, TM: transmembrane domain, CT: cytosolic tail. b) Loading of SOSIP into NVP confirmed by non-reducing PAGE, followed by silver staining. c) Blue native PAGE showing intact SOSIP trimer before or after physical disruption as well as after loading in NVP using an optimized formulation condition. d-e) To examine SOSIP display on NVP, human anti-SOSIP antibodies, b6 and PGV04, were incubated with SOSIP-NVP and PE-labeled anti-human IgG antibody, followed by quantification of antibody binding by d) plate-based fluorescence measurement and e) individual nanoparticle-based flow cytometry. Data are presented as mean ± SEM. *p < 0.05, ***p < 0.001, analyzed by one-way ANOVA, followed by Tukey's HSD multiple comparison post hoc test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To further examine whether SOSIP was displayed on the surface of NVP with its epitopes remaining intact, we performed immunofluorescence directly on SOSIP-NVP. For this, we employed Env-specific human antibodies, PGV04 and b6, which recognize the CD4 binding site of Env. PGV04 and b6 are HIV-1 broadly neutralizing antibody and non-neutralizing antibody, respectively. PGV04 and b6 were incubated with SOSIP-NVP, followed by washing and another round of incubation with fluorophore-tagged anti-human IgG antibody. Then fluorescence signal on SOSIP-NVP was quantified to assess antibody binding (Fig. 4d, e). SOSIP-NVP was readily recognized and bound by PGV04, a broadly neutralizing antibody, on a whole population level, as shown by a plate-based fluorescence measurement (Fig. 4d). SOSIP-NVP was also bound by b6, a non-neutralizing antibody, but to a lesser extent than PGV04. We recently reported that antibody-binding on nanoparticles could be quantified on an individual nanoparticle-basis using “NanoFACS” [33]. Using NanoFACS, we confirmed that PGV04 was bound to individual SOSIP-NVPs (Fig. 4e), thus showing the homogenous display of SOSIP on the surfaces of SOSIP-NVPs. These results indicated the preservation of SOSIP epitopes after loading in NVP.
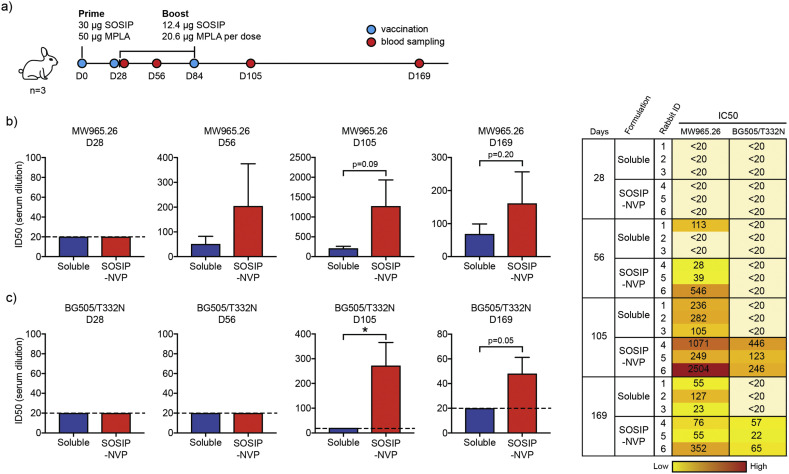
3.5. SOSIP-NVP vaccination study in rabbits
We performed immunization studies with SOSIP-NVP and examined their potency to generate neutralizing antibody response in rabbits. White New Zealand rabbits were immunized on day 0 with 30 μg SOSIP and 50 μg MPLA, followed by two boost immunizations on days 28 and 84, each with 12.4 μg SOSIP and 20.6 μg MPLA (Fig. 5a). SOSIP and MPLA were administered subcutaneously either in SOSIP-NVP or soluble formulations. Sera samples collected on days 28, 56, 105 and 169 were assessed for neutralization against HIV-1 viral entry to TZM-bl cells using heterologous tier 1A virus (MW965.26, clade C) and autologous tier 2 virus (BG505/T332N) [41,42]. On Day 56 and more noticeably on Day 105, immune sera from the SOSIP-NVP vaccine group showed strong neutralizing activity against heterologous titer 1A MW965.26 virus (Fig. 5b), with a trend for increasing neutralizing activity compared with the soluble vaccine group. Day 169 immune sera also showed neutralizing activity, although dampened compared with day 105, against MW965.26 (Fig. 5b). As BG505.SOSIP.664 trimer is derived from HIV-1 clade A, our results showing neutralizing activity against MW965.26 HIV-1 clade C virus indicated that SOSIP-NVP elicited a cross-neutralizing antibody response. Furthermore, day 105 immune sera from SOSIP-NVP immunized rabbits exhibited high neutralizing antibody titers against neutralization-resistant tier 2 BG505/T332N virus, whereas the soluble vaccine group induced no neutralizing antibody response (Fig. 5c). Moreover, there was also a trend for higher neutralizing antibody titers against autologous BG505/T332N up to day 169 (Fig. 5c). As it has been challenging to produce vaccines capable of neutralizing against a tier 2 virus, even an autologous one such as BG505/T332N virus, these results show the promise of SOSIP-NVP for vaccination against HIV-1.
Fig. 5.
Vaccination of rabbits using SOSIP-NVP enhances neutralizing antibody titers against HIV-1. (a) Rabbits were prime vaccinated on D0 and boost vaccinated on D28 and D84, followed by blood sampling on D28, D56, D105 and D169. (b-c) Immune sera were analyzed for neutralization of (b) tier 1A and (c) autologous tier 2 viruses in vitro. Data are presented as mean ± SEM. *p < 0.05, analyzed by unpaired student's t-test.
3.6. NVP carrying receptor-binding domain (RBD) of SARS-CoV-2
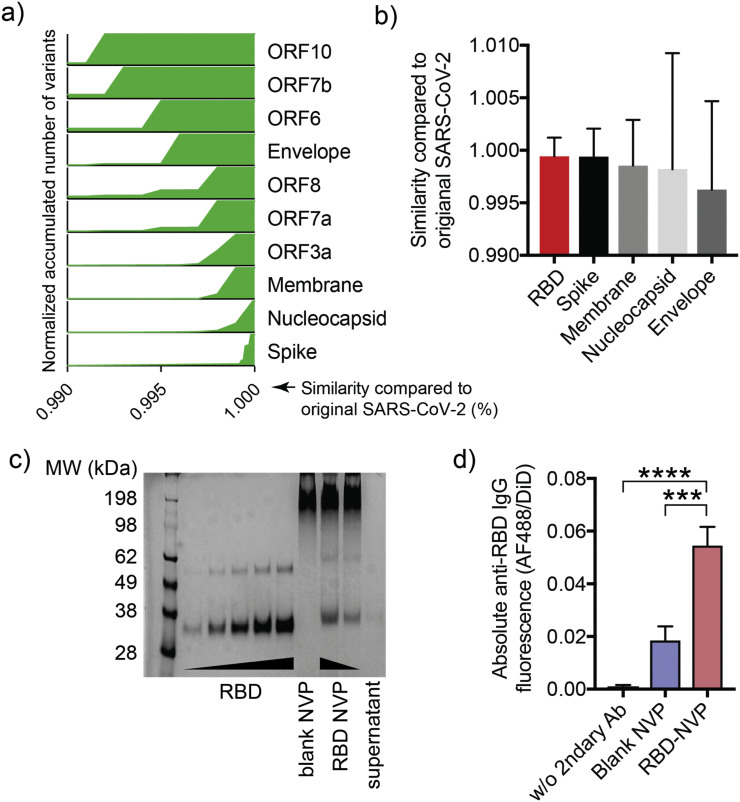
Motivated by the promising results of SOSIP-NVP, we sought to apply the NVP technology for COVID-19 vaccine development. According to previous studies, the spike protein (S protein) of SARS-CoV-2 is responsible for the interaction with ACE2 receptors on the human cells which leads to viral infection. Inducing antibody response against S protein therefore is an effective strategy to neutralize SARS-CoV-2 [26,27]. The receptor-binding domain (RBD) is the functional region within the S protein that engages ACE2, and has been suggested as a great target for vaccines against SARS-CoV-2 [28,29]. In addition, our analysis on genetic sequence comparison between SARS-CoV-2 variants reported on National Center for Biotechnology Information (NCBI) as of August 2020 and the original SARS-CoV-2 that appeared in Wuhan in 2019 indicated high conservation of RBD genetic sequence, compared with other genetic regions (Fig. 6a-b), thus highlighting RBD as a promising target for COVID-19 vaccine development.
Fig. 6.
Genomic deviation of SARS-CoV-2 by coding region and characterization of RBD-NVP. (a) Spike protein is the most genetically conserved region within the genetic sequence of SARS-CoV-2, based on variants appearing with near 100% similarity to the original SARS-CoV-2 in this region. (b) RBD of SARS-CoV-2 variants has the highest sequence similarity to that of the original SASR-CoV-2 with the smallest deviation, compared with other domains. (c) RBD loading in NVP was confirmed by SDS-PAGE analysis. (d) RBD display on NVP surface was assessed by incubation with human anti-SARS-CoV-2 neutralizing antibody, followed by incubation with Alexa Fluor 488-labeled anti-human IgG1 Fc secondary antibody. Antibody bound to NVP was quantified by fluorometry. Data are presented as mean ± SEM. ***p < 0.001, ****p < 0.0001, analyzed by one-way ANOVA, followed by Tukey's HSD multiple comparison post hoc test.
We synthesized RBD-loaded NVP as described above. Loading of RBD in NVP was quantified by PAGE analysis, which showed ~21% loading efficiency (Fig. 6c). The resulting RBD-loaded NVP (RBD-NVP) had a hydrodynamic size of ~240 nm as measured by DLS (Fig. 1b). Surface-display of RBD on RBD-NVP was examined by direct immunofluorescence as described. RBD-NVP was incubated with human RBD neutralizing antibodies, followed by washing and addition of AF488-labeled anti-human IgG secondary antibody. RBD-NVP exhibited a significantly higher fluorescence signal, compared with blank NVP control (Fig. 6d), indicating the proper display of RBD and preservation of epitopes in RBD-NVP.
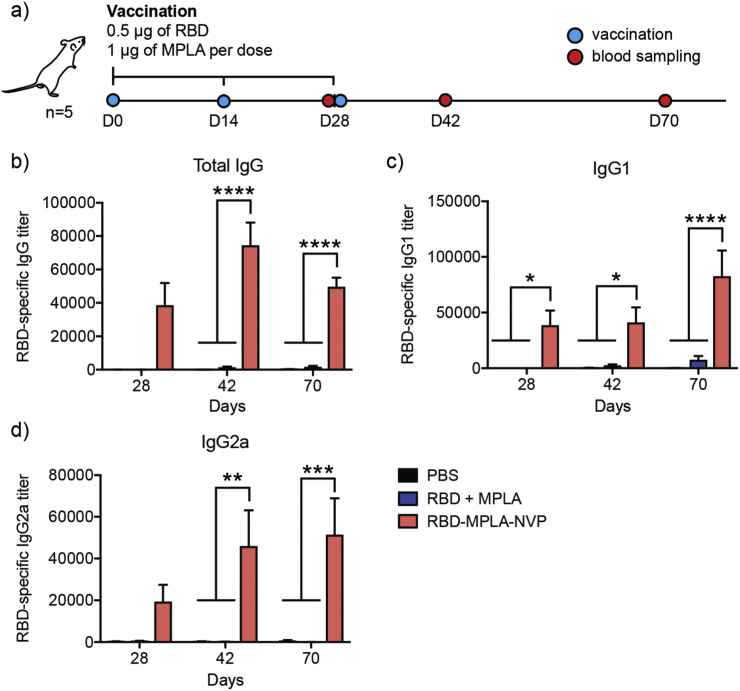
3.7. RBD-NVP vaccination study in mice
Lastly, we examined the potency of RBD-NVP to generate anti-RBD antibody response in mice. BALB/c mice were vaccinated three times with 2 weeks interval between each injection (days 0, 14, and 28) using 0.5 μg of RBD and 1 μg of MPLA either in NVP or soluble formulation (Fig. 7a). Sera samples were collected on days 28, 42, and 70 and assessed for RBD-specific serum IgG, IgG1, and IgG2a titers. RBD-NVP generated significantly higher RBD-specific antibody titers, compared with RBD + MPLA soluble vaccine (Fig. 7b-d). Specifically, by day 42 (2 weeks after 3rd vaccination), RBD-NVP elicited 55-fold, 17-fold, and 284-fold higher RBD-specific IgG, IgG1, and IgG2a titers, respectively, compared with the soluble vaccine (Fig. 7b-d). By day 70 (6 weeks after 3rd vaccination), mice immunized with RBD-NVP still maintained 30-fold, 13-fold, and 671-fold higher RBD-specific IgG, IgG1, and IgG2a titers, respectively, compared with the soluble vaccine group (Fig. 7b-d). While the examination of functionality and neutralizing activities of these antibodies are beyond the scope of our current studies, these initial results indicated that RBD-NVP induced robust, long-lasting, Th1/Th2-balanced antibody responses against RBD.
Fig. 7.
RBD-NVP elicits robust RBD-specific antibody responses in mice. (a) BALB/c mice were vaccinated three times, with 2 weeks intervals between each injection. Blood was sampled on the indicated days. RBD-NVP significantly increased serum antibody titers of RBD-specific (b) IgG, (c) IgG1, and (d) IgG2a, compared to the soluble vaccine. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001, analyzed by two-way ANOVA, followed by Tukey's HSD multiple comparison post hoc test.
Overall, we have developed NVP for the delivery of protein antigens and demonstrated the versatility of NVP for protein-based vaccination against infectious pathogens. Protein antigens incorporated into NVP maintained the configuration of the epitopes, as shown by the recognition and binding of neutralizing antibodies on the surfaces of antigen-displaying NVP. NVP was readily taken up by DCs in vitro, leading to greater DC activation and antigen presentation, compared with soluble vaccine formulation. We have successfully prepared NVP carrying SOSIP derived from HIV-1 and RBD derived from SARS-CoV-2. Animals immunized with NVP generated strong antigen-specific antibody responses. While these initial proof-of-concept studies have shown the promise of NVP, more studies are warranted to delineate the immunological mechanisms of action and to assess protective immunity against viral challenge (e.g., HIV-1 or SHIV challenge in non-human primates; and SARS-CoV-2 challenge in mice engineered to express human ACE2).
Credit author statement
KSP and JDB contributed conceptualization, investigation, formal analysis, and writing. SS, JN, SWS, LJO, JLM, LC, JAS, JS, DCM, CCL, JLS, and JX contributed methodology and resources. JJM contributed conceptualization, formal analysis, funding acquisition, supervision, and writing.
Declaration of Competing Interest
There is no conflict of interest to declare.
Acknowledgments
This work was supported by NIH (R01AI127070, R01CA210273, R01DK125087, U01CA210152, and HHSN272201800004C). J.J.M. is supported by NSF CAREER Award (1553831). K.S.P. acknowledges financial support from the UM TEAM Training Program (DE007057 from NIDCR). The Center for Structural Biology of Life Sciences Institute, University of Michigan, was supported by the Open Philanthropy Project Fund.
References
- 1.Pandey S.C., Pande V., Sati D., Upreti S., Samant M. Vaccination strategies to combat novel corona virus SARS-CoV-2. Life Sci. 2020;256:117956. doi: 10.1016/j.lfs.2020.117956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Thanh T., Andreadakis Z., Kumar A., Gomez Roman R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 3.Vetter V., Denizer G., Friedland L.R., Krishnan J., Shapiro M. Understanding modern-day vaccines: what you need to know. Ann. Med. 2018;50:110–120. doi: 10.1080/07853890.2017.1407035. [DOI] [PubMed] [Google Scholar]
- 4.Fox C.B., Kramer R.M., Barnes L., Dowling Q.M., Vedvick T.S. Working together: interactions between vaccine antigens and adjuvants. Ther. Adv. Vaccin. 2013;1:7–20. doi: 10.1177/2051013613480144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan Y., Moon J.J. Particulate delivery systems for vaccination against bioterrorism agents and emerging infectious pathogens. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017;9 doi: 10.1002/wnan.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva A.L., Soema P.C., Slütter B., Ossendorp F., Jiskoot W. PLGA particulate delivery systems for subunit vaccines: linking particle properties to immunogenicity. Hum. Vaccin. Immunother. 2016;12:1056–1069. doi: 10.1080/21645515.2015.1117714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu P., Wusiman A., Zhang Y., Liu Z., Bo R., Hu Y., Liu J., Wang D. Rational design of PLGA nanoparticle vaccine delivery systems to improve immune responses. Mol. Pharm. 2019;16:5000–5012. doi: 10.1021/acs.molpharmaceut.9b00860. [DOI] [PubMed] [Google Scholar]
- 8.Nam J., Son S., Ochyl L.J., Kuai R., Schwendeman A., Moon J.J. Chemo-photothermal therapy combination elicits anti-tumor immunity against advanced metastatic cancer. Nat. Commun. 2018;9:1074. doi: 10.1038/s41467-018-03473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carabineiro S.A.C. Applications of gold nanoparticles in nanomedicine: recent advances in vaccines. Molecules. 2017;22 doi: 10.3390/molecules22050857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu C., Nam J., Hong H., Xu Y., Moon J.J. Positron emission tomography-guided photodynamic therapy with biodegradable mesoporous silica nanoparticles for personalized cancer immunotherapy. ACS Nano. 2019;13:12148–12161. doi: 10.1021/acsnano.9b06691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong X., Zhong X., Du G., Hou Y., Zhang Y., Zhang Z., Gong T., Zhang L., Sun X. The pore size of mesoporous silica nanoparticles regulates their antigen delivery efficiency. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aaz4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Halifa S., Gauthier L., Arpin D., Bourgault S., Archambault D. Nanoparticle-based vaccines against respiratory viruses. Front. Immunol. 2019;10:22. doi: 10.3389/fimmu.2019.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory A.E., Titball R., Williamson D. Vaccine delivery using nanoparticles. Front. Cell. Infect. Microbiol. 2013;3:13. doi: 10.3389/fcimb.2013.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mucker E.M., Karmali P.P., Vega J., Kwilas S.A., Wu H., Joselyn M., Ballantyne J., Sampey D., Mukthavaram R., Sullivan E., Chivukula P., Hooper J.W. Lipid nanoparticle formulation increases efficiency of DNA-vectored vaccines/immunoprophylaxis in animals including transchromosomic bovines. Sci. Rep. 2020;10:8764. doi: 10.1038/s41598-020-65059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobernik D., Bros M. DNA vaccines-how far from clinical use? Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fotoran W.L., Santangelo R., de Miranda B.N.M., Irvine D.J., Wunderlich G. DNA-loaded cationic liposomes efficiently function as a vaccine against malarial proteins. Mol. Ther. Methods Clin. Dev. 2017;7:1–10. doi: 10.1016/j.omtm.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuai R., Ochyl L.J., Bahjat K.S., Schwendeman A., Moon J.J. Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat. Mater. 2017;16:489–496. doi: 10.1038/nmat4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalgleish A.G., Beverley P.C., Clapham P.R., Crawford D.H., Greaves M.F., Weiss R.A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 20.Mascola J.R., Haynes B.F. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol. Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders R.W., Derking R., Cupo A., Julien J.P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Pena A.T., Korzun J., Golabek M., de Los Reyes K., Ketas T.J., van Gils M.J., King C.R., Wilson I.A., Ward A.B., Klasse P.J., Moore J.P. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Taeye S.W., Ozorowski G., de la Peña A. Torrents, Guttman M., Julien J.P., van den Kerkhof T.L., Burger J.A., Pritchard L.K., Pugach P., Yasmeen A., Crampton J., Hu J., Bontjer I., Torres J.L., Arendt H., DeStefano J., Koff W.C., Schuitemaker H., Eggink D., Berkhout B., Dean H., LaBranche C., Crotty S., Crispin M., Montefiori D.C., Klasse P.J., Lee K.K., Moore J.P., Wilson I.A., Ward A.B., Sanders R.W. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders R.W., van Gils M.J., Derking R., Sok D., Ketas T.J., Burger J.A., Ozorowski G., Cupo A., Simonich C., Goo L., Arendt H., Kim H.J., Lee J.H., Pugach P., Williams M., Debnath G., Moldt B., van Breemen M.J., Isik G., Medina-Ramirez M., Back J.W., Koff W.C., Julien J.P., Rakasz E.G., Seaman M.S., Guttman M., Lee K.K., Klasse P.J., LaBranche C., Schief W.R., Wilson I.A., Overbaugh J., Burton D.R., Ward A.B., Montefiori D.C., Dean H., Moore J.P. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349 doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goo L., Chohan V., Nduati R., Overbaugh J. Early development of broadly neutralizing antibodies in HIV-1-infected infants. Nat. Med. 2014;20:655–658. doi: 10.1038/nm.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., Hong W., Yang Y., Zhao Y., Ye F., Lin S., Deng W., Chen H., Lei H., Zhang Z., Luo M., Gao H., Zheng Y., Gong Y., Jiang X., Xu Y., Lv Q., Li D., Wang M., Li F., Wang S., Wang G., Yu P., Qu Y., Yang L., Deng H., Tong A., Li J., Wang Z., Shen G., Zhao Z., Li Y., Luo J., Liu H., Yu W., Yang M., Xu J., Wang J., Li H., Wang H., Kuang D., Lin P., Hu Z., Guo W., Cheng W., He Y., Song X., Chen C., Xue Z., Yao S., Chen L., Ma X., Chen S., Gou M., Huang W., Wang Y., Fan C., Tian Z., Shi M., Wang F.S., Dai L., Wu M., Li G., Peng Y., Qian Z., Huang C., Lau J.Y., Yang Z., Wei Y., Cen X., Peng X., Qin C., Zhang K., Lu G., Wei X. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 29.Tai W., Zhang X., Drelich A., Shi J., Hsu J.C., Luchsinger L., Hillyer C.D., Tseng C.K., Jiang S., Du L. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 2020;30:932–935. doi: 10.1038/s41422-020-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 31.Georgiev I.S., Joyce M.G., Yang Y., Sastry M., Zhang B., Baxa U., Chen R.E., Druz A., Lees C.R., Narpala S., Schön A., Van Galen J., Chuang G.Y., Gorman J., Harned A., Pancera M., Stewart-Jones G.B., Cheng C., Freire E., McDermott A.B., Mascola J.R., Kwong P.D. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J. Virol. 2015;89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon J.J., Suh H., Bershteyn A., Stephan M.T., Liu H.P., Huang B., Sohail M., Luo S., Um S.H., Khant H., Goodwin J.T., Ramos J., Chiu W., Irvine D.J. Interbilayer-crosslinked multilamellar vesicles as synthetic vaccines for potent humoral and cellular immune responses. Nat. Mater. 2011;10:243–251. doi: 10.1038/nmat2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bazzill J.D., Ochyl L.J., Giang E., Castillo S., Law M., Moon J.J. Interrogation of antigen display on individual vaccine nanoparticles for achieving neutralizing antibody responses against hepatitis C virus. Nano Lett. 2018;18:7832–7838. doi: 10.1021/acs.nanolett.8b03601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bazzill J.D., Stronsky S.M., Kalinyak L.C., Ochyl L.J., Steffens J.T., van Tongeren S.A., Cooper C.L., Moon J.J. Vaccine nanoparticles displaying recombinant Ebola virus glycoprotein for induction of potent antibody and polyfunctional T cell responses. Nanomedicine. 2019;18:414–425. doi: 10.1016/j.nano.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan Y., Stronsky S.M., Xu Y., Steffens J.T., van Tongeren S.A., Erwin A., Cooper C.L., Moon J.J. Multilamellar vaccine particle elicits potent immune activation with protein antigens and protects mice against Ebola virus infection. ACS Nano. 2019;13:11087–11096. doi: 10.1021/acsnano.9b03660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montefiori D.C. HIV Protocols. Springer; 2009. Measuring HIV neutralization in a luciferase reporter gene assay; pp. 395–405. [DOI] [PubMed] [Google Scholar]
- 37.Sarzotti-Kelsoe M., Bailer R.T., Turk E., Lin C.L., Bilska M., Greene K.M., Gao H., Todd C.A., Ozaki D.A., Seaman M.S., Mascola J.R., Montefiori D.C. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J., Ochyl L.J., Yang E., Moon J.J. Cationic liposomes promote antigen cross-presentation in dendritic cells by alkalizing the lysosomal pH and limiting the degradation of antigens. Int. J. Nanomedicine. 2017;12:1251–1264. doi: 10.2147/IJN.S125866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beddows S., Franti M., Dey A.K., Kirschner M., Iyer S.P., Fisch D.C., Ketas T., Yuste E., Desrosiers R.C., Klasse P.J., Maddon P.J., Olson W.C., Moore J.P. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology. 2007;360:329–340. doi: 10.1016/j.virol.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 40.Graham B.S., Mascola J.R. Lessons from failure—preparing for future HIV-1 vaccine efficacy trials. J. Infect. Dis. 2005;191:647–649. doi: 10.1086/428406. [DOI] [PubMed] [Google Scholar]
- 41.Qualls Z.M., Choudhary A., Honnen W., Prattipati R., Robinson J.E., Pinter A. Identification of novel structural determinants in MW965 Env that regulate the neutralization phenotype and conformational masking potential of primary HIV-1 isolates. J. Virol. 2018;92 doi: 10.1128/JVI.01779-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders R.W., van Gils M.J., Derking R., Sok D., Ketas T.J., Burger J.A., Ozorowski G., Cupo A., Simonich C., Goo L., Arendt H., Kim H.J., Lee J.H., Pugach P., Williams M., Debnath G., Moldt B., van Breemen M.J., Isik G., Medina-Ramírez M., Back J.W., Koff W.C., Julien J.P., Rakasz E.G., Seaman M.S., Guttman M., Lee K.K., Klasse P.J., LaBranche C., Schief W.R., Wilson I.A., Overbaugh J., Burton D.R., Ward A.B., Montefiori D.C., Dean H., Moore J.P., HIV-1 VACCINES HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349 doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]