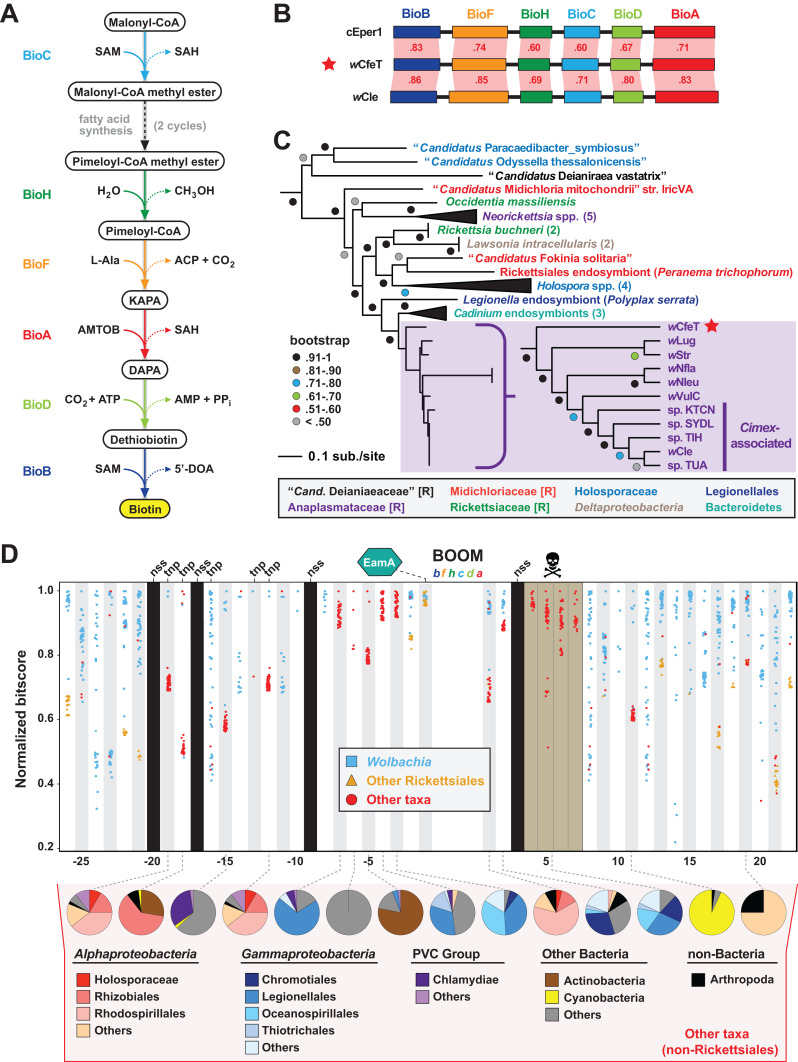
Figure 2. wCfeT carries the Biotin synthesis Operon of Obligate intracellular Microbes (BOOM).
(A) Metabolism of biotin from malonyl-CoA (Lin & Cronan, 2011). (B) Comparisons of wCfeT BOOM to equivalent loci in Cardinium endosymbiont of Encarsia pergandiella (cEper1, CCM10336–CCM10341) and Wolbachia endosymbiont of Cimex lectularius (wCle, BAP00143–BAP00148). Red shading and numbers indicate % identity across pairwise protein alignments (Blastp). Gene colors correspond to enzymatic steps depicted in panel A, with all proteins drawn to scale (as a reference, wCfeT BioB is 316 aa). (C) Phylogeny of BOOM and other bio gene sets from diverse bacteria estimated from the concatenation of six bio enzymes (BioC, BioH, BioF, BioA, BioD, and BioB); see Table S2 for all sequence information, and Fig. S1A for complete tree estimated under Maximum Likelihood. In taxon color scheme, [R] denotes Rickettsiales, with Holosporaceae as a revised family of Rhodospirillales (Muñoz-Gómez et al., 2019). (D) Analysis of genes flanking wCfeT BOOM. See Table S3 for comparison with other BOOM-containing wolbachiae. Graph depicts top 50 Blastp subjects (e-value < 1, BLOSUM45 matrix) using the wCfeT proteins as queries against the NCBI nr database (January 9, 2020). Colors distinguish three taxonomic groups (see inset). Black columns: no significant similarity (nss) in the NCBI nr database. Tnp, predicted transposase. Brown, pseudogenization. See Fig. S1B for EamA phylogeny estimation. Pie charts across the bottom delineate taxonomic diversity of hits to “Other taxa,” for wCfeT proteins with significant Blastp matches outside of the Rickettsiales.