Abstract
Macrophages are the first immune cells in the developing embryo and have a central role in organ development, homeostasis, immunity and repair. Over the last century, our understanding of these cells has evolved from being thought of as simple phagocytic cells to master regulators involved in governing a myriad of cellular processes. A better appreciation of macrophage biology has been matched with a clearer understanding of their diverse origins and the flexibility of their metabolic and transcriptional machinery. The understanding of the classical mononuclear phagocyte system in its original form has now been expanded to include the embryonic origin of tissue‐resident macrophages. A better knowledge of the intrinsic similarities and differences between macrophages of embryonic or monocyte origin has highlighted the importance of ontogeny in macrophage dysfunction in disease. In this review, we provide an update on origin and classification of tissue macrophages, the mechanisms of macrophage specialisation and their role in health and disease. The importance of the macrophage niche in providing trophic factors and a specialised environment for macrophage differentiation and specialisation is also discussed.
Keywords: innate immunity, macrophage diversity, macrophage functions, macrophage niche, macrophage origins
In this review, we provide a clear, comprehensive update on monocyte/ macrophage ontogeny, differentiation and function.

Introduction
Macrophages were first identified by Metchnikoff in 1882 when he observed phagocytes surrounding and trying to devour a rose thorn introduced into the transparent body of a starfish larva. 1 , 2 Metchnikoff also identified major roles for these phagocytes in host resistance against infections, phagocytosis of unwanted cells during development, injury and repair. Macrophages have subsequently been shown to initiate and shape the adaptive immune system and in general acting as an inflammation rheostat. Macrophages achieve this by processing and presenting antigens to T cells 3 and by integrating multiple signals from a repertoire of cell surface and cytoplasmic pattern recognition receptors. 4
Macrophages are the first immune cells to appear in an organism’s development and are essential during the early stages of development. 5 Tissue macrophages also play a crucial role in homeostasis, 6 , 7 wound healing 8 and tissue regeneration. 9 , 10 The wide variety of macrophage functions partly arise because of their ability to sense and sample the local tissue environment and via expression of specific transcription factors and enhancer‐associated histone modifications unique to a local microenvironment. 11 , 12 Macrophages are also able to make extensive changes to their intracellular metabolism in response to environmental and inflammatory cues. 13 Unfortunately, aberrant macrophage function is strongly associated in the pathogenesis of disease states such as fibrosis, obesity and cancer. 14 In this review, we discuss our current understanding of the ontogeny of tissue‐resident macrophages, the interaction of macrophages with components of the tissue niche and how these interactions shape macrophage function. We also discuss the links between cellular metabolism and macrophage phenotype, the contribution of monocytes to the maintenance of tissue macrophage populations and how monocyte‐derived macrophages differ from embryo‐derived macrophages.
Macrophage ontogeny and the mononuclear phagocyte system
Macrophages were classified as part of mononuclear phagocyte system (MPS) along with monocytes and dendritic cells (DCs) in the mid‐1970s. 15 According to the description of the MPS, tissue macrophages were considered fully differentiated cells that were constantly being replenished by circulating monocytes. 16 The concept of MPS is supported by in vitro studies showing monocyte differentiation into macrophages and in vivo adoptive transfer of monocytes under inflammatory conditions showing recruitment and conversion to macrophages in the peritoneal cavity. 17 However, several studies in humans and mice have contradicted the non‐dividing, terminally differentiated, circulation‐dependent ontogeny of tissue macrophages. In congenic parabiotic mice, which share the same circulation and have a mixed population of lymphocytes and monocytes in the blood, the macrophage populations in the brain 18 , 19 and epidermis 20 do not mix even after a year of parabiosis. Moreover, histological approaches have demonstrated the presence of macrophages before the establishment of definitive haematopoiesis that gives rise to monocytes. 21 , 22 , 23 Several human studies have further supported the circulation‐independent origin of tissue macrophages. For example, patients with severe monocytopenia have normal numbers of macrophages in the epidermis (Langerhans cell, LC) 24 , 25 and host LCs remained in patients who received sex‐mismatched allogeneic bone marrow transplants. 26 , 27 Donor LCs can also be detected for years in recipients of human limb graft. 28 Donor macrophages also self‐maintain for years in the transplanted heart, 29 liver 30 and lungs. 31 , 32 , 33 Despite these findings, more work is needed to understand the origin of tissue macrophages in humans. Much of our current knowledge of tissue macrophage ontogeny comes from mouse models. It should be noted that whilst these models are extremely useful, they have inherent limitations around life span and environmental exposure that may not reflect the situation in humans.
The embryonic origin of tissue macrophages has also been confirmed by Cre‐LoxP approaches. The chemokine receptor, CX3CR1, is prominently expressed in the MPS. 34 Using CX3CR1Cre:R26‐YFP reporter mice that display constitutive Cre activity in CX3CR1+ cells and drug‐induced activation of Cre in CX3CR1CreER:R26‐YFP mice, it has been established that most tissue macrophages are generated prenatally that self‐renew in peripheral tissues during adulthood at least in the absence of challenge. 17 , 35 These observations led to the conclusions that tissue‐resident macrophages are not solely derived from haematopoietic stem cells (HSCs) or BM‐derived progenitors but also derived from local or embryonic precursors. 36 This has led investigators to more thoroughly explore the embryonic origin of macrophages.
Embryonic macrophages
Myeloid cells including macrophages arise from three successive haematopoietic waves, referred to as primitive, pro‐definitive and definitive phases, respectively 37 (Figure 1). The primitive programme is independent of the transcription factor c‐Myb and starts at embryonic days 6.5 (E6.5)–E8.5 in the blood islands of the extraembryonic yolk sac (YS). This phase gives rise to bipotent progenitors for nucleated erythrocytes and megakaryocytes and a progenitor restricted to the macrophage lineage (Mac‐CFC). 38 , 39 , 40 , 41 , 42 The c‐Myb‐independent, pro‐definitive wave occurs in different sites of the embryo (YS, allantois and embryo proper) and gives rise to erythroid and myeloid progenitors (EMPs) between E8.5 and E10.5. 43 , 44 Unlike long‐term haematopoietic stem cells (LT‐HSCs), EMPs do not have long‐term repopulating capacity and develop into macrophages through a CX3CR1 expressing intermediate population called p‐Macs. 45 EMPs give rise to p‐Macs without passing through the monocyte stage as evidenced by the lack of peroxidase activity (a signature feature of monocytes) and the presence of a core macrophage transcriptional programme occurring in p‐Macs. 45 EMPs and p‐Macs expand in the YS and then traffic towards foetal liver up until E14.5, where they serve as a reservoir for macrophages throughout embryogenesis. 43 , 44 EMP progeny seed different tissues in the embryo and may become life‐long tissue‐resident macrophages. 37 , 46 , 47 , 48 Brain microglia are a prototypical primitive macrophage generated in the YS, which are maintained throughout adult life by virtue of their longevity and limited self‐renewal capability without input from definitive haematopoiesis. 18 , 19 , 49 EMP‐derived macrophages contribute to embryonic development and tissue remodelling through phagocytosis of unwanted and obsolete cell structures and cells. 50 , 51
Figure 1.
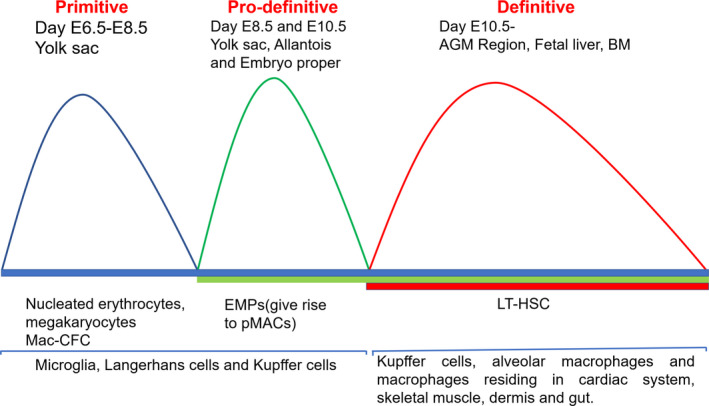
Origin of macrophages. Myeloid cells including macrophages arise from three successive haematopoietic waves, referred to as primitive, pro‐definitive and definitive. The primitive programme starts at embryonic days 6.5 (E6.5)‐E8.5 in the blood islands of the extraembryonic yolk sac (YS) and gives rise to nucleated erythrocytes, megakaryocytes and Mac‐CFCs. The pro‐definitive wave starts at E8.5 and E10.5 in the yolk sac, allantois and embryo proper and gives rise to erythroid and myeloid progenitors (EMPs). Primitive and pro‐definitive phases contribute to microglia, Langerhans and Kupffer cells. The third wave of definitive haematopoiesis starts at E10.5 from the aorta–gonad–mesonephros region (AGM) region and gives rise to LT‐HSC. They migrate to the foetal liver and definitive haematopoiesis shifts to BM around E17.5. Definitive haematopoietic stem cells give rise to Kupffer cells and alveolar macrophage tissue residing in cardiac system, skeletal muscle, dermis and gut.
Macrophages also act as cellular chaperones for tissue vascularisation. 52 Mouse mutants lacking macrophages during embryonic development, because of deficiency of colony‐stimulating factor 1 receptor (CSF‐1R also known as CD115) or the transcription factor PU.1, display growth retardation and perinatal mortality. 53 , 54 The transcription factor c‐Myb is not needed for primitive haematopoiesis but is required for definitive haematopoiesis. 55 This was shown in Myb mutant mouse embryos, where impairment in definitive haematopoiesis was seen, 56 , 57 but tissue‐resident macrophages in the brain (microglia), skin (LCs) and liver (Kupffer cells; KC) were unaffected. 58 Similarly, Myb mutant zebrafish develop tissue macrophage populations in the absence of definitive haematopoiesis. 55 The third wave of c‐Myb‐dependent definitive haematopoiesis starts at E10.5 from the aorta–gonad–mesonephros region (AGM) region and gives rise to LT‐HSC. They migrate to the foetal liver and definitive haematopoiesis shifts to the bone marrow (BM) at around E17.5. 38 , 59 , 60 Definitive HSCs arising from the AGM region at around E10.5 61 give rise to KCs and alveolar macrophages. HSC activity then peaks in the foetal liver at around E16.5 and gives rise to tissue macrophages residing in the cardiac system, skeletal muscle, dermis and the gut before shifting to BM. 62 The BM then remains the major site of haematopoiesis in adult life.
Primitive vs. definitive origin of macrophages
Despite the general agreement that most tissue macrophages have an embryonic origin, the exact contributions of primitive and definitive haematopoiesis to embryo‐derived adult tissue macrophage populations remain unclear. All tissue macrophages may arise from Myb and HSC‐independent lineage without going through a monocyte intermediate. 63 These macrophages can seed various locations and give rise to bonafide long‐lived tissue macrophages. For example, the BM contains precursor cells that give rise to LC and microglia. 64 , 65 Macrophages may also arise from definitive haematopoiesis in foetal liver through a monocyte intermediate. 19 Pulse labelling of myeloid precursors in the Runx1CreER mouse has enabled researchers to determine the relative contributions of YS and foetal liver to tissue macrophage populations. Runt‐related transcription factor 1 (Runx1) expression is restricted to the extraembryonic YS between E6.5 and E8. 19 , 66 Inducible CreER reporter gene expression driven by Runx1 has established considerable input from foetal liver‐resident precursors to lung, dermis and spleen macrophages 19 with the exception of microglia that originate solely from the yolk sac. Most tissue macrophages except microglia lose their Runx1+ labelling in adult tissues suggesting that they are replaced by non‐labelled precursors before birth. 19 , 67 , 68 Contributions of YS versus foetal liver‐derived precursors, however, vary between tissue macrophage compartments. For example, heart‐resident, cardiac macrophages are derived from both YS‐derived and foetal liver‐derived progenitors, 69 while adult LC 68 and adult lung alveolar macrophages 70 mainly originate from foetal liver‐derived monocytes. Despite the results from the Runx1+ mice, the origin of cells arising from foetal liver is less well‐defined because foetal liver is itself seeded by YS precursors. 38 , 39
The revised concept of MPS now accommodates two independent origins of tissue macrophages. Embryonic macrophages are established prenatally and self‐maintain independent of any haematopoietic input, 17 , 19 , 58 , 69 , 70 , 71 , 72 whereas adult‐derived macrophages develop from tissue‐infiltrating monocytes, have a limited lifespan and are associated with pathological inflammatory reactions. Both types of macrophages seem to co‐exist in tissues, whether they have different behaviour based on ontogeny or are made functionally homogenous by the tissue environment remains to be seen.
Macrophage subsets
Macrophages are a highly heterogenous population of cells. The initial classification of macrophages into M1 and M2 subsets was based on macrophages isolated from C57BL/6 mice and Balb/c mice. Macrophages from C57BL/6 mice have a Th1‐dominated immune response and, when challenged with LPS and IFN‐γ, produce nitric oxide (NO) from arginine via iNOS. 73 Macrophages from Balb/c mice have a Th2‐dominated immune response and, when challenged by LPS and IFN‐γ, produce ornithine via arginase. 73 C57BL/6 mice carry a deletion in the promoter of Slc7a2, the key arginine transporter in macrophages causing large differences in arginine utilisation between C57BL/6 and BALB/c mice. 74 Categorisation of macrophages into M1 and M2 subsets based on arginine metabolism fits neatly with the inflammation vs. resolution functions of macrophages. Macrophages producing NO inhibit/kill pathogens or nearby cells, while ornithine promotes cell proliferation and wound healing. M1/M2 classification has also been used to define macrophage polarisation states. LPS and IFN‐γ induce M1 macrophages in a STAT‐1 and aerobic glycolysis‐dependent manner, 75 while IL‐4 induces M2 macrophages in a STAT6 and fatty acid oxidation (FAO)‐dependent manner. 76 , 77 Currently, M1/M2 macrophages are divided based on the expression of specific markers; M1 macrophages express CD68, TNF‐α, iNOS, IL‐1β and IL‐12, while M2 macrophages express arginase 1, transforming growth factor (TGF)‐β, CD163 (cluster of differentiation 163), mannose receptor 1, CD206, Rtnla, IL‐10, VEGF and Ym1. 78 , 79 M1 macrophages produce pro‐inflammatory cytokines (TNF‐α, IL‐12, IL‐27 and IL‐23), chemokines (CXCL11, CXCL9 and CXCL10) and matrix‐metalloproteinases (MMP‐1, 2, 7, 9, 12) and demonstrate enhanced antigen presentation and generation of reactive oxygen species. In contrast, macrophages stimulated with IL‐4 and IL‐13 show an anti‐inflammatory and reparative profile. 80 M2 macrophages produce anti‐inflammatory cytokines (IL‐10), chemokines (CCL17) 81 , 82 and growth factors (VEGF, TGF‐β). Together, these mediators promote tissue remodelling and repair by stimulating extracellular matrix production by fibroblasts, cell proliferation and angiogenesis.
The classification of macrophages into M1/M2 groups based on well‐defined stimuli does not model the infinitely more complex tissue milieu where macrophages (potentially of different origin) would be exposed to multiple signals in different sequential order. Nevertheless, macrophages have been classified into subgroups within the M1‐M2 range as M2a, M2b, M2c and Mox macrophages. 79 , 83 , 84 , 85 Given the phenotypic diversity of macrophage populations in vivo, the relevance of the M1‐M2 paradigm may be minimal. For example, one study acquired a data set of 299 macrophage transcriptomes in response to diverse activation signals. 85 In another study, CyTOF analysis of renal cancer macrophages identified 17 different subsets. 86 A plethora of recent publications have used single‐cell RNA‐seq to identify previously unrecognised macrophage populations with unique gene expression signatures. 87 , 88 , 89 , 90 , 91 These new subtypes may represent macrophage adaptation to unique microenvironments within organs. Macrophage subtypes are also classified based on the expression of few cell surface markers, but M1 or M2 macrophages can acquire canonical markers of the other subset in vitro. 92 , 93 In spite of the M1/ M2 classification model being accepted as an over‐simplification, it continues to be widely used. 94 A standardised experimental framework for macrophage subtype classification based on the source of macrophages, activators used and macrophage markers has been proposed. 95 Single‐cell RNA sequencing, mass cytometry and advanced clustering algorithms should shed more light on macrophage heterogeneity in the future. 96 , 97
The macrophage niche
Tissue‐resident macrophages develop with the organ they reside in and adapt to perform not only immune functions but also homeostatic functions essential for the particular organ they inhabit. 12 , 98 Circulating monocytes taking up residence in tissues also adopt a tissue‐specific identity very similar to resident macrophages, if not completely similar. 99 The existence of a niche for macrophages in individual tissues has been postulated. These niches may nurture and modify macrophages by providing them with a physical scaffold and trophic factors for survival and proliferation. The type of physical scaffold may affect the differentiation and function of macrophages by inducing specific transcription factors to suit the temporal homeostatic function of a tissue.
Niche adaptation of macrophages
All tissue macrophages, after going through a programme of lineage determination directed by a unique set of transcription factors such as PU.1 and MafB, 54 , 100 , 101 , 102 acquire a common set of functions (e.g. phagocytosis, immune surveillance) and cell surface markers (F4/80, CD64, Mertk). Ultimately, the tissue microenvironment customises the local macrophage population to suit its homeostatic needs (Figure 2). As organogenesis proceeds, the differentiating milieu of an organ guides the resident macrophages to acquire the phenotype and functions appropriate to that organ. Expression of a limited set of transcription factors confers a tissue‐specific character on macrophages. For example, nuclear factor of activated T cells 1 (NFATC1) is necessary for osteoclast differentiation and functional specialisation. 103 Similarly, transforming growth factor‐ β (TGF‐β) signalling, 104 , 105 , 106 , 107 , 108 notch signalling 109 , 110 , 111 , 112 and bone morphogenetic protein (BMP) signalling drive the specialisation of multiple subsets. It appears that all macrophage subsets are active phagocytes and the material they ingest appears to dictate their fate. Tissue macrophages are exposed to specific metabolites in different organs. For example, haem, 113 oxysterol 113 , 114 , 115 and retinoic acid 98 , 116 , 117 can induce functional polarisation of macrophages. Macrophage crosstalk with other immune cells also plays a role in defining their differentiation. For example, alveolar macrophage development involves crosstalk with pulmonary innate lymphoid cell 2s (ILC2s) and basophils producing CSF2 and IL‐13, 118 whereas LC replenishment requires CSF1 produced by neutrophils. 119
Figure 2.
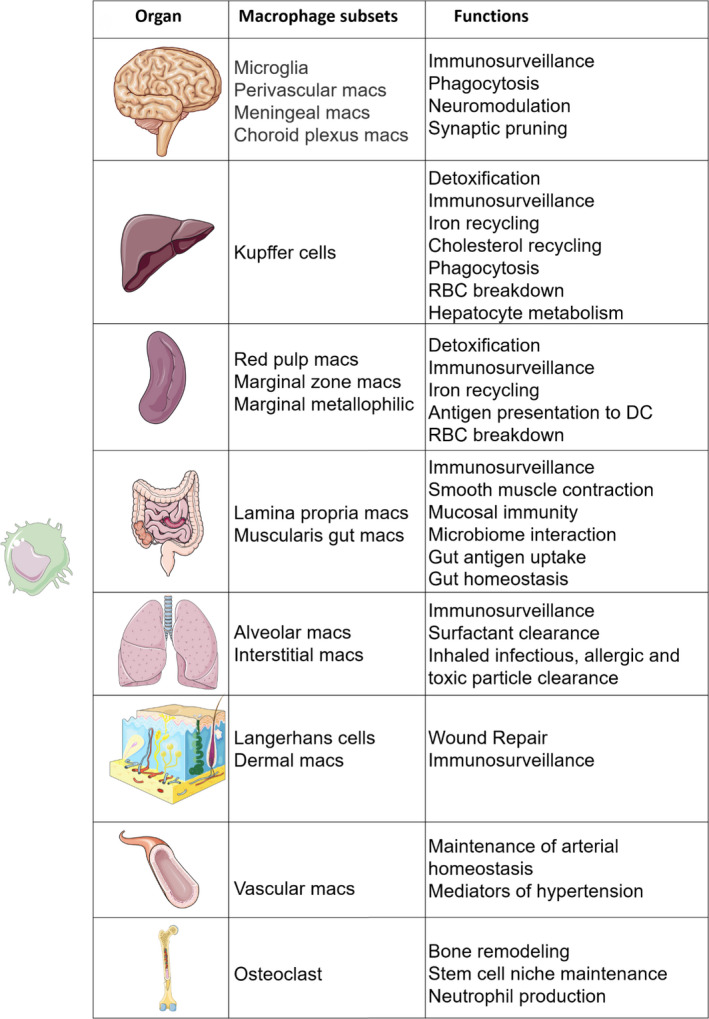
The heterogenous functions of tissue macrophages. All tissue macrophages go through a process of lineage determination via expression of limited set of transcription factors to acquire functions and cell surface markers common to all macrophages (phagocytosis, F4/80, MertK). The tissue microenvironment customises the macrophage to take over organ‐specific functions by inducing expression of unique set of transcription factors. Multiple signals specific to a tissue in a sequential combination are required to prime the macrophage and prepare the epigenetic landscape for macrophages to take up a tissue‐specific identity.
Even though a restricted set of transcription factors decides macrophage identity, multiple signals in a specific sequence are required to prime macrophages and prepare the epigenetic landscape for macrophages to adopt a tissue‐specific identity. Predictably, these multiple signals are specific to a location or a macrophage niche. For example, monocyte engraftment to liver requires interaction with endothelial cells, hepatocytes and stellate cells with key roles for TGF‐β and desmosterol. 109 , 112 Another example of multistep imprinting of macrophage identity by the niche is mucosal Langerhans cell differentiation. In this particular example, precursor cells have to be first exposed to BMP7 in lamina propria and then TGF‐β from endothelial cells to complete their differentiation. 120 , 121 Thus, a unique combination of tissue niche factors can induce reversible activation of gene expression programmes that is responsible for functional polarisation of macrophages in tissues.
Nurture in the niche
Macrophages require a continuous supply of trophic factors IL‐34, CSF‐1(M‐CSF) and CSF‐2 (GM‐CSF) for normal maintenance and development. IL‐34 and CSF‐1 share a similar tertiary structure and bind to a common receptor, CSF1R. 5 , 122 , 123 , 124 , 125 CSF‐1 is produced in three different forms by alternate splicing (secreted form, secreted proteoglycan form and a membrane form), and all three forms have a common active N‐terminal and distinct but overlapping functions. 126 , 127 , 128 CSF‐1‐deficient mice have normal number of Langerhans and microglial cells but display a deficiency in most other macrophage types. 53 Reconstitution of CSF‐1‐deficient mice with the soluble form of CSF‐1 rescues most resident macrophage populations, the membrane form corrects for macrophages in most tissues except in liver, spleen and kidneys. 127 , 128 The proteoglycan form of CSF‐1 integrates into the matrix of local tissues and regulates local macrophage numbers. Mice lacking CSF1R, the common receptor for IL‐34 and CSF‐1, have reduced numbers of macrophages throughout the body. 53 , 129 Administration of anti‐CSFR1 blocking antibody depletes most of the macrophage populations in embryonic 67 and adult tissues. 130 conversely, administration of CSF‐1 produces a massive expansion of blood monocytes and tissue macrophages in mice. 131 , 132 , 133 , 134 The secreted form of CSF‐1 when injected into CSF‐1‐deficient mice rescues most of the resident macrophage population. 127 , 128 In fact, administration of CSF‐1 leads to increased macrophage numbers in the liver and a rapid increase in the size of liver. 131 , 134 This may indicate a role for macrophages in homeostatic regulation of organ size. CSF‐1 consumption by Ly6Chi monocytes can regulate the generation of the Ly6Clo subpopulation 17 , 125 , 132 and depletion of monocytes can lead to an increase in circulating CSF‐1 levels which, in turn, will promote an increase in tissue macrophage numbers. Bioavailability of CSF‐1 may also be regulated by post‐translational modification. For example, tumor necrosis factor‐α (TNF‐α)‐converting enzyme (TACE) can convert the membrane‐bound isoform of CSF‐1 to the soluble form of CSF‐1. 135 IL34 KO mice are deficient in LC and brain microglia. 136 , 137 , 138 , 139 , 140 IL‐34 is not detected in blood, probably because it acts locally near the tissue where it is produced. 141 , 142 In summary, IL‐34 and CSF‐1 are the most important trophic factors produced by the macrophage niche and are essential for the maintenance and survival of macrophages.
Regulation of macrophage density in tissues
Resident macrophages are abundant in every organ of the body and have similar relative densities and are arranged with regular spacing. 143 , 144 The regular spacing of macrophages in tissues has been explained by self‐avoidance or self‐repulsion. 145 Macrophages may actively surveil large areas 146 of their environment through highly motile filopodia 145 and actively repel neighbouring macrophages when encountered. Thus, macrophages may establish territories in a cell‐autonomous manner. The mutual repulsion theory may not be an entirely sufficient explanation, as macrophages are densely packed in splenic red pulp and the subcapsular sinus of the lymph node, 113 , 147 , 148 compared to the T‐cell zone of these two organs where macrophages are regularly patterned. 149 Thus, the repulsion hypothesis may not be sufficient to explain macrophage density in some organs, with other variables such as tissue‐specific factors or inflammatory status playing a role in macrophage density. Zhou et al. 150 used the concept of carrying capacity from evolutionary biology to postulate that each tissue has an abundant population of cells like fibroblasts whose numbers are regulated by the carrying capacity of that tissue (the carrying capacity of a tissue is influenced by the availability of glucose, oxygen, space and other growth factors). The abundant tissue fibroblast population can then in turn negatively regulate an accessory population of cells, such as macrophages. Fibroblasts form a cell‐circuit based on growth factor exchange with macrophages. 150 , 151 Fibroblasts produce the macrophage survival factor CSF‐1, whilst macrophages provide the fibroblast growth factor PDGFs. Both CSF‐1 and its receptor (CSF1R) are rapidly internalised upon binding allowing for negative feedback regulation of macrophage numbers. This reductionist explanation may also provide a template for complex models involving multiple cell types, secreted factors and physical interactions coming together to regulate macrophage density in tissues. Also, since macrophage numbers are well below the carrying capacity of tissues, inflammation may transiently change the status quo and lead to increases in the number of macrophages.
During inflammation, apoptotic macrophages may produce chemotactic factors to attract monocytes which would then clear the dying cell and occupy the vacant site. 49 , 152 , 153 Another possibility is that an increase in local concentration of tropic factors (e.g. CSF‐1) after macrophage death may cause neighbouring macrophages to divide and occupy the available space. 154 Yet, another possibility is that inflammatory conditions may lead to downregulation of macrophage‐repulsive function and increase their number in tissues. The degree of inflammation is correlated with the engraftment efficiency of infiltrating monocytes. 109 During Listeria infection, the recruitment and differentiation of monocytes into KCs are regulated by the release of IL‐1 from dying KCs. 155 The time window of inflammation may also affect infiltrating monocyte engraftment vs. repopulation by dividing tissue macrophages, as the infiltrating monocytes are at a disadvantage because they must differentiate into macrophages before they can engraft. This may be the reason why tissues in a state of constant inflammation witness the highest turnover rate of tissue macrophages. For example, infiltrating monocytes replace gut macrophages only after the establishment of gut microbiota 156 and the contribution of infiltrating monocytes to gut macrophages is very low in antibiotic‐treated and germ‐free mice. 156 , 157 Monocyte‐derived macrophages also gradually replace tissue macrophages in organs (kidney, heart, liver) that are subject to continuous low‐grade inflammation because of mechanical or metabolic inflammation 158 , 159 and levels of monocyte‐derived macrophages in tissues may be an indicator of the inflammatory state of specific tissue. 160 Thus, in summary each tissue has a certain macrophage density under homeostatic conditions that can be substantially altered by inflammatory conditions.
We are what we eat: immunometabolism of macrophages
Metabolic pathways contribute to the development, fate and behaviour of macrophages and are critical for induction of inflammatory responses and initiation of tissue healing. 13 , 161 , 162 , 163 The plastic nature of macrophages is reflected in the ability of macrophages to make dramatic changes to their intracellular metabolism in response to environmental and inflammatory cues. In macrophages treated with lipopolysaccharide (LPS), prototypical of inflamed macrophages, the Warburg effect is observed with a preference towards glycolysis over oxidative phosphorylation. 164 Inflammatory activation of macrophages by LPS hampers pyruvate transport to the mitochondria and inhibits the TCA cycle. Pyruvate generated during glycolysis is preferentially converted to lactate instead of being shuttled into mitochondria to be converted into acetyl‐CoA to fuel the tricarboxylic acid cycle (TCA) cycle. The many inflammatory stimuli [e.g. pathogen‐associated molecular patterns (PAMPs) and damage‐associated molecular patterns (DAMPs)] that activate NF‐κB also lead to activation of HIF1α which in turn causes macrophages to switch to glycolysis and inhibit TCA cycle. 165
The preference for glycolysis is conducive for an inflammatory response in macrophages. Glycolysis is not only a faster source of ATPs but also has other roles during inflammation. For example, lactate produced by glycolysis is involved in termination of inflammation. 166 Increased lactate promotes histone acetylation that leads to arginase 1 expression and resolution of inflammation. 166 , 167 The pentose phosphate pathway which is highly activated in a glycolytic cell provides ribose sugars and NADPH for biosynthetic pathways essential for macrophage inflammatory response. 75 , 168 LPS inhibits the expression of SHPK (sedoheptulose kinase) that controls the non‐oxidative phase of the pentose phosphate pathway. This inhibition increases the availability of ribose to be used for fatty acid and sterol synthesis pathways. The enhanced commitment to glycolysis in activated macrophages also supports the production of inflammatory mediators (e.g. TNF‐α, CCL2, IL‐12 and nitric oxide), and these mediators in turn have an inhibitory effect on critical steps of the TCA cycle. 168 In macrophages, LPS boosts the expression of several rate‐limiting enzymes in glycolysis, including hexokinase 3, 169 PFKFB3 (6‐phosphofructo‐2‐kinase/ fructose‐2,6‐biphosphatase 3) and pyruvate kinase isozymes 2 (PKM2). 169 , 170 These changes are coupled to the inhibition of the key TCA cycle enzymes, isocitrate dehydrogenase and succinate dehydrogenase leading to accumulation of citrate and succinate. 168 , 171 , 172 The autocrine type I IFN pathway is responsible for the inhibition of isocitrate dehydrogenase in LPS‐stimulated macrophages. 173 Nitric oxide (NO) produced by M1 macrophages can also lead to suppression and loss of mitochondrial electron transport chain (ETC) complexes and rerouting of pyruvate away from pyruvate dehydrogenase (PDH) to promoting glutamine‐based anaplerosis. 174 These TCA cycle intermediates get diverted to other biosynthetic reactions specific to inflammatory metabolism. The full spectrum of inflammatory activation by macrophages requires increased expression of glycolytic enzymes and accumulation of TCA cycle intermediates. For example, hexokinase is needed for inflammasome activation and release of IL‐1β 175 , 176 ; similarly, PKM2 serves to increases glycolytic flux by induction of GLUT‐1 in the nucleus and serves as a co‐activator for HIF1α. 177 PKM2/HIF1α complexes bind to the Il1β promoter and induce IL‐1β expression. 178 The accumulation of certain metabolites can support macrophage activation or restore homeostasis. Succinate, can drive IL‐1β production via stabilisation of HIF1α, 172 whereas citrate accumulation can, via malonylation of GAPDH, promote TNF‐α translation. 179
In addition to LPS, the effect of other factors on the metabolism of macrophages and their inflammatory status has been studied. For example, insulin has been shown to enhance glycolysis and IL‐1β secretion in intraperitoneal macrophages. 180 IL‐1β is also known to activate macrophages in the pancreas leading to β‐cell dysfunction and death, 181 explaining a link between chronic elevation of IL‐1β signalling and type 2 diabetes. 182 Indeed, in patients with type 2 diabetes, blockade of interleukin‐1 with IL‐1 receptor antagonist anakinra improved glycemia and β‐cell secretory function and reduced markers of systemic inflammation. 183 Macrophages exposed to oxidised phospholipids in hyperlipidemic states use mitochondrial respiration, feeding the Krebs cycle with glutamine and causing the accumulation of oxaloacetate in the cytoplasm. This subsequently leads to increased IL‐1β production, resulting in hyperinflammation. 184 Oxidised LDL also can bind to CD36 on macrophages and suppress oxidative phosphorylation leading to mitochondrial ROS production, which drives chronic inflammation. 185 Macrophages exposed to extracellular pathogenic lipids can activate a triggering receptor expressed on myeloid cells 2 (TREM2)‐dependent gene response involved in phagocytosis and lipid catabolism. 186 , 187 TREM2 expression is required for a metabolic switch towards glycolysis and is essential for the maintenance of healthy energy metabolism under conditions of stress. 90 , 188 TREM2 signalling also drives the formation of lipid‐associated macrophages (LAM) in adipose tissue. LAMs regulate systemic lipid homeostasis in obesity 90 and may also be involved in suppression of obesity‐induced inflammation. 90 TREM2 macrophages are also reported to play a role in neurodegenerative disease 189 and atherosclerosis. 87 Hypoxia can induce glycolysis in macrophages, for example tumor‐associated macrophages (TAMs) present in the hypoxic regions of tumors express HIF‐1α inducing a switch to glycolytic fermentation. High amounts of lactic acid present in the tumor microenvironment also stabilise the expression of HIF‐1α and cause M1 to M2 polarisation. 167 Hypoxia also promotes pro‐tumoral activities of TAMs by increasing the availability of iron for tumor cell proliferation and by causing upregulation of DNA damage‐inducible transcript 4 (DDT4), which inhibits the mechanistic target of rapamycin (mTOR) pathway to promote OXPHOS and reduced glucose intake in TAMs. 190
In contrast to LPS‐treated macrophages, IL‐4‐treated M2 macrophages are more dependent on OXPHOS and have an intact TCA cycle. 191 The elevated OXPHOS in M2 macrophages is supported by increased FAO. 77 There is some debate as to the role of glycolysis in M2 macrophages. 192 However, both glucose and glutamine seem to support OXPHOS and M2 polarisation. 193 Macrophage activation by IL‐4 stimulates the Akt‐mTORC1 pathway which regulates ATP citrate lyase (ACLY), a transferase that catalyses the conversion of citrate and coenzyme A to acetyl‐CoA, leading to increased histone acetylation and M2 gene induction. 194 In comparison, the impaired OXPHOS in LPS‐treated macrophages can reduce acetyl‐CoA levels and alter histone acetylation, leading to impaired expression of inflammatory genes and tolerance. 195 LPS stimulation of macrophages also results in reduction of FAO, 168 whereas IL‐4 can induce FAO through transcription factors STAT‐6 and PGC1β. 76 In summary, macrophage plasticity is most likely supported by their remarkable ability to remodel their core metabolic pathways in response to a range of signals. This rewiring of metabolism provides a faster source of energy, activates biosynthetic pathways needed for inflammation, stimulates the production of inflammatory mediators such as IL‐1β and TNF‐α and sets the ground for shutdown of inflammation in a time‐delayed manner.
The relationship between monocytes and macrophages
No discussion of macrophages is complete without an understanding of the origin and function of monocytes. Monocytes can give rise to macrophages under pathological conditions and can support near‐complete reconstitution of tissue macrophages after depletion. Human monocytes are divided into three groups based on the expression of CD14 and CD16 on HLA‐DR+ cells. CD14+CD16− monocytes are referred to as classical monocytes, CD14+CD16+ cells as intermediate cells and CD14‐CD16+ monocytes are referred as non‐classical monocytes. Mouse monocytes are divided into Ly6Chi monocytes (also defined as CX3CR1int CCR2+ CD62L+ CD43lo) and Ly6Clo monocytes (CX3CR1hi CCR2− CD62L− CD43hi). 34 , 72 , 81 , 196 Transcriptional comparisons correlate mouse Ly6Chi monocytes with ‘classical’ CD14+CD16− monocytes in humans and Ly6Clo monocytes with ‘non‐classical’ CD14lo CD16+ monocytes. Despite similar transcriptional profiles and cell surface marker expression, differences exist between human and mouse monocytes. For example, major histocompatibility complex (MHC II) is expressed on Ly6Clo monocytes and absent on mouse Ly6Chi monocytes, but human monocytes overall are positive for MHC II. 197
Monocyte origins and egress from BM
According to the classical model of monocyte development, monocytes arise from haematopoietic stem cell‐derived common myeloid progenitor (CMP) with granulocyte–macrophage progenitors (GMPs), macrophage (monocyte)/ dendritic cell precursor (MDP) and common monocyte progenitor (cMoP) acting as intermediates. 198 Yanez et al. showed that MDPs arise directly from CMPs directly and give rise to monocytes via cMoPs. 199 Also recently, Liu et al. 200 used Ms4a3 reporter mice (a specific gene reporter for GMPs) and showed that MDPs do not arise from GMPs and that monocytes arise from both GMP and MDPs. Emergency monopoiesis can also give rise to granulocyte like segregated nucleus containing Ly6Clo monocytes (SatM). 201 Ly6Chi monocytes egress out of BM in a CCR2/CCL2/CCL7. 202 , 203 , 204 and CXCR4 ‐dependent manner. 205 , 206 , 207 CCL2 and CCR2‐deficient mice show increased number of monocytes in the BM but fewer numbers in the periphery. 202 Ly6Chi monocytes in the BM parenchyma are juxtaposed to nestin+ stromal cells. 206 , 208 CCL2 binding to CCR2 leads to desensitisation of monocyte response to CXCL12 because of internalisation of CCR2‐CXCR4 complex, which weakens CXCR4 binding and causes egress of monocytes out of BM. 206 , 209 The release of Ly6Chi monocytes from BM is also regulated by circadian rhythm. Ly6Chi monocyte egress from BM peaks between 4 and 8 hours after light onset and is controlled by the circadian rhythm transcription factor, Bmal1. 210 The number of circulating monocytes is strongly linked to the physiological status of an organism 211 and depends on monocyte production and release from BM and peripheral reservoirs. Exercise, age and a host of other pathophysiological conditions (e.g. chronic inflammatory disorders) can also influence the number and ratio of monocyte subsets. 212 , 213 , 214 , 215
Monocyte reprogramming or conversion to macrophages
Classical monocytes (Ly6Chi monocytes in mice) have a diverse differentiation potential because of their plastic transcriptional profile which allows them to take on different roles under homeostatic conditions. Classical monocytes comprise over 90% of circulating monocytes, 81 and upon extravasation into tissues, they contribute to the innate immune response via production of TNF‐α and NO, or by differentiating into macrophages and dendritic cells. 72 Ly6Chi monocytes can replace embryo‐derived tissue‐resident macrophages by differentiating into macrophages. 69 , 222 Conversion of monocyte to tissue macrophages is accompanied by extensive transcriptional changes to mirror the transcriptome of resident macrophages. Even though monocyte‐derived macrophages adopt most of the functions associated with the tissue‐resident macrophages that they are replacing, some epigenetic, transcriptional and functional differences remain. 11 , 99 , 223 , 224 Some monocytes can also remain within tissues, show minimal transcriptional change and act as a local monocyte reservoir. 225 These monocytes can survey resident tissues and transport antigen to lymph nodes. 226 Once in the lymph nodes, they can either differentiate into dendritic cells or remain as monocytes while losing their ability to recirculate. 72 Thus, monocytes as macrophage precursor cells that mirror the flexibility and plastic nature of macrophages can readily replace tissue macrophages.
Tissue‐Resident Macrophages and the Relevance of MPS Classification
Monocytes are rapidly recruited to sites of inflammation/injury and depending on the situation they encounter they undergo different cell fates. Under conditions of inflammation, tissue injury or macrophage depletion, embryonically derived macrophages undergo death and are replaced by monocyte‐derived macrophages. 69 , 71 , 155 , 227 , 228 Long‐term integration of monocyte‐derived macrophages depends on the type of tissue and conditions encountered. For example, monocyte‐derived macrophages do not integrate into the CNS after injury, 64 but do integrate into the heart with ageing and after a myocardial infarction (MI). 220 , 227 They also integrate into the peritoneal cavity after thioglycolate challenge 17 and in the liver after KC depletion. 224 Under inflammatory conditions, they take on pro‐inflammatory effector functions and DC‐like functions such as antigen presentation and migration to LNs. In addition to monocyte‐derived macrophages, peritoneal cavity macrophages and pericardial macrophages can also be recruited to sites of inflammation. For example, Gata6+ peritoneal cavity macrophages are recruited to help resolve inflammation in the setting of sterile liver injury, 229 and Gata6+ macrophages in mouse pericardial fluid contribute to reparative immune response in heart following experimental MI. 230 Since these macrophages do not have to take a vascular route to get to the sites of injury or undergo differentiation into macrophages, they may represent rapid responders to the site of injury.
Tissue‐resident macrophages are imprinted to have a higher tolerance to stimuli associated with acute inflammation, while macrophages derived from infiltrating monocytes may be more inflammatory. In experimental autoimmune encephalomyelitis (EAE) which is a commonly used murine model for multiple sclerosis, infiltrating monocytes trigger EAE progression. 18 Monocyte‐derived macrophages in EAE are highly phagocytic, express pro‐inflammatory genes such as IL‐1β and TNF‐α 231 , 232 and initiate demyelination, whereas microglia are inert and appear to be dedicated to the clearance of debris. 231 CCR2‐deficient animals (deficient for recruitment of Ly6Chi monocytes) and mice depleted for Ly6Chi monocytes are relatively protected from EAE. 233 Monocyte‐derived cells and microglia remain distinct entities during disease progression. Following recovery, recruited monocytes vanish and do not integrate into the resident microglial pool, while the microglia can enter the cell cycle and return to quiescence following remission from EAE. 64 Such a scenario, where tissue‐resident macrophages have higher inflammatory signal threshold, is also supported by studies focusing on acutely inflamed gut 218 , 234 and liver. 235 Monocyte‐derived macrophages also replace the Kupffer cells lost because of inflammation in nonalcoholic steatohepatitis (NASH) a form of nonalcoholic fatty liver disease. A NASH diet was found to induce significant changes in resident Kupffer cell gene expression and result in cell death, while monocyte‐derived macrophages replacing the lost Kupffer cells exhibited convergent epigenomes, transcriptomes and functions. 187
Gut macrophages are the largest macrophage population in the mouse, and macrophages in the intestinal lamina propria are continuously replaced by blood monocytes in the adult mouse. 157 TGF‐β‐dependent monocyte differentiation in the colonic lamina propria causes rapid downregulation of inflammatory signalling molecules and rapid upregulation of receptors involved in apoptotic cell recognition. 107 However, the intestine has a population of TIM4+CD4+ macrophages that can self‐maintain for months. 157
The liver and lung macrophage populations are seeded primarily from foetal liver‐derived monocytes and maintained by self‐renewal. 43 , 67 Depletion of KCs in adult mice would result in BM‐derived monocytes occupying the vacant sinusoidal location and adopting the transcriptomic profile and clearance functions of the cells they replaced. 224 , 236 , 237 YS macrophages, foetal liver monocytes or adult BM monocytes when transplanted to Csf2r −/−mice can acquire the differentiated alveolar macrophage phenotype. 99 When all the three subtypes were mixed and transferred to Csf2r −/− mice, preferential outgrowth of foetal monocytes was observed, correlating with better GM‐CSF sensitivity. When transferred separately, however, all precursors colonised the alveolar niche and generated AMs that were transcriptionally almost identical and self‐maintained. 99
Brain macrophage populations are established during embryonic development and are maintained independently of monocytes. Microglia are constantly replaced by proliferation in the adult mouse brain, 152 and the perivascular macrophages have also been shown to be maintained independently of monocytes. 153 However, donor monocytes could replace brain macrophages in irradiated mouse chimeras, 49 , 153 macrophage‐deficient, PU.1 knockout mice 238 and Csf1r –/– mice at birth. 239 As in liver, subtle transcriptome differences are detected between resident microglia and the engrafted macrophages. 223
Cardiac macrophages originate from YS and foetal monocyte progenitors, and four different types of macrophages have been reported. 69 , 220 Monocyte‐derived macrophages increase in the heart with age, and Ly6Chi monocytes were able to differentiate into long‐lasting populations of cardiac macrophages after macrophage depletion. 220 F4/80hi peritoneal macrophages are also slowly replaced by differentiation of F4/80lo MHCII+ monocyte‐derived progenitors. All the above evidence points to the continued relevance of the MPS model where blood monocytes can and do enter tissues to progressively replace tissue macrophages.
Conclusion
Macrophages are key players in the immune system, but beyond their role as sentinels, macrophages play a crucial role during development and homeostasis. After starting out with a relatively homogenous gene expression profile in the embryo, macrophages become specialised for disparate functions in different tissues. This diversity of function makes us rethink their classification as a single‐cell type. Despite functional specialisation in different locations, macrophages are amazingly plastic with a fluid identity. Monocyte‐derived macrophages further add to the complexity by functionally replacing embryo‐derived macrophages but still retaining a lower threshold for inflammatory activation and not quite taking over the reparative function of tissue‐resident macrophages. Much still needs to be understood regarding the origin and maintenance of tissue‐resident macrophages. We need to fully understand the reparative properties of embryo‐resident macrophages and why they are progressively lost with age and why monocyte‐derived macrophages are lacking in their reparative capability. Exploiting these diverse macrophage functions for therapeutic benefit is a promising strategy in a range of pathologies.
Funding
This work was supported by NIH funding to PRN (HL137799).
Conflicts of interest
The authors declare no conflicts of interest.
Author Contribution
Andrew Fleetwood: Writing‐review & editing. Andrew Murphy: Conceptualization; Writing‐review & editing. G Sreejit: Conceptualization; Writing‐review & editing. P Nagareddy: Conceptualization; Writing‐review & editing.
References
- 1. Metchnikoff O. Life of Elie Metchnikoff, 1845–1916. London: Houghton Mifflin Company, 1921. [Google Scholar]
- 2. Yona S, Gordon S. From the reticuloendothelial to mononuclear phagocyte system ‐ the unaccounted years. Front Immunol 2015; 6: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davis MM, Boniface JJ, Reich Z et al Ligand recognition by αβ T cell receptors. Annu Rev Immunol 1998; 16: 523–544. [DOI] [PubMed] [Google Scholar]
- 4. Stuart LM, Ezekowitz RA. Phagocytosis: elegant complexity. Immunity 2005; 22: 539–550. [DOI] [PubMed] [Google Scholar]
- 5. Chitu V, Stanley ER. Regulation of Embryonic and Postnatal Development by the CSF‐1 Receptor. Curr Top Dev Biol 2017; 123: 229–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hulsmans M, Clauss S, Xiao L et al Macrophages Facilitate Electrical Conduction in the Heart. Cell 2017; 169: 510–522 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nicolás‐Ávila JA, Lechuga‐Vieco AV, Esteban‐Martínez L et al A Network of Macrophages Supports Mitochondrial Homeostasis in the Heart. Cell 2020; 183: 94–109.e23. 10.1016/j.cell.2020.08.031 [DOI] [PubMed] [Google Scholar]
- 8. Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity 2016; 44: 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proc Natl Acad Sci USA 2013; 110: 9415–9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lavine KJ, Epelman S, Uchida K et al Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. PNAS 2014; 111: 16029–16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lavin Y, Winter D, Blecher‐Gonen R et al Tissue‐resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014; 159: 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gautier EL, Shay T, Miller J et al Gene‐expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 2012; 13: 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tabas I, Bornfeldt KE. Intracellular and intercellular aspects of macrophage immunometabolism in atherosclerosis. Circ Res 2020; 126: 1209–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013; 496: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Furth R, Cohn ZA, Hirsch JG, Humphrey JH, Spector WG, Langevoort HL. The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells. Bull World Health Organ 1972; 46: 845–852. [PMC free article] [PubMed] [Google Scholar]
- 16. van Furth R, Cohn ZA. The origin and kinetics of mononuclear phagocytes. J Exp Med 1968; 128: 415–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yona S, Kim KW, Wolf Y et al Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 2013; 38: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self‐renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci 2007; 10: 1538–1543. [DOI] [PubMed] [Google Scholar]
- 19. Ginhoux F, Greter M, Leboeuf M et al Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010; 330: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Merad M, Manz MG, Karsunky H et al Langerhans cells renew in the skin throughout life under steady‐state conditions. Nat Immunol 2002; 3: 1135–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mizoguchi S, Takahashi K, Takeya M, Naito M, Morioka T. Development, differentiation, and proliferation of epidermal Langerhans cells in rat ontogeny studied by a novel monoclonal antibody against epidermal Langerhans cells, RED‐1. J Leukoc Biol 1992; 52: 52–61. [DOI] [PubMed] [Google Scholar]
- 22. Naito M, Takahashi K. The role of Kupffer cells in glucan‐induced granuloma formation in the liver of mice depleted of blood monocytes by administration of strontium‐89. Lab Invest 1991; 64: 664–674. [PubMed] [Google Scholar]
- 23. Sorokin SP, Hoyt RF Jr. Macrophage development: I. Rationale for using Griffonia simplicifolia isolectin B4 as a marker for the line. Anat Rec 1992; 232: 520–526. [DOI] [PubMed] [Google Scholar]
- 24. Bigley V, Haniffa M, Doulatov S et al The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J Exp Med 2011; 208: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hambleton S, Salem S, Bustamante J et al IRF8 mutations and human dendritic‐cell immunodeficiency. N Engl J Med 2011; 365: 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collin MP, Hart DN, Jackson GH et al The fate of human Langerhans cells in hematopoietic stem cell transplantation. J Exp Med 2006; 203: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mielcarek M, Kirkorian AY, Hackman RC et al Langerhans cell homeostasis and turnover after nonmyeloablative and myeloablative allogeneic hematopoietic cell transplantation. Transplantation 2014; 98: 563–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanitakis J, Petruzzo P, Dubernard JM. Turnover of epidermal Langerhans' cells. N Engl J Med 2004; 351: 2661–2662. [DOI] [PubMed] [Google Scholar]
- 29. Bajpai G, Schneider C, Wong N et al The human heart contains distinct macrophage subsets with divergent origins and functions. Nat Med 2018; 24: 1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bittmann I, Bottino A, Baretton GB et al The role of graft‐resident Kupffer cells and lymphocytes of donor type during the time course after liver transplantation–a clinico‐pathological study. Virchows Arch 2003; 443: 541–548. [DOI] [PubMed] [Google Scholar]
- 31. Bittmann I, Dose T, Baretton GB et al Cellular chimerism of the lung after transplantation. An interphase cytogenetic study. Am J Clin Pathol 2001; 115: 525–533. [DOI] [PubMed] [Google Scholar]
- 32. Byrne AJ, Powell JE, O'Sullivan BJ et al Dynamics of human monocytes and airway macrophages during healthy aging and after transplant. J Exp Med 2020; 217: e20191236 https://rupress.org/jem/article/217/3/e20191236/133575/Dynamics‐of‐human‐monocytes‐and‐airway‐macrophages [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nayak DK, Zhou F, Xu M et al Long‐Term Persistence of Donor Alveolar Macrophages in Human Lung Transplant Recipients That Influences Donor‐Specific Immune Responses. Am J Transplant 2016; 16: 2300–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jung S, Aliberti J, Graemmel P et al Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 2000; 20: 4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salter MW, Beggs S. Sublime microglia: expanding roles for the guardians of the CNS. Cell 2014; 158: 15–24. [DOI] [PubMed] [Google Scholar]
- 36. Varol C, Mildner A, Jung S. Macrophages: development and tissue specialization. Annu Rev Immunol 2015; 33: 643–675. [DOI] [PubMed] [Google Scholar]
- 37. Dzierzak E, Bigas A. Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell 2018; 22: 639–651. [DOI] [PubMed] [Google Scholar]
- 38. Bertrand JY, Jalil A, Klaine M, Jung S, Cumano A, Godin I. Three pathways to mature macrophages in the early mouse yolk sac. Blood 2005; 106: 3004–3011. [DOI] [PubMed] [Google Scholar]
- 39. Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 1999; 126: 5073–5084. [DOI] [PubMed] [Google Scholar]
- 40. Tober J, Koniski A, McGrath KE et al The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood 2007; 109: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ji RP, Phoon CK, Aristizabal O, McGrath KE, Palis J, Turnbull DH. Onset of cardiac function during early mouse embryogenesis coincides with entry of primitive erythroblasts into the embryo proper. Circ Res 2003; 92: 133–135. [DOI] [PubMed] [Google Scholar]
- 42. McGrath KE, Koniski AD, Malik J, Palis J. Circulation is established in a stepwise pattern in the mammalian embryo. Blood 2003; 101: 1669–1676. [DOI] [PubMed] [Google Scholar]
- 43. Gomez Perdiguero E, Klapproth K, Schulz C et al Tissue‐resident macrophages originate from yolk‐sac‐derived erythro‐myeloid progenitors. Nature 2015; 518: 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McGrath KE, Frame JM, Fegan KH et al Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep 2015; 11: 1892–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mass E, Ballesteros I, Farlik M et al Specification of tissue‐resident macrophages during organogenesis. Science 2016; 353: aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Naito M, Takahashi K, Nishikawa S. Development, differentiation, and maturation of macrophages in the fetal mouse liver. J Leukoc Biol 1990; 48: 27–37. [DOI] [PubMed] [Google Scholar]
- 47. Naito M, Umeda S, Yamamoto T et al Development, differentiation, and phenotypic heterogeneity of murine tissue macrophages. J Leukoc Biol 1996; 59: 133–138. [DOI] [PubMed] [Google Scholar]
- 48. Takahashi K, Yamamura F, Naito M. Differentiation, maturation, and proliferation of macrophages in the mouse yolk sac: a light‐microscopic, enzyme‐cytochemical, immunohistochemical, and ultrastructural study. J Leukoc Biol 1989; 45: 87–96. [DOI] [PubMed] [Google Scholar]
- 49. Mildner A, Schmidt H, Nitsche M et al Microglia in the adult brain arise from Ly‐6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci 2007; 10: 1544–1553. [DOI] [PubMed] [Google Scholar]
- 50. Lang RA, Bishop JM. Macrophages are required for cell death and tissue remodeling in the developing mouse eye. Cell 1993; 74: 453–462. [DOI] [PubMed] [Google Scholar]
- 51. Stevens B, Allen NJ, Vazquez LE et al The classical complement cascade mediates CNS synapse elimination. Cell 2007; 131: 1164–1178. [DOI] [PubMed] [Google Scholar]
- 52. Fantin A, Vieira JM, Gestri G et al Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF‐mediated endothelial tip cell induction. Blood 2010; 116: 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dai XM, Ryan GR, Hapel AJ et al Targeted disruption of the mouse colony‐stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 2002; 99: 111–120. [DOI] [PubMed] [Google Scholar]
- 54. McKercher SR, Torbett BE, Anderson KL et al Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 1996; 15: 5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 55. Soza‐Ried C, Hess I, Netuschil N, Schorpp M, Boehm T. Essential role of c‐myb in definitive hematopoiesis is evolutionarily conserved. Proc Natl Acad Sci USA 2010; 107: 17304–17308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lieu YK, Reddy EP. Conditional c‐myb knockout in adult hematopoietic stem cells leads to loss of self‐renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci USA 2009; 106: 21689–21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sumner R, Crawford A, Mucenski M, Frampton J. Initiation of adult myelopoiesis can occur in the absence of c‐Myb whereas subsequent development is strictly dependent on the transcription factor. Oncogene 2000; 19: 3335–3342. [DOI] [PubMed] [Google Scholar]
- 58. Schulz C, Gomez Perdiguero E et al A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012; 336: 86–90. [DOI] [PubMed] [Google Scholar]
- 59. Boisset JC, van Cappellen W, Andrieu‐Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 2010; 464: 116–120. [DOI] [PubMed] [Google Scholar]
- 60. Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 2010; 464: 112–115. [DOI] [PubMed] [Google Scholar]
- 61. Kumaravelu P, Hook L, Morrison AM et al Quantitative developmental anatomy of definitive haematopoietic stem cells/long‐term repopulating units (HSC/RUs): role of the aorta‐gonad‐mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development 2002; 129: 4891–4899. [DOI] [PubMed] [Google Scholar]
- 62. Cumano A, Godin I. Ontogeny of the hematopoietic system. Annu Rev Immunol 2007; 25: 745–785. [DOI] [PubMed] [Google Scholar]
- 63. Gomez Perdiguero E, Geissmann F. Myb‐independent macrophages: a family of cells that develops with their tissue of residence and is involved in its homeostasis. Cold Spring Harb Symp Quant Biol 2013; 78: 91–100. [DOI] [PubMed] [Google Scholar]
- 64. Ajami B, Bennett JL, Krieger C, McNagny KM, Rossi FM. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat Neurosci 2011; 14: 1142–1149. [DOI] [PubMed] [Google Scholar]
- 65. Sere K, Baek JH, Ober‐Blobaum J et al Two distinct types of Langerhans cells populate the skin during steady state and inflammation. Immunity 2012; 37: 905–916. [DOI] [PubMed] [Google Scholar]
- 66. Samokhvalov IM, Samokhvalova NI, Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature 2007; 446: 1056–1061. [DOI] [PubMed] [Google Scholar]
- 67. Hoeffel G, Chen J, Lavin Y et al C‐Myb+ erythro‐myeloid progenitor‐derived fetal monocytes give rise to adult tissue‐resident macrophages. Immunity 2015; 42: 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hoeffel G, Wang Y, Greter M et al Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac‐derived macrophages. J Exp Med 2012; 209: 1167–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Epelman S, Lavine KJ, Beaudin AE et al Embryonic and adult‐derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 2014; 40: 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Guilliams M, De Kleer I, Henri S et al Alveolar macrophages develop from fetal monocytes that differentiate into long‐lived cells in the first week of life via GM‐CSF. J Exp Med 2013; 210: 1977–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hashimoto D, Chow A, Noizat C et al Tissue‐resident macrophages self‐maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 2013; 38: 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jakubzick C, Gautier EL, Gibbings SL et al Minimal differentiation of classical monocytes as they survey steady‐state tissues and transport antigen to lymph nodes. Immunity 2013; 39: 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M‐1/M‐2 macrophages and the Th1/Th2 paradigm. J Immunol 2000; 164: 6166–6173. [DOI] [PubMed] [Google Scholar]
- 74. Sans‐Fons MG, Yeramian A, Pereira‐Lopes S et al Arginine transport is impaired in C57Bl/6 mouse macrophages as a result of a deletion in the promoter of Slc7a2 (CAT2), and susceptibility to Leishmania infection is reduced. J Infect Dis 2013; 207: 1684–1693. [DOI] [PubMed] [Google Scholar]
- 75. Haschemi A, Kosma P, Gille L et al The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab 2012; 15: 813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vats D, Mukundan L, Odegaard JI et al Oxidative metabolism and PGC‐1β attenuate macrophage‐mediated inflammation. Cell Metab 2006; 4: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Huang SC, Everts B, Ivanova Y et al Cell‐intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol 2014; 15: 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008; 8: 958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 2011; 11: 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 2012; 122: 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 2003; 19: 71–82. [DOI] [PubMed] [Google Scholar]
- 82. Hanna RN, Carlin LM, Hubbeling HG et al The transcription factor NR4A1 (Nur77) controls bone marrow differentiation, the survival of Ly6C‐ monocytes. Nat Immunol 2011; 12: 778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 2004; 25: 677–686. [DOI] [PubMed] [Google Scholar]
- 84. Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 2014; 6: e13 https://facultyopinions.com/prime/reports/pubmed/24669294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xue J, Schmidt SV, Sander J et al Transcriptome‐based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014; 40: 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chevrier S, Levine JH, Zanotelli VRT et al An immune atlas of clear cell renal cell carcinoma. Cell 2017; 169: 736–749 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cochain C, Vafadarnejad E, Arampatzi P et al Single‐Cell RNA‐Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res 2018; 122: 1661–1674. [DOI] [PubMed] [Google Scholar]
- 88. Lin JD, Nishi H, Poles J et al Single‐cell analysis of fate‐mapped macrophages reveals heterogeneity, including stem‐like properties, during atherosclerosis progression and regression. JCI Insight 2019; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mould KJ, Jackson ND, Henson PM, Seibold M, Janssen WJ. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight 2019; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jaitin DA, Adlung L, Thaiss CA et al Lipid‐Associated Macrophages Control Metabolic Homeostasis in a Trem2‐Dependent Manner. Cell 2019; 178: 686–698 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hill DA, Lim HW, Kim YH et al Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc Natl Acad Sci USA 2018; 115: E5096–E5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Czapla J, Matuszczak S, Wisniewska E et al human cardiac mesenchymal stromal cells with CD105+CD34− phenotype enhance the function of post‐infarction heart in mice. PLoS One 2016; 11: e0158745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Martinez FO, Helming L, Milde R et al Genetic programs expressed in resting and IL‐4 alternatively activated mouse and human macrophages: similarities and differences. Blood 2013; 121: e57–e69. [DOI] [PubMed] [Google Scholar]
- 94. Nahrendorf M, Swirski FK. Abandoning M1/M2 for a Network Model of Macrophage Function. Circ Res 2016; 119: 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Murray PJ, Allen JE, Biswas SK et al Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 2014; 41: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Adlung L, Amit I. From the Human Cell Atlas to dynamic immune maps in human disease. Nat Rev Immunol 2018; 18: 597–598. [DOI] [PubMed] [Google Scholar]
- 97. Rozenblatt‐Rosen O, Stubbington MJT, Regev A, Teichmann SA. The Human Cell Atlas: from vision to reality. Nature 2017; 550: 451–453. [DOI] [PubMed] [Google Scholar]
- 98. Gosselin D, Link VM, Romanoski CE et al Environment drives selection and function of enhancers controlling tissue‐specific macrophage identities. Cell 2014; 159: 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. van de Laar L, Saelens W, De Prijck S et al Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue‐resident macrophages. Immunity 2016; 44: 755–768. [DOI] [PubMed] [Google Scholar]
- 100. Heinz S, Benner C, Spann N et al Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. MafB is an inducer of monocytic differentiation. EMBO J 2000; 19: 1987–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Soucie EL, Weng Z, Geirsdottir L et al Lineage‐specific enhancers activate self‐renewal genes in macrophages and embryonic stem cells. Science 2016; 351: aad5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Takayanagi H, Kim S, Koga T et al Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell 2002; 3: 889–901. [DOI] [PubMed] [Google Scholar]
- 104. Butovsky O, Jedrychowski MP, Moore CS et al Identification of a unique TGF‐β‐dependent molecular and functional signature in microglia. Nat Neurosci 2014; 17: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fuller K, Lean JM, Bayley KE, Wani MR, Chambers TJ. A role for TGFβ1 in osteoclast differentiation and survival. J Cell Sci 2000; 113(Pt 13): 2445–2453. [DOI] [PubMed] [Google Scholar]
- 106. Kaplan DH, Li MO, Jenison MC, Shlomchik WD, Flavell RA, Shlomchik MJ. Autocrine/paracrine TGFβ1 is required for the development of epidermal Langerhans cells. J Exp Med 2007; 204: 2545–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Schridde A, Bain CC, Mayer JU et al Tissue‐specific differentiation of colonic macrophages requires TGFβ receptor‐mediated signaling. Mucosal Immunol 2017; 10: 1387–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yu X, Buttgereit A, Lelios I et al The Cytokine TGF‐β Promotes the Development and Homeostasis of Alveolar Macrophages. Immunity 2017; 47: 903–912 e4. [DOI] [PubMed] [Google Scholar]
- 109. Bonnardel J, T'Jonck W, Gaublomme D et al Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 2019; 51: 638–654.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chakrabarti R, Celia‐Terrassa T, Kumar S et al Notch ligand Dll1 mediates cross‐talk between mammary stem cells and the macrophageal niche. Science 2018; 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Outtz HH, Tattersall IW, Kofler NM, Steinbach N, Kitajewski J. Notch1 controls macrophage recruitment and Notch signaling is activated at sites of endothelial cell anastomosis during retinal angiogenesis in mice. Blood 2011; 118: 3436–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sakai M, Troutman TD, Seidman JS et al Liver‐derived signals sequentially reprogram myeloid enhancers to initiate and maintain kupffer cell identity. Immunity 2019; 51: 655–670.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Haldar M, Kohyama M, So AY et al Heme‐mediated SPI‐C induction promotes monocyte differentiation into iron‐recycling macrophages. Cell 2014; 156: 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR α. Nature 1996; 383: 728–731. [DOI] [PubMed] [Google Scholar]
- 115. A Gonzalez N, Guillen JA, Gallardo G et al The nuclear receptor LXRα controls the functional specialization of splenic macrophages. Nat Immunol 2013; 14: 831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Buechler MB, Kim KW, Onufer EJ et al A stromal niche defined by expression of the transcription factor WT1 mediates programming and homeostasis of cavity‐resident macrophages. Immunity 2019; 51: 119–130.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Okabe Y, Medzhitov R. Tissue‐specific signals control reversible program of localization and functional polarization of macrophages. Cell 2014; 157: 832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Cohen M, Giladi A, Gorki AD et al Lung single‐cell signaling interaction map reveals basophil role in macrophage imprinting. Cell 2018; 175: 1031–1044 e18. [DOI] [PubMed] [Google Scholar]
- 119. Wang Y, Bugatti M, Ulland TK, Vermi W, Gilfillan S, Colonna M. Nonredundant roles of keratinocyte‐derived IL‐34 and neutrophil‐derived CSF1 in Langerhans cell renewal in the steady state and during inflammation. Eur J Immunol 2016; 46: 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Capucha T, Koren N, Nassar M et al Sequential BMP7/TGF‐β1 signaling and microbiota instruct mucosal Langerhans cell differentiation. J Exp Med 2018; 215: 481–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Yasmin N, Bauer T, Modak M et al Identification of bone morphogenetic protein 7 (BMP7) as an instructive factor for human epidermal Langerhans cell differentiation. J Exp Med 2013; 210: 2597–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lin H, Lee E, Hestir K et al Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science 2008; 320: 807–811. [DOI] [PubMed] [Google Scholar]
- 123. Ma X, Lin WY, Chen Y et al Structural basis for the dual recognition of helical cytokines IL‐34 and CSF‐1 by CSF‐1R. Structure 2012; 20: 676–687. [DOI] [PubMed] [Google Scholar]
- 124. Stanley ER, Heard PM. Factors regulating macrophage production and growth. Purification and some properties of the colony stimulating factor from medium conditioned by mouse L cells. J Biol Chem 1977; 252: 4305–4312. [PubMed] [Google Scholar]
- 125. Jenkins SJ, Hume DA. Homeostasis in the mononuclear phagocyte system. Trends Immunol 2014; 35: 358–367. [DOI] [PubMed] [Google Scholar]
- 126. Chitu V, Stanley ER. Colony‐stimulating factor‐1 in immunity and inflammation. Curr Opin Immunol 2006; 18: 39–48. [DOI] [PubMed] [Google Scholar]
- 127. Dai XM, Zong XH, Sylvestre V, Stanley ER. Incomplete restoration of colony‐stimulating factor 1 (CSF‐1) function in CSF‐1‐deficient Csf1op/Csf1op mice by transgenic expression of cell surface CSF‐1. Blood 2004; 103: 1114–1123. [DOI] [PubMed] [Google Scholar]
- 128. Nandi S, Akhter MP, Seifert MF, Dai XM, Stanley ER. Developmental and functional significance of the CSF‐1 proteoglycan chondroitin sulfate chain. Blood 2006; 107: 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Pridans C, Raper A, Davis GM et al Pleiotropic impacts of macrophage and microglial deficiency on development in rats with targeted mutation of the Csf1r locus. J Immunol 2018; 201: 2683–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. MacDonald KP, Palmer JS, Cronau S et al An antibody against the colony‐stimulating factor 1 receptor depletes the resident subset of monocytes and tissue‐ and tumor‐associated macrophages but does not inhibit inflammation. Blood 2010; 116: 3955–3963. [DOI] [PubMed] [Google Scholar]
- 131. Gow DJ, Sauter KA, Pridans C et al Characterisation of a novel Fc conjugate of macrophage colony‐stimulating factor. Mol Ther 2014; 22: 1580–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hawley CA, Rojo R, Raper A et al Csf1r‐mApple transgene expression and ligand binding in vivo reveal dynamics of CSF1R expression within the mononuclear phagocyte system. J Immunol 2018; 200: 2209–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Jenkins SJ, Ruckerl D, Thomas GD et al IL‐4 directly signals tissue‐resident macrophages to proliferate beyond homeostatic levels controlled by CSF‐1. J Exp Med 2013; 210: 2477–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Stutchfield BM, Antoine DJ, Mackinnon AC et al CSF1 restores innate immunity after liver injury in mice and serum levels indicate outcomes of patients with acute liver failure. Gastroenterology 2015; 149: 1896–1909 e14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 135. Horiuchi K, Miyamoto T, Takaishi H et al Cell surface colony‐stimulating factor 1 can be cleaved by TNF‐α converting enzyme or endocytosed in a clathrin‐dependent manner. J Immunol 2007; 179: 6715–6724. [DOI] [PubMed] [Google Scholar]
- 136. Blevins G, Fedoroff S. Microglia in colony‐stimulating factor 1‐deficient op/op mice. J Neurosci Res 1995; 40: 535–544. [DOI] [PubMed] [Google Scholar]
- 137. Greter M, Lelios I, Pelczar P et al Stroma‐derived interleukin‐34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity 2012; 37: 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kondo Y, Duncan ID. Selective reduction in microglia density and function in the white matter of colony‐stimulating factor‐1‐deficient mice. J Neurosci Res 2009; 87: 2686–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wang Y, Szretter KJ, Vermi W et al IL‐34 is a tissue‐restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol 2012; 13: 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wiktor‐Jedrzejczak W, Bartocci A, Ferrante AW Jr et al Total absence of colony‐stimulating factor 1 in the macrophage‐deficient osteopetrotic (op/op) mouse. Proc Natl Acad Sci USA 1990; 87: 4828–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Hwang SJ, Choi B, Kang SS et al Interleukin‐34 produced by human fibroblast‐like synovial cells in rheumatoid arthritis supports osteoclastogenesis. Arthritis Res Ther 2012; 14: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Tian Y, Shen H, Xia L, Lu J. Elevated serum and synovial fluid levels of interleukin‐34 in rheumatoid arthritis: possible association with disease progression via interleukin‐17 production. J Interferon Cytokine Res 2013; 33: 398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hume DA, Irvine KM, Pridans C. The Mononuclear Phagocyte System: The Relationship between Monocytes and Macrophages. Trends Immunol 2019; 40: 98–112. [DOI] [PubMed] [Google Scholar]
- 144. Uderhardt S, Martins AJ, Tsang JS, Lammermann T, Germain RN. Resident macrophages cloak tissue microlesions to prevent neutrophil‐driven inflammatory damage. Cell 2019; 177: 541–555 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Avetisyan M, Rood JE, Huerta Lopez S et al Muscularis macrophage development in the absence of an enteric nervous system. Proc Natl Acad Sci USA 2018; 115: 4696–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Poche RA, Hsu CW, McElwee ML, Burns AR, Dickinson ME. Macrophages engulf endothelial cell membrane particles preceding pupillary membrane capillary regression. Dev Biol 2015; 403: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Mondor I, Baratin M, Lagueyrie M et al Lymphatic endothelial cells are essential components of the subcapsular sinus macrophage niche. Immunity 2019; 50: 1453–1466 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Guilliams M, Thierry GR, Bonnardel J, Bajenoff M. Establishment and maintenance of the macrophage niche. Immunity 2020; 52: 434–451. [DOI] [PubMed] [Google Scholar]
- 149. Baratin M, Simon L, Jorquera A et al T Cell Zone Resident Macrophages Silently Dispose of Apoptotic Cells in the Lymph Node. Immunity 2017; 47: 349–362.e5. [DOI] [PubMed] [Google Scholar]
- 150. Zhou X, Franklin RA, Adler M et al Circuit design features of a stable two‐cell system. Cell 2018; 172: 744–757.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Adler M, Mayo A, Zhou X et al Principles of cell circuits for tissue repair and fibrosis. iScience 2020; 23: 100841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Askew K, Li K, Olmos‐Alonso A et al Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep 2017; 18: 391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Goldmann T, Wieghofer P, Jordao MJ et al Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol 2016; 17: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Hume DA, Pavli P, Donahue RE, Fidler IJ. The effect of human recombinant macrophage colony‐stimulating factor (CSF‐1) on the murine mononuclear phagocyte system in vivo. J Immunol 1988; 141: 3405–3409. [PubMed] [Google Scholar]
- 155. Bleriot C, Dupuis T, Jouvion G, Eberl G, Disson O, Lecuit M. Liver‐resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type‐2‐mediated tissue repair during bacterial infection. Immunity 2015; 42: 145–158. [DOI] [PubMed] [Google Scholar]
- 156. Bain CC, Bravo‐Blas A, Scott CL et al Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol 2014; 15: 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Shaw TN, Houston SA, Wemyss K et al Tissue‐resident macrophages in the intestine are long lived and defined by Tim‐4 and CD4 expression. J Exp Med 2018; 215: 1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Berry MR, Mathews RJ, Ferdinand JR et al Renal sodium gradient orchestrates a dynamic antibacterial defense zone. Cell 2017; 170: 860–874.e19. [DOI] [PubMed] [Google Scholar]
- 159. Lambrecht B, Guilliams M. Monocytes find a new place to dwell in the niche of heartbreak hotel. J Exp Med 2014; 211: 2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Medzhitov R. Origin and physiological roles of inflammation. Nature 2008; 454: 428–435. [DOI] [PubMed] [Google Scholar]
- 161. Murray PJ, Rathmell J. Pearce E. SnapShot: Immunometabolism. Cell Metab 2015; 22: 190.e1. [DOI] [PubMed] [Google Scholar]
- 162. O'Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol 2016; 16: 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Russell DG, Huang L, VanderVen BC. Immunometabolism at the interface between macrophages and pathogens. Nat Rev Immunol 2019; 19: 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annu Rev Immunol 2014; 32: 609–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Majmundar AJ, Wong WJ, Simon MC. Hypoxia‐inducible factors and the response to hypoxic stress. Mol Cell 2010; 40: 294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Zhang D, Tang Z, Huang H et al Metabolic regulation of gene expression by histone lactylation. Nature 2019; 574: 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Colegio OR, Chu NQ, Szabo AL et al Functional polarization of tumour‐associated macrophages by tumour‐derived lactic acid. Nature 2014; 513: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Van den Bossche J, O'Neill LA, Menon D. Macrophage Immunometabolism: Where Are We (Going)? Trends immunol 2017; 38: 395–406. [DOI] [PubMed] [Google Scholar]
- 169. Nishizawa T, Kanter JE, Kramer F et al Testing the role of myeloid cell glucose flux in inflammation and atherosclerosis. Cell Rep 2014; 7: 356–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Ruiz‐Garcia A, Monsalve E, Novellasdemunt L et al Cooperation of adenosine with macrophage Toll‐4 receptor agonists leads to increased glycolytic flux through the enhanced expression of PFKFB3 gene. J Biol Chem 2011; 286: 19247–19258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. O'Neill LA. A broken krebs cycle in macrophages. Immunity 2015; 42: 393–394. [DOI] [PubMed] [Google Scholar]
- 172. Ryan DG, Murphy MP, Frezza C et al Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat Metab 2019; 1: 16–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. De Souza DP, Achuthan A, Lee MK et al Autocrine IFN‐I inhibits isocitrate dehydrogenase in the TCA cycle of LPS‐stimulated macrophages. J Clin Invest 2019; 129: 4239–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Palmieri EM, Gonzalez‐Cotto M, Baseler WA et al Nitric oxide orchestrates metabolic rewiring in M1 macrophages by targeting aconitase 2 and pyruvate dehydrogenase. Nat Commun 2020; 11: 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Hughes MM, O'Neill LAJ. Metabolic regulation of NLRP3. Immunol Rev 2018; 281: 88–98. [DOI] [PubMed] [Google Scholar]
- 176. Moon JS, Hisata S, Park MA et al mTORC1‐Induced HK1‐Dependent Glycolysis Regulates NLRP3 Inflammasome Activation. Cell Rep 2015; 12: 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 177. Fukuzumi M, Shinomiya H, Shimizu Y, Ohishi K, Utsumi S. Endotoxin‐induced enhancement of glucose influx into murine peritoneal macrophages via GLUT1. Infect Immun 1996; 64: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Palsson‐McDermott EM, Curtis AM, Goel G et al Pyruvate kinase M2 regulates Hif‐1α activity and IL‐1β induction and is a critical determinant of the warburg effect in LPS‐activated macrophages. Cell Metab 2015; 21: 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Galvan‐Pena S, Carroll RG, Newman C et al Malonylation of GAPDH is an inflammatory signal in macrophages. Nat Commun 2019; 10: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Dror E, Dalmas E, Meier DT et al Postprandial macrophage‐derived IL‐1β stimulates insulin, and both synergistically promote glucose disposal and inflammation. Nat Immunol 2017; 18: 283–292. [DOI] [PubMed] [Google Scholar]
- 181. Haneklaus M, O'Neill LA. NLRP3 at the interface of metabolism and inflammation. Immunol Rev 2015; 265: 53–62. [DOI] [PubMed] [Google Scholar]
- 182. Dinarello CA. Interleukin‐1 in the pathogenesis and treatment of inflammatory diseases. Blood 2011; 117: 3720–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Larsen CM, Faulenbach M, Vaag A et al Interleukin‐1‐receptor antagonist in type 2 diabetes mellitus. N Engl J Med 2007; 356: 1517–1526. [DOI] [PubMed] [Google Scholar]
- 184. Di Gioia M, Spreafico R, Springstead JR et al Endogenous oxidized phospholipids reprogram cellular metabolism and boost hyperinflammation. Nat Immunol 2020; 21: 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Chen Y, Yang M, Huang W et al Mitochondrial Metabolic Reprogramming by CD36 Signaling Drives Macrophage Inflammatory Responses. Circ Res 2019; 125: 1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Deczkowska A, Keren‐Shaul H, Weiner A, Colonna M, Schwartz M, Amit I. Disease‐Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018; 173: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 187. Seidman JS, Troutman TD, Sakai M et al Niche‐Specific Reprogramming of Epigenetic Landscapes Drives Myeloid Cell Diversity in Nonalcoholic Steatohepatitis. Immunity 2020; 52: 1057–1074.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Piers TM, Cosker K, Mallach A et al A locked immunometabolic switch underlies TREM2 R47H loss of function in human iPSC‐derived microglia. FASEB J 2020; 34: 2436–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Keren‐Shaul H, Spinrad A, Weiner A et al A unique microglia type associated with restricting development of Alzheimer's Disease. Cell 2017; 169: 1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- 190. Puthenveetil A, Dubey S. Metabolic reprograming of tumor‐associated macrophages. Ann Transl Med 2020; 8: 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Jha AK, Huang SC, Sergushichev A et al Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015; 42: 419–430. [DOI] [PubMed] [Google Scholar]
- 192. Viola A, Munari F, Sanchez‐Rodriguez R, Scolaro T, Castegna A. The metabolic signature of macrophage responses. Front Immunol 2019; 10: 1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Liu PS, Wang H, Li X et al α‐ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 2017; 18: 985–994. [DOI] [PubMed] [Google Scholar]
- 194. Covarrubias AJ, Aksoylar HI, Yu J et al Akt‐mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. Elife 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Langston PK, Nambu A, Jung J et al Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses. Nat Immunol 2019; 20: 1186–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Palframan RT, Jung S, Cheng G et al Inflammatory chemokine transport and presentation in HEV: a remote control mechanism for monocyte recruitment to lymph nodes in inflamed tissues. J Exp Med 2001; 194: 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Ingersoll MA, Spanbroek R, Lottaz C et al Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 2010; 115: e10–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Hettinger J, Richards DM, Hansson J et al Origin of monocytes and macrophages in a committed progenitor. Nat Immunol 2013; 14: 821–830. [DOI] [PubMed] [Google Scholar]
- 199. Yanez A, Coetzee SG, Olsson A et al Granulocyte‐monocyte progenitors and monocyte‐dendritic cell progenitors independently produce functionally distinct monocytes. Immunity 2017; 47: 890–902.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Liu Z, Gu Y, Chakarov S et al Fate mapping via ms4a3‐expression history traces monocyte‐derived cells. Cell 2019; 178: 1509–1525.e19. [DOI] [PubMed] [Google Scholar]
- 201. Satoh T, Nakagawa K, Sugihara F et al Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 2017; 541: 96–101. [DOI] [PubMed] [Google Scholar]
- 202. Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol 2006; 7: 311–317. [DOI] [PubMed] [Google Scholar]
- 203. Tsou CL, Peters W, Si Y et al Critical roles for CCR2 and MCP‐3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest 2007; 117: 902–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Shi C, Jia T, Mendez‐Ferrer S et al Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll‐like receptor ligands. Immunity 2011; 34: 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Chong SZ, Evrard M, Devi S et al CXCR4 identifies transitional bone marrow premonocytes that replenish the mature monocyte pool for peripheral responses. J Exp Med 2016; 213: 2293–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Jung H, Mithal DS, Park JE, Miller RJ. Localized CCR2 Activation in the Bone Marrow Niche Mobilizes Monocytes by Desensitizing CXCR4. PLoS One 2015; 10: e0128387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Liu Q, Li Z, Gao JL et al CXCR4 antagonist AMD3100 redistributes leukocytes from primary immune organs to secondary immune organs, lung, and blood in mice. Eur J Immunol 2015; 45: 1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol 2011; 11: 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Evrard M, Chong SZ, Devi S et al Visualization of bone marrow monocyte mobilization using Cx3cr1 gfp/+ Flt3L ‐/ ‐ reporter mouse by multiphoton intravital microscopy. J Leukoc Biol 2015; 97: 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6Chi inflammatory monocytes. Science 2013; 341: 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Selimoglu‐Buet D, Wagner‐Ballon O, Saada V et al Characteristic repartition of monocyte subsets as a diagnostic signature of chronic myelomonocytic leukemia. Blood 2015; 125: 3618–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Heimbeck I, Hofer TP, Eder C et al Standardized single‐platform assay for human monocyte subpopulations: Lower CD14+CD16++ monocytes in females. Cytometry Part A 2010; 77: 823–830. [DOI] [PubMed] [Google Scholar]
- 213. Seidler S, Zimmermann HW, Bartneck M, Trautwein C, Tacke F. Age‐dependent alterations of monocyte subsets and monocyte‐related chemokine pathways in healthy adults. BMC Immunol 2010; 11: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Verschoor CP, Johnstone J, Millar J et al Alterations to the frequency and function of peripheral blood monocytes and associations with chronic disease in the advanced‐age, frail elderly. PLoS One 2014; 9: e104522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Sreejit G, Abdel‐Latif A, Athmanathan B et al Neutrophil‐Derived S100A8/A9 amplify granulopoiesis after myocardial infarction. Circulation 2020; 141: 1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Bain CC, Scott CL, Uronen‐Hansson H et al Resident and pro‐inflammatory macrophages in the colon represent alternative context‐dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol 2013; 6: 498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Tamoutounour S, Guilliams M, Montanana Sanchis F et al Origins and functional specialization of macrophages and of conventional and monocyte‐derived dendritic cells in mouse skin. Immunity 2013; 39: 925–938. [DOI] [PubMed] [Google Scholar]
- 218. Zigmond E, Varol C, Farache J et al Ly6Chi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen‐presenting cells. Immunity 2012; 37: 1076–1090. [DOI] [PubMed] [Google Scholar]
- 219. Tamoutounour S, Henri S, Lelouard H et al CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1‐inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol 2012; 42: 3150–3166. [DOI] [PubMed] [Google Scholar]
- 220. Molawi K, Wolf Y, Kandalla PK et al Progressive replacement of embryo‐derived cardiac macrophages with age. J Exp Med 2014; 211: 2151–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Calderon B, Carrero JA, Ferris ST et al The pancreas anatomy conditions the origin and properties of resident macrophages. J Exp Med 2015; 212: 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Mossadegh‐Keller N, Gentek R, Gimenez G, Bigot S, Mailfert S, Sieweke MH. Developmental origin and maintenance of distinct testicular macrophage populations. J Exp Med 2017; 214: 2829–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Cronk JC, Filiano AJ, Louveau A et al Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J Exp Med 2018; 215: 1627–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Scott CL, Zheng F, De Baetselier P et al Bone marrow‐derived monocytes give rise to self‐renewing and fully differentiated Kupffer cells. Nat Commun 2016; 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Swirski FK, Nahrendorf M, Etzrodt M et al Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 2009; 325: 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Randolph GJ, Inaba K, Robbiani DF, Steinman RM, Muller WA. Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo . Immunity 1999; 11: 753–761. [DOI] [PubMed] [Google Scholar]
- 227. Heidt T, Courties G, Dutta P et al Differential contribution of monocytes to heart macrophages in steady‐state and after myocardial infarction. Circ Res 2014; 115: 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Theurl I, Hilgendorf I, Nairz M et al On‐demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat Med 2016; 22: 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Wang J, Kubes P. A Reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell 2016; 165: 668–678. [DOI] [PubMed] [Google Scholar]
- 230. Deniset JF, Belke D, Lee WY et al Gata6+ pericardial cavity macrophages relocate to the injured heart and prevent cardiac fibrosis. Immunity 2019; 51: 131–140.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Yamasaki R, Lu H, Butovsky O et al Differential roles of microglia and monocytes in the inflamed central nervous system. J Exp Med 2014; 211: 1533–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Vainchtein ID, Vinet J, Brouwer N et al In acute experimental autoimmune encephalomyelitis, infiltrating macrophages are immune activated, whereas microglia remain immune suppressed. Glia 2014; 62: 1724–1735. [DOI] [PubMed] [Google Scholar]
- 233. Mildner A, Mack M, Schmidt H et al CCR2+Ly‐6Chi monocytes are crucial for the effector phase of autoimmunity in the central nervous system. Brain 2009; 132: 2487–2500. [DOI] [PubMed] [Google Scholar]
- 234. Weber B, Saurer L, Schenk M, Dickgreber N, Mueller C. CX3CR1 defines functionally distinct intestinal mononuclear phagocyte subsets which maintain their respective functions during homeostatic and inflammatory conditions. Eur J Immunol 2011; 41: 773–779. [DOI] [PubMed] [Google Scholar]
- 235. Zigmond E, Bernshtein B, Friedlander G et al Macrophage‐restricted interleukin‐10 receptor deficiency, but not IL‐10 deficiency, causes severe spontaneous colitis. Immunity 2014; 40: 720–733. [DOI] [PubMed] [Google Scholar]
- 236. Beattie L, Sawtell A, Mann J et al Bone marrow‐derived and resident liver macrophages display unique transcriptomic signatures but similar biological functions. J Hepatol 2016; 65: 758–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. David BA, Rezende RM, Antunes MM et al Combination of mass cytometry and imaging analysis reveals origin, location, and functional repopulation of liver myeloid cells in mice. Gastroenterology 2016; 151: 1176–1191. [DOI] [PubMed] [Google Scholar]
- 238. Beers DR, Henkel JS, Xiao Q et al Wild‐type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA 2006; 103: 16021–16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Bennett FC, Bennett ML, Yaqoob F et al A Combination of Ontogeny and CNS Environment Establishes Microglial Identity. Neuron 2018; 98: 1170–1183.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]


