Abstract
Background
Stenotrophomonas species are multi-resistant bacteria with ability to cause opportunistic infections
Objective
We isolated 45 Stenotrophomonas species from soil, sewage and the clinic with the aim of investigating their susceptibility to commonly used antimicrobial agents.
Methodology
The identities of isolates were confirmed with 16S rRNA gene sequence and MALDI-TOF analysis. Anti-microbial resistance, biofilm production and clonal diversity were also evaluated. The minimum inhibitory concentration technique as described by Clinical & Laboratory Standards Institute: CLSI Guidelines (CLSI) was employed for the evaluation of isolate susceptibility to antibiotics.
Result
Forty-five Stenotrophomonas species which include 36 environmental strains and 9 clinical strains of S. maltophilia were considered in this study. 32 (88.9 %) environmental strains were identified to be S. maltophilia, 2 (5.6 %) were Stenotrophomonas nitritireducens, and 2 (5.6 %) cluster as Stenotrophomonas spp. Stenotrophomonas isolates were resistant to at least six of the antibiotics tested, including Trimethoprim/Sulfamethoxazole (SXT).
Conclusion
Environmental isolates from this study were resistant to SXT which is commonly used for the treatment of S. maltophilia infections. This informs the need for good public hygiene as the environment could be a reservoir of multi-resistant bacteria. It also buttresses the importance of surveillance study in the management of bacterial resistance.
Keywords: Stenotrophomonas, environmental, biochemical characterization, clonal diversity, anti-microbial susceptibility
Introduction
The genus Stenotrophomonas is made up of ubiquitous motile Gram-negative bacteria. The most commonly isolated species both in the hospital and from the environment is Stenotrophomonas maltophilia. Environmental strains of Stenotrophomonas have been reported to be beneficial such as helping in plant growth, bioremediation and biodegradation, and secretion of siderophores1–4. However, S. maltophilia has been implicated in many nosocomial infections as an opportunistic pathogen in individuals with various conditions, such as compromised immune system, cystic fibrosis, chronic pulmonary obstructive disease, cancer, meningitis, soft tissue infections, and wound infections in diabetic patients, keratitis, biliary sepsis, and endocarditis5. One major characteristic that contributes to the nosocomial importance of Stenotrophomonas strains is their high rate of antimicrobial resistance such that the treatment of Stenotrophomonas infection is often very difficult. Stenotrophomonas are opportunistic pathogens which have been regarded as member of the multi-drug resistance (MDR) pathogens because of their resistance to many antimicrobial agents.6–8. MDR pathogen associated infections has resulted in long hospital stay, increased cost of treatment and may even lead to death in affected patients9.
There are many mechanisms involved in resistance to antimicrobial agents, including the presence of some multi efflux pumps, inactivating enzymes, outer membrane modification and the alteration of target sites10. The resistant strains harbor both inherent and acquired antibiotic resistant genes which either could be chromosomally borne or from other vectors11,12. For example, beta-lactamase genes are located on the chromosome10. Similarly, the Sul genes which has been found responsible for resistance to trimethoprim sulfamethoxazole, are related to a class I integron13. Also, Alonso & Martinez, 2000, and Sanchez et al., 200914,15 showed that the resistance of S. maltophilia D457 to fluoroquinolones was due to the presence of a multi-drug resistant efflux pump in its genome. Stenotrophomonas resistance to major antibiotics has been associated with the SmrA. Al-Hamad, Upton & Burnie, 200916 reported an increase in resistance to some antibiotics (including fluoroquinolones, tetracycline, doxorubicin and multiple dyes) following the overexpression of S. maltophilia SmrA gene in acrAB mutant Escherichia coli strain SM1411 with an increased rate extrusion of norfloxacin thus reducing the uptake of norfloxacin by the mutant strain. Similarly, the overexpression of FUA (ABC-type) multidrug efflux pump in S. maltophilia K279a confer on them resistance to fusidic acid17. Another type of ABC-type tripartite efflux pump. MacABCsm was found to influence the ejection of macrolides, aminoglycosides and polymyxins and enhanced the formation of biofilm in S. maltophilia18. Likewise, the presence of two inducible β-lactamases L1 and L2 encoding genes in their genomes19. L1 is a Zn+ dependent β-lactamases which ensures that S. maltophilia is resistant to cephalosporines, carbapenem, penicillin's, and not monobactam, L2 however is an acid sensitive cephalosporine which confers resistance on S. maltophilia to cephalosporines, monobactam and penicillin20,21. Other mechanisms that have been reported in Stenotrophomonas species include plasmid borne resistant gene such as bla PER born on IncA/C Plasmid and bla NDM which conferred extended beta-lactam resistance on a strain of S. maltophilia isolated from the soil in Brazil22. Resistant mechanisms described above in Stenotrophomonas species might be due to the possession of extra-chromosomal genes acquired from other bacteria. A comparison of 19 genomes of Stenotrophomonas showed that they still have an open pangenome23. Hence, the probable reason for gene alteration in the genomes could be through horizontal gene transfer (HGT). Thus, they could acquire virulence, biofilm formation and other resistance genes through HGT. Gene acquisition in Stenotrophomonas species can also confer on them new ecological features such as obligate intra-cellular behavior, survival in extreme conditions, unique metabolic capabilities for the degradation of xenobiotic, and bioremediation of organophosphate contaminated soils.
Bacteria can evolve characteristics which can enhance their survival in different environments. It is common to find bacteria of the same species surviving in different ecological niche. Vidigal-Golcalves, (2014) reported possible niche adaptation in Stenotrophomonas species isolated from cystic fibrosis (CF) Patient. They noted that Stenotrophomonas species from CF patient lack fatty acid structural isomers (11: 0 anteiso, 12: 0 ISO 3OH and 17: 0 anteiso), which were present in the CF non-chronic colonized group24. Thus, it is possible for the environment to influence the susceptibility pattern of Stenotrophomonas species to antimicrobial agents. Most studies have focused on the susceptibility pattern of nosocomial S. maltophilia and reported the trend in their resistant patterns6,14,25 but it is worth mentioning that the environment plays important role in shaping the characteristics of bacteria. Etinosa et al., (2014)11 and Ochoa-Sánchez & Vinuesa (2017)12 reported strains of multi-resistant Stenotrophomonas from different environmental settings. Stenotrophomonas spp. are diverse in the natural communities and the level of diversity may be the source for the multi-resistant strains of infection and the strains with special abilities. Microbial resistance to antimicrobial agents remains a major health care problem contributing a higher percentage of death in reported cases19. One of the appropriate mechanisms for managing the microbial resistance to drug is via susceptibility studies. It usually gives current information about bacteria behavior to antimicrobial agents. Susceptibility study will also give information on the appropriate antimicrobial agents for infection control. Hence, in this study, we characterized environmental and a few clinical Stenotrophomonas strains from Mexico using phenotypic and molecular methods. We also evaluated their susceptibility pattern to antimicrobial agents with a view towards understanding the impact of the environment on Stenotrophomonas species' resistance to antimicrobial agents. We focused on extreme environments such as sanitary sewage, textile effluent polluted sewage, petroleum polluted soil and other distinct types of soils, and water bodies where Stenotrophomonas could develop new adaptive characteristics.
Materials and methods
Sample collection
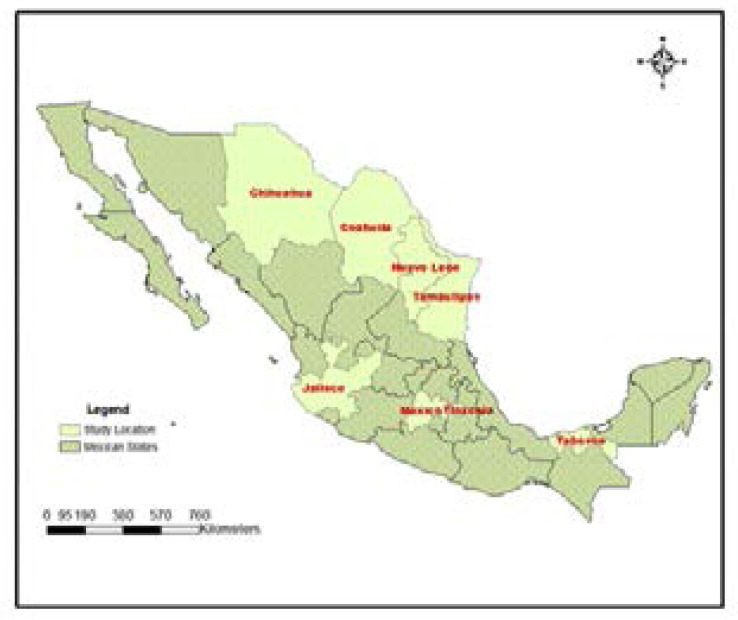
Sewage and soil samples were collected from eight states of Mexico which are Tabasco, Tamaulipas, Tlaxcala, Coahuila, Chihuahua, Mexico state, Jalisco, Nuevo Leon and Mexico City between April 2015 and June 2017. The states were purposively selected because they possess characteristics that can stimulate new adaptation in bacteria. These include crude oil contamination, Textile effluent pollution, animal fecal contamination high temperature climate regions, and the presence of ancient sea larvae (Cuatrocienegas) among others. The criteria for the selection of sewage is the frequency of contamination in the region while different soil samples considered in the study were the soil from current agricultural soils and abandon soil as well highly polluted soils. The clinical strains were obtained from Hospital Civil de Guadalara, Mexico. They were isolated from Bronchoalveolar lavage (1), bile (1), respiratory (2), and tracheal secretion (3) samples, while clinical source of two strains were not indicated and one S. maltophilia ATCC 13637. Each clinical strain was isolated from different patients with underlying disease condition. A total of 51 sewage sites, 96 soils were sampled, and 9 clinical samples were collected for the screening of Stenotrophomonas species.
Isolation of Stenotrophomonas spp.
Two methods were employed for the isolation of environmental Stenotrophomonas spp in this study. One method involved the use of selective medium (Stenotrophomonas Vancomycin Imipenem Amphotericin (StenoVIA) agar, HIMEDIA, India) for the isolation of free-living Stenotrophomonas species from the environment. The second method employed double layer agar plating technique (DLAP) for the isolation of certain complexes of S. maltophilia which have been reported to require a host for their isolation or growth26
Isolation of Stenotrophomonas spp. using StenoVIA agar
Stenotrophomonas spp. were isolated by culturing collected samples on Stenotrophomonas Vancomycin Imipinem AmphotericinB (StenoVIA) Agar (HIMEDIA, India) supplemented with 0.2 mg/mL methionine. The sewage samples were first centrifuged at 3,000 rpm, at 4°C for 20 min and serially diluted to the factor of 10-8 before spreading 100 µL of each diluent on StenoVIA agar plate. Soil samples (100mg), on the other hand, were suspended of soil in 5 mL of Luria-Bertani broth and the suspension was incubated at 30°C overnight. 100 µL of this overnight broth culture was serially diluted as described above for sewage sample and 100 microliters of each dilution was spread on StenoVIA agar plate. The inoculated StenoVIA agar plates were incubated at 30°C for 48 – 72 h. Distinct yellow and dark green bacterial colony that appeared after 48 h were purified by sub-culturing on fresh StenoVIA and LB agar. The bacterial colonies suspected to be Stenotrophomonas spp. were selected for further analysis.
Isolation of endosymbiotic or Intracellularly borne Stenotrophomonas spp. with DLAP
This method was performed as described by27 with some modifications. Water samples collected from sewage channels were centrifuged at 3,000 rpm at 4°C for 20 min. The supernatant was filtered using 0.45 µm syringe filter and serially diluted to the factor of 10-8. Then, 400 µl of overnight grown Escherichia coli culture washed in phosphate buffer was mixed with 100 µl of serially diluted filtered sewage samples. The diluent from the filtrate and E. coli cells were vortexed and added to already prepared bottom dilute nutrient broth agar (DNB) as described previously27. The clean plaque forming unit (PFU) that developed after plate incubation was aseptically cut into DNB and incubated for 48 hrs. The bacteria which grew afterwards was streaked on stenoVIA and LB. Two to three colonies of presumptive Stenotrophomonas spp. colonies were selected for further analysis.
Species identification
Three steps were employed for species identification, including quick biochemical identification, MALTI-DOF check and 16S rRNA gene sequencing.
a. Quick Biochemical Identification
Gram-reaction, oxidase test, catalase, lactose fermentation and resistance to vancomycin, amphotericin and imipenem were employed for the quick identification of isolates. All biochemical test employed for the identification were carried out as previously described in literature28
b. Species Identification with MALTI-DOF
The species identification was firstly done by MALTI-DOF. Strains were prepared for MALDI-TOF mass spectrometry using the manufacturer's recommended direct transfer protocol. A single colony of each strain was inoculated directly onto the MSP 96 target plate spot (Bruker Daltonics, Bremen, Germany). Each spot was overlaid with α-cyano-4-hydroxycinnamic acid matrix (Sigma Aldrich, St. Louis, MO, USA). The target plate was analyzed by the Bruker Microflex LT system (Bruker Daltonics). The protein profile of each spot generated m/z values of 3000–15,000, which was then analyzed by the MALDI Biotyper V.3.1.66 with the most updated spectra library, V.7.0 (7311 spectra). The top 10 identification matches were generated along with confidence scores which ranged from 0.0 to 3.0. A score >2.0 indicated promising species-level identification. If two or more different species were shown within the top 10 matches, the species of the isolate would be considered as indeterminate and repeated29.
c. Molecular Identification of Isolated Stenotrophomonas spp.
The suspected Stenotrophomonas spp. were confirmed by PCR amplification, the 16S rRNA gene. Genomic DNA was extracted from 1 ml overnight grown Stenotrophomonas culture in Luria broth and DNB, using Promega wizard genomic DNA purification kit (Promega ®, Madison, USA) according to the manufacturer's instruction. The 16S rRNA gene was amplified using a pair of primer designed in our laboratory steno1-21F: 5′ AGG GAA ACTTAC GCT AAT ACC- 3′ and steno2-1200R: 5′ CTC TGT CCC TAC CAT TGT AG-3′). PCR reaction mixture was initially denatured at 95°C, for 3 min, following 25 cycles of denaturation at 95°C for 30 secs, annealing temperature at 62°C for 30 secs, extension at 72°C for 5 min, and allowed to cool to 4°C. The amplified PCR product was analyzed using 0.7 % agarose gel electrophoresis.
Sequencing and analysis of PCR products
The PCR amplified products were directly sequenced at the Centro de Biotecnología Genómica, Instituto Politécnico Nacional (IPN), Mexico with the Applied Biosystems 3130 Genetic Analyzers using the Sanger sequencing technology. The 16S rRNA gene sequences were analyzed using the Lasergene program Seqman® software (DNAstar Inc., Madison, Wisconsin, USA), and the Blastn program of the National Center for Biotechnology Institute (NCBI) (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The partial 16S rRNA gene sequences of the Stenotrophomonasstrains isolated have been submitted to the NCBI database with the accession numbers KX470411.1, KX500117.1, KX785139.1 - KX785155.1, KX863509.1, and KY454854.1 - KY454855.1). Details of data submission can be found at GenBank: www.ncbi/nlm.nih.gov.
Physiological characterization of isolates
Isolates identified to be Stenotrophomonas species were characterized by conventional biochemical tests such as sugar fermentation test, lysine decarboxylase test, bile aesculin hydrolysis, decarboxylase activity, urease activity, and gelatin hydrolysis starch and Tween 80 hydrolysis were evaluated as described previously30,31. Growth characteristics of the isolates including optimal temperature and pH were determined as described previously (Lee et al., 2011). E. coli ATCC (8739) and Stenotrophomonas ATCC 13637 were control in the experimental set up while an uninoculated sugar medium serves as the negative control and Pseudomonas aeruginosa (ATCC 27853) also serve as other control check.
Phylogenetic analysis
The 16S rRNA gene sequences were further aligned with other related sequences for Stenotrophomonas spp. retrieved from NCBI database using Mega 6.0. A phylogenetic tree was constructed using neighbor-joining algorithm and reliability of tree topologies was confirmed by bootstrap analysis using 1000 repeat alignment32.
Clonal diversity
The genetic relatedness of isolates was analyzed by amplifying the enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) as described previously33 with slight modifications. The DNA was extracted with Promega DNA Purification Kit according to the manufacturer's instructions. The following primers were used: 5′-ATG TAA GCT CCTGGGGAT TCA C-3′ and 5′-AAG TAA GTG ACT GGG GTG AGC CG-3′. The PCR conditions were 95°C for 3 min, 42 cycles of 95°C for 30 s, 58°C for 30 s, 2 min at 72°C, and 72°C for 10 min. The amplified products were checked by electrophoresis on 2% agarose gels. The band patterns were generated with bio-numeric software package with 1% tolerance (Applied-Maths, http://www.applied-maths.com/bionumerics). Similarity coefficients were generated from a similarity matrix calculated with the Jaccard coefficient using the SPSS 20.0 statistical software package (IBM Corporation, Somers, NY).
Antimicrobial susceptibility testing
Antibiotic susceptibility of the isolated Stenotrophomonas was tested by the broth microdilution method, using the CLSI guidelines for antimicrobial susceptibility testing28 Performance standards for antimicrobial susceptibility testing 2016). The broth microdilution method included levofloxacin, ceftazidime, doxycycline, chloramphenicol, ofloxacin, trimethoprim-sulfamethoxazole, ceftriaxone, imipenem and ampicillin. The minimum inhibitory concentration (MIC) of Stenotrophomonas spp. to essential antibiotics was determined as previously reported28,34. The multiple antibiotic resistance index (MAR= number of antibiotics to which the bacterium is resistant/number of antibiotics tested) for each isolate was determined as described previously35. Stenotrophomonas ATCC 13637 was used as the control strain
Biofilm formation assay
Biofilm formation assay was carried out as previously described36. The optical density (OD) reading at a wavelength of 500 nm was taken on a microtiter plate reader (IMAD plate reader BIO-RAD, USA). The OD value was used to determine the formation of biofilm by isolates. An uninoculated culture in the plate was used as a negative control. The low cut-off included three standard deviations above the mean OD of control wells37. The results were subtracted from the negative control (OD500 = 0.042) and expressed as means. Strains were classified as follows: non-biofilm producer (OD500 = 0.042); weak biofilm producer (≤0.0 OD500 ≤0.1); average biofilm producer (>0.1 OD500 ≤0.58); strong biofilm producer (OD500 ≥ 0.59).
Statistical analysis
Statistical analysis was performed using SPSS version 20.0 (IBM Corp., Somers, NY). Data were presented as numbers and percentages. Chi-square test was used to compare quantitative variables between groups.
Results
Sample Collection, Isolation and Identification of Stenotrophomonas
Samples were collected from selected part in 8 states from Mexico as shown in Figure 1. A total of 141 environmental samples from sewage and soil were collected in addition to 9 samples from the hospital. Only 101 of 151 bacterial strains (one strain/sample) which showed the color characteristic corresponding to that of Stenotrophomonas species in quick biochemical test were selected for further identification. However, 54 strains were confirmed as Stenotrophomonas species by MALTI-DOF. Finally, only 45 strains were identified as Stenotrophomonas spp. by the sequences of 16S rRNA genes.
Figure 1.
This figure represented the selected states in Mexico from which sampling were conducted
Characterization of isolated Stenotrophomonas species
The test for bacterial motility revealed that all isolates are motile except the Stenotrophomonas strain TEPEL. The colony size varies from one to another. For example, the strain A915 and B195 isolated by the method DLAP showed high rate of motility when observed under the light microscope. All isolates grew effectively at 30°C, 35°C and 37°C while only four Stenotrophomonas strains (A915, B915, C915 and D915) obtained from sewage at the distinct sites of the same channel grew at 42°C. The optimal pH for growth was estimated to be in the range 6 or 7–8
The biochemical characteristics observed in the isolated Stenotrophomonas strains are shown in (Table 1 and Table 2)
Table 1.
Categorical information about the utilization of sugars and other biochemical features in isolates
| Sugar | All isolates | Isolates from soil | Isolates from sewage | Clinical isolates |
| Mannitol | 22 (47.8%) | 11 (23.9%) | 10 (21.7%) | 1 (2.2%) |
| Arabinose | 24 (52.0%) | 12 (26.1%) | 11 (23.9%) | 1 (2.2%) |
| Glucose | 28 (60.9%) | 15 (32.6%) | 11 (23.9%) | 2 (4.4%) |
| Sucrose | 15 (32.6%) | 8 (17.4%) | 6 (13.0%) | 1 (2.2%) |
| Trehalose | 27 (58.7%) | 14 (30.4%) | 11 (23.9) | 2 (4.4%) |
| Lactose | 1 (2.2%) | 0 (0%) | 0 (0%) | 1(2.2%) |
| Maltose | 35 (76.1%) | 19 (41.3%) | 12 (26.1%) | 4 (8.7%) |
| Mannose | 27 (58.7%) | 15 (32.6%) | 11 (23.9%) | 1 (2.2%) |
| Fructose | 37 (80.4%) | 22 (47.8%) | 12 (26.1%) | 3 (6.5%) |
| Galactose | 20 (43.5%) | 9 (19.6%) | 11 (23.9%) | 0 (0%) |
| Dulcitol | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Lysine | 16 (34.8%) | 9 (19.6%) | 0 (0%) | 7 (15.2%) |
| Gelatin | 26 (56.2%) | 18 (39.1%) | 8 (17.4%) | 0 (0%) |
| Tween80 | 45 (98%) | 23 (50%) | 12 (26.1%) | 10 (21.7%) |
| Urease | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Starch | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Esculin | 36 (78.3%) | 20 (43.5%) | 7 (15.2%) | 9 (19.1%) |
| Citrate | 22 (47.8%) | 7 (15.2%) | 8 (17.4%) | 7 (15.2%) |
Table 2.
Phenotypic and Biochemical Characteristics of Stenotrophomonas Isolates.
| Strains | MAN | ARA | GLU | SUC | TRE | LAC | MAL | MNO | FRU | GAL | DUL | LYS | GEL | TWE | URE | STA | ESC | CIT | 30°C | 42°C |
| Environmental strains | ||||||||||||||||||||
| A915 | + | + | + | - | + | - | + | + | + | + | - | - | + | + | - | - | - | + | + | + |
| B915 | + | + | + | + | + | - | + | + | + | + | - | - | + | + | - | - | - | + | + | + |
| C915 | + | + | + | + | + | - | + | + | + | + | - | - | + | + | - | - | - | + | + | + |
| D915 | + | + | + | - | + | - | + | + | + | + | - | - | + | + | - | - | - | + | + | + |
| TEPE | + | + | + | - | + | - | + | + | + | + | - | - | + | + | - | - | - | - | + | - |
| TEPEL | - | + | + | - | + | - | + | + | + | + | - | - | + | + | - | - | + | - | + | - |
| SPM1 | - | - | - | + | - | - | + | - | + | - | - | - | + | + | - | - | + | - | + | - |
| SPM3 | - | + | - | - | - | - | + | - | - | - | - | - | + | + | - | - | + | - | + | - |
| ATCM1_1 | - | + | + | - | + | - | + | - | + | - | - | - | + | + | - | - | + | - | + | - |
| ATCM1_3 | + | - | - | - | + | - | + | + | + | - | - | + | + | + | - | - | + | - | + | - |
| ATCM1_4 | - | + | + | - | + | - | + | + | + | + | - | + | + | + | - | - | - | + | + | - |
| ATCM1_5 | - | - | + | - | + | - | + | + | + | + | - | - | + | + | - | - | + | - | + | - |
| ATCM2_1 | - | + | + | - | - | - | + | + | - | - | - | - | + | + | - | - | + | - | + | - |
| ATCM2_2 | + | - | + | + | + | - | + | - | + | - | - | + | + | + | - | - | + | - | + | - |
| ATCM2_3 | + | + | - | + | + | - | + | + | + | + | - | - | + | + | - | - | + | - | + | - |
| LLD1 | + | + | - | - | - | - | - | + | + | - | - | + | + | + | - | - | + | - | + | - |
| LLD2_1 | - | - | + | + | + | - | + | + | + | - | - | - | + | + | - | - | + | - | + | - |
| LLD2_2 | - | - | + | + | - | - | + | - | + | - | - | - | + | + | - | - | + | - | + | - |
| LLD2_3 | + | - | - | + | - | - | + | - | + | - | - | - | + | + | - | - | + | - | + | - |
| LLD2_4 | - | - | - | + | - | - | - | + | + | - | - | - | + | + | - | - | + | - | + | - |
| SDL1 | + | - | + | - | - | - | + | + | + | - | - | + | + | + | - | - | + | - | + | - |
| FTY | - | + | - | + | - | - | + | - | + | - | - | + | + | - | - | + | - | + | - | |
| PEMSOL | - | - | - | - | + | - | - | + | + | - | - | + | + | + | - | - | + | - | + | - |
| TEPEGREEN | + | - | + | - | + | - | + | + | + | + | - | - | + | + | - | - | + | - | + | - |
| LLD3_1 | - | - | + | - | - | - | - | - | + | - | - | - | + | - | - | - | + | - | + | - |
| LLD3_2 | - | - | - | - | - | - | - | - | + | - | - | - | + | + | - | - | + | - | + | - |
| SVIA1 | + | + | + | - | + | - | + | + | + | + | - | + | - | + | - | - | - | + | + | - |
| SVIA2 | + | + | + | - | + | - | + | + | + | + | - | - | - | + | - | - | - | + | + | - |
| ASS1 | + | + | + | - | + | - | + | + | + | + | - | + | - | + | - | - | + | + | + | - |
| ASS2 | - | + | + | + | + | - | + | - | + | + | - | + | - | + | - | - | + | + | + | - |
| J5 | + | + | + | - | + | - | + | + | + | + | - | - | - | + | - | - | + | + | + | - |
| JBL | + | + | + | + | + | - | + | + | + | + | - | - | - | + | - | - | + | + | + | - |
| SM6 | + | + | + | - | + | - | + | + | + | + | - | - | - | + | - | - | + | + | + | - |
| LM2 | + | + | + | + | + | - | + | + | + | + | - | - | - | + | - | - | + | + | + | - |
| RS2B | + | + | + | - | + | - | + | + | + | + | - | - | - | + | - | - | - | + | + | - |
| ASKF | + | + | + | + | + | - | + | + | + | + | - | - | - | + | - | - | + | + | + | - |
| Clinical strains | ||||||||||||||||||||
| 17/1078 | + | + | + | - | + | - | + | - | + | - | - | - | - | + | - | - | - | + | + | - |
| 17/1122 | - | - | - | - | - | + | + | - | + | - | - | - | - | + | - | - | + | - | + | - |
| 17/1129 | - | - | - | + | + | - | + | ND | + | - | ND | + | - | + | - | - | + | + | + | - |
| 17/1174 | - | - | - | - | - | - | - | - | - | - | - | + | - | + | - | - | + | + | + | - |
| 17/1177 | - | - | + | - | - | - | + | - | - | - | - | + | - | + | - | - | + | - | + | - |
| 17/1178 | - | - | - | - | - | - | - | - | - | - | - | + | - | + | - | - | + | + | + | - |
| 17/1184 | - | - | - | - | - | - | - | - | - | - | - | + | - | + | - | - | + | + | + | - |
| 17/1193 | - | - | - | - | - | - | - | - | - | - | - | + | - | + | - | - | + | - | + | - |
| 17/1259 | - | - | - | - | - | - | - | - | - | - | - | + | - | + | - | - | + | + | + | - |
| 17/1293 | - | - | - | - | - | - | - | - | - | - | - | - | - | + | - | - | + | + | + | - |
Note: The characteristics of all environmental isolates and all clinical isolates is shown. Optimal pH growth was 6.5–8.0 for all strains, including environmental and clinical strains. ARA: arabinose utilization; CIT: citrate utilization; DUL: dulcitol utilization; ESC: esculin hydrolysis; FRU: fructose utilization; GAL: galactose utilization; GEL: gelatin hydrolysis; GLU: glucose utilization; LAC: lactose utilization; LYS: lysine decarboxylase activity; MAL: maltose utilization; MAN: mannitol utilization; MNO: mannose utilization; STA: starch hydrolysis; SUC: sucrose utilization; TRE: trehalose utilization; TWE: tween 80 hydrolysis; URE: urease activity; 30°C: Growth at 30°C; 42°C: Growth at 42°C. ND: not determined.
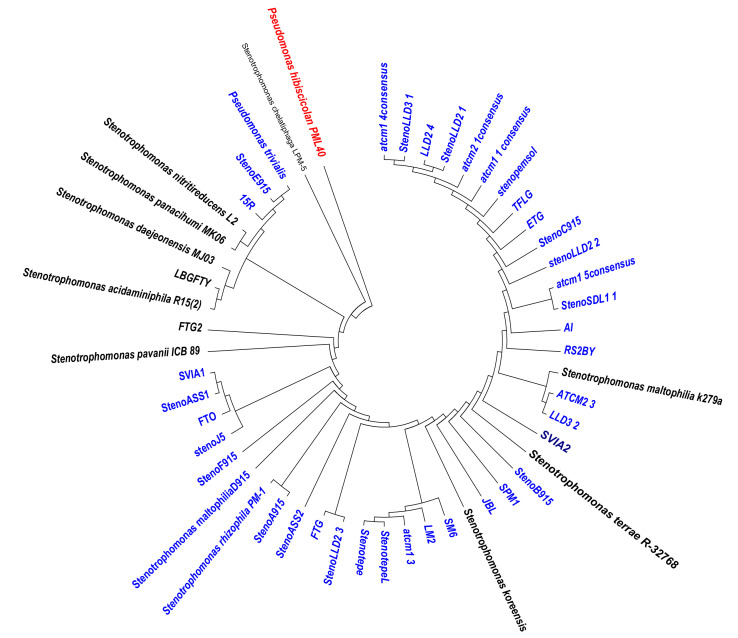
The phylogenetic analysis and clonal diversity
The phylogenetic analysis based on 16S RNA gene showed that the isolates formed two major clades and two singletons. The two major phylogenetic clades consisted of a branch with mostly environmental isolates from soil while the other branch is a mixture of strains isolated from the environments (soil, sewage) and patients (clinical isolates) (Figure 2).
Figure 2.
Phylogenetic tree showing the evolutionary relationship of the isolates with other Stenotrophomonas spp.
Environmental strains from soil were grouped into two main branches, but the strains recovered from sewage belong to only one branch, indicating that the soil strains are more diverse than sewage strains. All the clinical strains were only distributed in the branch with mixed strains, implying such strains in the branch could have some relationship with the clinical strains. We detected the diversity of 36 environmental isolates and all clinical isolates by ERIC method; 7 isolates could not be analyzed using this method due to technical issues. The analyzed isolates showed patterns containing 1 to 7 bands while the clinical isolates did not give any distinct typing pattern as they mostly produced only one band from the PCR amplicon. The percentage of similarity among analyzed isolates ranged from 75% to 100% and only two isolates shared the same pattern (Figure S1), further confirming that members of the genus have high genetic diversity either in intra or interspecies.
Biofilm formation and antimicrobial resistance susceptibility
The ability to form biofilm was examined in isolates and most Stenotrophomonas species isolated (93.1%) produced biofilm, of which one third (38.7%) were weak biofilm producers, 10.3% average biofilm producers, and about half (45.16%) were strong biofilm producers.
MIC based antimicrobial susceptibility test for the isolated Stenotrophomonas strains showed that isolates were resistant to most antimicrobial agents, such as doxycycline (100%), imipenem (100%), ampicillin (100%), trimethoprim-sulfamethoxazole (80.43%), ceftriaxone (71.7%) and chloramphenicol (54.35%). Low resistance to ceftazidime (26%) and none whatsoever to levofloxacin and ofloxacin were detected (Table 3). Meanwhile, the MAR index of all isolates was also tested. The MAR index for most isolates surpassed the 0.5 resistance index. The average MAR index for isolates from the hospital settings is 0.7 (0.67–-0.9) while the average indices are 0.6 (0.4–0.8) for isolates from sewage and 0.7 for those from soil (0.5–0.8) (Table S1).
Table 3.
Antimicrobial Susceptibility of Stenotrophomonas species. Values corresponding to 100% is from 46 isolates
| Antimicrobial agents | MIC (µg/ml) | Percentages (n) | ||||
| Range | MIC50 | MIC90 | Susceptible | Intermediate | Resistant | |
| Trimethoprimsulfamethoxazole | 0.125–4 | >4 | >4 | 19.6 (9) | 0.0 (0) | 80.4 (37) |
| Doxycycline | ≤2–>64 | >64 | >64 | 0.0 (0) | 0.0 (0) | 100.0 (46) |
| Chloramphenicol | ≤4 –>64 | >16 | >32 | 45.7 (21) | 0.0 (0) | 54.4 (25) |
| Imipenem | ≤4 –>64 | >64 | >64 | 0.0 (0) | 0.0 (0) | 100.0 (46) |
| Ceftriaxone | ≤8–>16 | >64 | >64 | 26.1 (12) | 2.2 (1) | 71.7 (33) |
| Ampicillin | ≤4–>16 | >16 | >16 | 0.0 (0) | 0.0 (0) | 100.0 (46) |
| Levofloxacin | ≤1–>8 | 1 | 2 | 100.0 (46) | 0.0 (0) | 0.0 (0) |
| Ofloxacin | ≤1–>8 | 0.87 | 2 | 100.0 (0) | 0.0 (0) | 0.0 (0) |
| Ceftazidime | ≤4–>16 | 4 | >16 | 63.0 (29) | 11.0 (5) | 26.0 (12) |
Discussion
We identified 45 strains as member of the genus Stenotrophomonas from a consensus identification technique involving the methods described earlier. Nine strains were from hospitalized patient samples at the Hospital Civil General Guadalajara, 12 were from sewage samples and 24 strains from soil samples. The 16S rRNA sequence analysis revealed that the isolates showed high similarity with S. maltophilia, S. nitritireducens and Stenotrophomonas spp and thus the isolated strains can be categorized into three species, S. maltophilia42 (91.3%), S. nitritireducen 2 (4.35%) and Stenotrophomonas spp, 2 (4.35%). The 9 clinical isolates were S. maltophilia while the isolates from sewage include 10 S. maltophilia strains, 1 S. nitritireducen (strain FTY) and 1 Stenotrophomonas sp. (strain TEPEL). Two out of the S. maltophilia (strains A915 and B915) obtained from sewage channels were recovered from the plaques obtained by a DLAP method where E. coli was used as the host cells. The isolates obtained from soil include 22 S. maltophilia, 1 S. nitritireducen (ATCM1_4) and 1 Stenotrophomonas sp. (LLD2_1). Stenotrophomonas species have been recovered from a wide range of environments including soil, sewage and clinic environments.1,38,39 S. maltophilia is the most encountered strains in the genus Stenotrophomonas2,40. Perhaps it is the reason for being the most recovered isolates in this study.
The phenotypic characteristics of the isolates conformed with previous studies5,40, however, we noted some deviations in some strains. For example, strain A915 and B915 showed the ability to grow at pH 5.0 and 5.5 and grew at 42°C. Similar, phenomenon was observed in Stenotrophomonas daejeonensis isolated from sewage39. These features could be a form of biological fitness for continuous adaptation in their environment24,41. Likewise, an isolate (S. maltophilia strain Pemsol49) from contaminated regions showed features that could enhance the degradation of different pollutants. Thus, it may possess attribute which could be employed for bioremediation. And such characteristics can be used for biodegradation or bioremediation purposes.
The biochemical behavior of the isolates was tested in 11 sugar and other compounds. All isolates were catalase-positive and oxidase-negative. Most environmental isolates could use fructose (94.4%), maltose (86.1%), glucose (72.2%), mannose (72.2%), trehalose (69.4%), arabinose (63.9%), sucrose (39.0%), mannitol (58.3%), galactose (55.5%) and sucralose (38.9%) as a sole carbon source. None of the isolates used dulcitol and lactose. Most of the isolates hydrolyzed Tween 80 (97.2%) and gelatin (72.2%) but could not hydrolyze starch. Lysine decarboxylase activity was shown in 25% of the isolates. The use of citrate was positive in 41.6% of the isolates. None of the strains could produce urease enzyme. In comparison with the environmental isolates, the clinical strains have less proportion in use of these sugars, fructose (30%), maltose (40%), glucose (20%), trehalose (20%), arabinose (10%), sucrose (10%), lactose (10%) and mannitol (10%) as a sole source of carbon. The use of galactose, mannose and citrate is common for the environmental isolates but not for the clinical strains. Gelatin hydrolysis exists in most environmental strains, whereas no clinical strains have this trait. Aesculin hydrolysis (90%) were shown in higher proportion in the clinical strains. It may be necessary to characterize more clinical strain in the future to be able to ascertain the varied features in the clinical strains as compared with the environmental strains. Assimilation of carbohydrates is an important aspect in the identification of Stenotrophomonas species. Conventionally, S. maltophilia, for example, cannot use some sugars such as arabinose, mannitol, dulcitol, mannose, rhamnose, and salicin, as a sole carbon source5,40. Thus, any bacteria possessing positive features for the use of any of these sugars may not be regarded as S. maltophilia in identification. However, in this study, some Stenotrophomonas isolates could use some of these sugars as sole carbon source (Table 1). For example, 9 S. maltophilia strains isolated from sewage, 11 S. maltophilia from soil and one S. maltophilia from the clinical isolates can use arabinose as sole carbon source for growth. This observation corroborates the findings of Urszula et al., 200942 on the isolation of a novel strains of S. maltophilia KB12, which possess the capability to use arabinose as sole carbon source. The observed variation in sugar utilization by S. maltophilia has led to a proposal for the reassessment of their phenotype43. The result from our study emphasizes the need to re-examine the criteria for the biochemical identification of S. maltophilia, or even for this genus.
The phylogenetic analysis of the isolated strains confirmed that isolates are member of the genus Stenotrophomonas species. The distribution of the isolates in the phylogenetic tree confirmed the heterogenous nature of the genus Stenotrophomonas sp.40. Although all isolates are of three origins, the phylogeny did not categorize them solely according to these origins. Although isolates from soil occupied one part of the phylogeny, the mixed population of isolates from different sources made it difficult to establish niche specificity. The complex heterogeneity of the genus Stenotrophomonas was further confirmed by the ERIC finger printing of the isolates. This is because only two isolates from the group produced similar typing pattern. Previous studies have reported similar observation19.
Biofilm can be considered as a biological system that coordinates the bacterial population to resist or protect from environmental factors, such as antibiotics, harmful chemical compounds or host immune system9,19. Thus, high rate of biofilm formation could contribute to this bacterium for better adaptation. Biofilm formation in isolates seems to be stronger in strain from sewage because all the strain from sewage except TepeL showed capacity to form biofilm. This property may be due fluctuating ecological features of the sewage. The treatment of Stenotrophomonas infection is difficult worldwide because of their ability to resist many antimicrobial agents including carbapenem15,44. In this study we isolated Stenotrophomonas from three different sources which are Soil, Sewage and Hospital. We noted that all the S. maltophilia species obtained from the different sources were resistance to at least four of the antimicrobial agents tested including trimethoprim-sulfamethoxazole (Table 3). The resistance pattern observed in Stenotrophomonas isolated strains, especially S. maltophilia, revealed high resistance to most of the antimicrobial agents analyzed (Table 3). A comparative look at the pattern of resistance to antimicrobial reagents revealed that clinical isolates were more resistance to the tested antimicrobial agent with MAR index which ranged from 0.7–0.9, they are closely followed by isolates from soil with MAR index which ranged between 0.68–0.8, Isolates from sewage showed least resistance to the tested antimicrobial agents with MAR index which ranged from 0.4–0.8. The lead role of hospital isolates to more antimicrobial agents than those from other source in this study might be due to previous exposure to antibiotics since they were isolated from patients with underlining clinical conditions45. Similarly, environmental variation might have influenced the difference in resistance per isolate between soil isolates and sewage isolates in this study. The highest rate of resistance among isolates to antimicrobial agents was to doxycycline (100%), imipenem (100%) and ampicillin (100%) and the lowest level of resistance was to levofloxacin and ofloxacin (0%). Isolates were less resistant to chloramphenicol (54.4%), ceftazidime (26%) Isolates showed high rate of resistance to Trimethoprim-sulfamethoxazole. SXT is the first clinical choice for the treatment of Stenotrophomonas associated infections7,40. The increase in the resistance of S. maltophilia to SXT has been reported in previous studies as well as the variation of this resistance with geographical distribution. For instance, SXT resistance has been reported to be 3.8% on a surveillance study of three continents46; 10% in Europe47, 4.5% in Latin America, and 32.8% in Mexico19. Both environmental and clinical strains showed similar trends of antimicrobial resistance, including SXT resistance. Thus, there may be a need to review the use of this drug for the treatment of S. maltophilia infections. Other antimicrobial agents such as ofloxacin and levofloxacin may be considered as alternatives.
Stenotrophomonas species like other multi-resistant bacteria have arsenals of mechanisms through which they resist antibiotics9,48, some of which may be responsible for the observed resistance to antibiotics in our study. Biofilm formation has been reported to contribute to antimicrobial resistance in Stenotrophomonas and given that most of our isolates showed the ability to produce biofilm, it may be involved in their resistance behaviour. Further studies are needed to confirm this.
Conclusion
The genus Stenotrophomonas is highly heterogeneous both at intra and interspecific level2. Environmental adaptation is common among them and may be responsible for the high degree of diversity in the genus as observed in our study. The variation observed in Stenotrophomonas maltophilia phenotypic characteristics suggests the need for a review in their characterization. The observed resistance patterns emphasize the importance of continuous surveillance study on the antimicrobial susceptibility pattern of Stenotrophomonas species. This study reveals that S. maltophilia are increasingly becoming resistance to SXT and may no longer be the best drug of choice for the treatment of S. maltophilia infection. Similarly, the statistical analysis of the relationship between environmental isolates (soil and sewage) and clinical isolates (p>0.05) revealed that there is no significance difference in the rate at which isolates develop resistance to SXT. This may infer that resistance to SXT may not only be associated with hospital exposure but due to some innate abilities that can be stimulated by external influences such as found in the environment. The main findings of this study can guide in the selection of antimicrobial agents for the treatment of Stenotrophomonas infections.
Acknowledgement
We acknowledge Dra Rayo Morfin-Otero of the Guadalajara civic hospital for providing us the clinical isolates used in this study
This work was supported by the Consejo Nacional de Ciencia y Tecnología (CONACyT)-México (Grant No. 168541), Secretaría de Investigación y Posgrado del Instituto Politécnico Nacional, México (No. 20171762 and 20171793). Temidayo Oluyomi Elufisan holds a scholarship from CONACyT.
Ethical statement
As patient information was anonymized, and only microbiological data were analyzed, informed consent was not required. The study was reviewed and approved by the Local Ethics Committee (Approval: 011/14).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol [Internet] 2009 Apr;68(1):1–13. doi: 10.1111/j.1574-6941.2009.00654.x. Available from: https://academic.oup.com/femsec/article-lookup/doi/10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 2.Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol [Internet] 2009 Jul 1;7(7):514–525. doi: 10.1038/nrmicro2163. Available from: http://www.nature.com/articles/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 3.Tebyanian H, Hassanshahian M, Kariminik A. Hexadecane-degradation by Teskumurella and StenotrophomonasStrains Isolated From Hydrocarbon Contaminated Soils. Jundishapur J Microbiol [Internet] 2013 Sep 1;6(7) Available from: http://jjmicrobiol.com/en/articles/72616.html. [Google Scholar]
- 4.Iyer R, Iken B, Leon A. Characterization and comparison of putative Stenotrophomonasmaltophiliamethyl parathion hydrolases. Bioremediat J. 2016 [Google Scholar]
- 5.Kerr KG, Denton M, Todd N, Corps CM, Kumari P, Hawkey PM. A new selective differential medium for isolation of Stenotrophomonasmaltophilia. Eur J Clin Microbiol Infect Dis. 1996 doi: 10.1007/BF01709373. [DOI] [PubMed] [Google Scholar]
- 6.Alonso A, Martínez JL. Multiple antibiotic resistance in Stenotrophomonasmaltophilia. Antimicrob Agents Chemother [Internet] 1997;41(5):1140–1142. doi: 10.1128/aac.41.5.1140. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9145884%5Cn http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC163865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooke JS. New strategies against Stenotrophomonasmaltophilia: A serious worldwide intrinsically drug-resistant opportunistic pathogen. Expert Review of Anti-Infective Therapy. 2014 doi: 10.1586/14787210.2014.864553. [DOI] [PubMed] [Google Scholar]
- 8.WHO, author. WHO's first global report on antibiotic resistance reveals serious, worldwide threat to public health [Internet] World Health Organization; 2014. News Release Available from: http://www.who.int/mediacentre/news/releases/2014/amr-report/en/%5Cn http://www.who.int/mediacentre/news/releases/2014/amr-report/en/ [Google Scholar]
- 9.Elufisan T O. Updates on microbial resistance to drugs. African J Microbiol Res [Internet] 2012 Jun 21;6(23) Available from: http://www.academicjournals.org/ajmr/abstracts/abstracts/abstract 2012/21Jun/Elufisan et al.htm. [Google Scholar]
- 10.Fukuda A, Usui M, Wakao H, Boonla C, Tamura Y. Stenotrophomonasmaltophiliais highly prevalent among houseflies (Musca domestica) J Med Microbiol [Internet] 2017 Aug 4;66(8):1202–1206. doi: 10.1099/jmm.0.000556. Available from: http://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.000556. [DOI] [PubMed] [Google Scholar]
- 11.Igbinosa E, And FO-J of AS, 2014 U Multiple antibiotics resistant among environmental isolates of Stenotrophomonasmaltophilia. ajol.info [Internet]. [cited 2018 Jun 27]; Available from: https://www.ajol.info/index.php/jasem/article/view/105445.
- 12.Ochoa-Sánchez LE, Vinuesa P. Evolutionary Genetic Analysis Uncovers Multiple Species with Distinct Habitat Preferences and Antibiotic Resistance Phenotypes in the Stenotrophomonasmaltophilia Complex. Front Microbiol [Internet] 2017 Aug 17;8:1548. doi: 10.3389/fmicb.2017.01548. Available from: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01548/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung H, Lieberman TD, Vargas SO, Flett KB, McAdam AJ, Priebe GP, et al. Nat Commun [Internet] Vol. 8. Nature Publishing Group; 2017. Global and local selection acting on the pathogen Stenotrophomonasmaltophiliain the human lung; pp. 1–7. PubMed Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alonso A, Sanchez P, Martínez JL. StenotrophomonasmaltophiliaD457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob Agents Chemother. 2000;44(7):1778–1782. doi: 10.1128/aac.44.7.1778-1782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez MB, Hernandez A, Martinez JL. Stenotrophomonasmaltophiliadrug resistance. Future Microbiol [Internet] 2009 Aug;4(6):655–660. doi: 10.2217/fmb.09.45. Available from: https://www.futuremedicine.com/doi/10.2217/fmb.09.45. [DOI] [PubMed] [Google Scholar]
- 16.Al-Hamad A, Burnie J, Upton M. Enhancement of antibiotic susceptibility of Stenotrophomonasmaltophiliausing a polyclonal antibody developed against an ABC multidrug efflux pump. Can J Microbiol [Internet] 2011 Oct;57(10):820–828. doi: 10.1139/w11-076. Available from: http://www.nrcresearchpress.com/doi/abs/10.1139/w11-076. [DOI] [PubMed] [Google Scholar]
- 17.Hu R-M, Liao S-T, Huang C-C, Huang Y-W, Yang T-C. An inducible fusaric acid tripartite efflux pump contributes to the fusaric acid resistance in Stenotrophomonasmaltophilia. PLoS One [Internet] 2012;7(12):e51053. doi: 10.1371/journal.pone.0051053. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23236431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin YT, Huang YW, Liou RS, Chang YC, Yang TC. MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonasmaltophiliainvolved in drug resistance, oxidative and envelope stress tolerances and biofilm formation. J Antimicrob Chemother [Internet] 2014 Dec 1;69(12):3221–3226. doi: 10.1093/jac/dku317. Available from: https://academic.oup.com/jac/article-lookup/doi/10.1093/jac/dku317. [DOI] [PubMed] [Google Scholar]
- 19.Flores-Treviño S, Gutiérrez-Ferman JL, Morfín-Otero R, Rodríguez-Noriega E, Estrada-Rivadeneyra D, Rivas-Morales C, et al. Stenotrophomonasmaltophiliain Mexico: Antimicrobial resistance, Biofilm formation and clonal diversity. J Med Microbiol. 2014 doi: 10.1099/jmm.0.074385-0. [DOI] [PubMed] [Google Scholar]
- 20.Walsh TR, MacGowan AP, Bennett PM. Sequence analysis and enzyme kinetics of the L2 serine β-lactamase from Stenotrophomonasmaltophilia. Antimicrob Agents Chemother [Internet] 1997;41(7):1460–1464. doi: 10.1128/aac.41.7.1460. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=163940&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. Overexpression, purification, and characterization of the cloned metallo-beta-lactamase L1 from Stenotrophomonasmaltophilia. Antimicrob Agents Chemother [Internet] 1998 Apr;42(4):921–926. doi: 10.1128/aac.42.4.921. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=105568&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furlan JPR, Pitondo-Silva A, Stehling EG. Detection of blaNDM-1 in Stenotrophomonasmaltophiliaisolated from Brazilian soil. Mem Inst Oswaldo Cruz [Internet] 2018 May 14;113(6) doi: 10.1590/0074-02760170558. Available from: http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0074-02762018000600400&lng=en&tlng=en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patil PP, Midha S, Kumar S, Patil PB. Genome Sequence of Type Strains of Genus Stenotrophomonas. Front Microbiol [Internet] 2016 Mar 10;7:309. doi: 10.3389/fmicb.2016.00309. Available from: http://journal.frontiersin.org/Article/10.3389/fmicb.2016.00309/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidigal PG, Dittmer S, Steinmann E, Buer J, Rath P-M, Steinmann J. Adaptation of Stenotrophomonasmaltophiliain cystic fibrosis: Molecular diversity, mutation frequency and antibiotic resistance. Int J Med Microbiol [Internet] 2014 Jul;304(5–6):613–619. doi: 10.1016/j.ijmm.2014.04.002. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1438422114000381. [DOI] [PubMed] [Google Scholar]
- 25.Coenye T. Stenotrophomonasafricana Drancourt et al. 1997 is a later synonym of Stenotrophomonasmaltophilia(Hugh 1981) Palleroni and Bradbury 1993. Int J Syst Evol Microbiol [Internet] 2004 Jul 1;54(4):1235–1237. doi: 10.1099/ijs.0.63093-0. Available from: http://ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.63093-0. [DOI] [PubMed] [Google Scholar]
- 26.Corsaro D, Müller K-D, Michel R. Molecular characterization and ultrastructure of a new amoeba endoparasite belonging to the Stenotrophomonasmaltophiliacomplex. Exp Parasitol [Internet] 2013 Apr;133(4):383–390. doi: 10.1016/j.exppara.2012.12.016. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0014489413000027. [DOI] [PubMed] [Google Scholar]
- 27.Jurkevitch E. Isolation and classification of bdellovibrio and like organisms. Curr Protoc Microbiol [Internet] 2006 Jan; doi: 10.1002/9780471729259.mc07b01s00. Chapter 7:Unit 7B.1 Available from: http://www.ncbi.nlm.nih.gov/pubmed/18770593. [DOI] [PubMed] [Google Scholar]
- 28.CLSI, author. Performance standards for antimicrobial susceptibility testing. CLSI supplement M100S. CLSI supplement M100S. Wayne, PA: Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- 29.Sogawa K, Watanabe M, Sato K, Segawa S, Ishii C, Miyabe A, et al. Use of the MALDI BioTyper system with MALDI-TOF mass spectrometry for rapid identification of microorganisms. Anal Bioanal Chem [Internet] 2011 Jun 26;400(7):1905–1911. doi: 10.1007/s00216-011-4877-7. Available from: http://link.springer.com/10.1007/s00216-011-4877-7. [DOI] [PubMed] [Google Scholar]
- 30.Sierra G. A simple method for the detection of lipolytic activity of micro-organisms and some observations on the influence of the contact between cells and fatty substrates. Antonie Van Leeuwenhoek [Internet] 1957 Dec;23(1):15–22. doi: 10.1007/BF02545855. Available from: http://link.springer.com/10.1007/BF02545855. [DOI] [PubMed] [Google Scholar]
- 31.Tindall BJ. A Comparative Study of the Lipid Composition of Halobacterium saccharovorum from Various Sources. Syst Appl Microbiol [Internet] 1990 May;13(2):128–130. Available from: http://linkinghub.elsevier.com/retrieve/pii/S072320201180158X. [Google Scholar]
- 32.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fülöp V, Szeltner Z, Renner V, Polgár L. Structures of Prolyl Oligopeptidase Substrate/Inhibitor Complexes. J Biol Chem [Internet] 2001 Jan 12;276(2):1262–1266. doi: 10.1074/jbc.M007003200. Available from: http://www.jbc.org/lookup/doi/10.1074/jbc.M007003200. [DOI] [PubMed] [Google Scholar]
- 34.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc [Internet] 2008 Feb 1;3(2):163–175. doi: 10.1038/nprot.2007.521. Available from: http://www.nature.com/articles/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 35.Titilawo Y, Sibanda T, Obi L, Okoh A. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of water. Environ Sci Pollut Res [Internet] 2015 Jul 18;22(14):10969–10980. doi: 10.1007/s11356-014-3887-3. Available from: http://link.springer.com/10.1007/s11356-014-3887-3. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole G, Kaplan HB, Kolter R. Biofilm Formation as Microbial Development. Annu Rev Microbiol [Internet] 2000 Oct;54(1):49–79. doi: 10.1146/annurev.micro.54.1.49. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11018124. [DOI] [PubMed] [Google Scholar]
- 37.Christensen BE. The role of extracellular polysaccharides in biofilms [Internet] Journal of Biotechnology. 1989;10:181–202. Available from: http://linkinghub.elsevier.com/retrieve/pii/0168165689900643. [Google Scholar]
- 38.Finkmann W, Altendorf K, Stackebrandt E, Lipski A. Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonasnitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int J Syst Evol Microbiol. 2000 doi: 10.1099/00207713-50-1-273. [DOI] [PubMed] [Google Scholar]
- 39.Lee M, Woo S-G, Chae M, Shin M-C, Jung H-M, Ten LN. Stenotrophomonasdaejeonensis sp. nov., isolated from sewage. Int J Syst Evol Microbiol [Internet] 2011 Mar 1;61(3):598–604. doi: 10.1099/ijs.0.017780-0. Available from: http://ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.017780-0. [DOI] [PubMed] [Google Scholar]
- 40.Brooke JS. Stenotrophomonasmaltophilia: An emerging global opportunistic pathogen. Clin Microbiol Rev. 2012 doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turrientes MC, Baquero MR, Sánchez MB, Valdezate S, Escudero E, Berg G, et al. Polymorphic mutation frequencies of clinical and environmental Stenotrophomonasmaltophiliapopulations. Appl Environ Microbiol. 2010 doi: 10.1128/AEM.02817-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urszula G, Izabela G, Danuta W, Sylwia L. Isolation and characterization of a novel strain of Stenotrophomonasmaltophiliapossessing various dioxygenases for monocyclic hydrocarbon degradation. Braz J Microbiol [Internet] 2009 Apr;40(2):285–291. doi: 10.1590/S1517-838220090002000014. Available from: http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1517-83822009000200014&lng=en&nrm=iso&tlng=en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carmody LA, Spilker T, LiPuma JJ. Reassessment of Stenotrophomonasmaltophiliaphenotype. J Clin Microbiol. 2011 doi: 10.1128/JCM.02204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lockhart SR, Abramson MA, Beekmann SE, Gallagher G, Riedel S, Diekema DJ, et al. Antimicrobial resistance among gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J Clin Microbiol. 2007 doi: 10.1128/JCM.01284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baek J-E, Jung E-Y, Kim H-J, Lee G-W, Hahm J-R, Kang K-R, et al. Stenotrophomonasmaltophiliainfection in patients receiving continuous ambulatory peritoneal dialysis. Korean J Intern Med [Internet] 2004 Jun 30;19(2):104–108. doi: 10.3904/kjim.2004.19.2.104. Available from: http://kjim.org/journal/view.php?doi=10.3904/kjim.2004.19.2.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fedler KA, Biedenbach DJ, Jones RN. Assessment of pathogen frequency and resistance patterns among pediatric patient isolates: Report from the 2004 SENTRY Antimicrobial Surveillance Program on 3 continents. Diagn Microbiol Infect Dis [Internet] 2006 Dec;56(4):427–436. doi: 10.1016/j.diagmicrobio.2006.07.003. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0732889306002501. [DOI] [PubMed] [Google Scholar]
- 47.Gales AC, Jones RN, Forward KR, Liñ J, Sader HS, Verhoef J. Emerging Importance of Multidrug-Resistant Acinetobacter Species and Stenotrophomonasmaltophiliaas Pathogens in Seriously Ill Patients: Geographic Patterns, Epidemiological Features, and Trends in the SENTRY Antimicrobial Surveillance Program. Clin Infect Dis. 2001 doi: 10.1086/320183. [DOI] [PubMed] [Google Scholar]
- 48.Alonso A, Martinez JL. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonasmaltophilia. Antimicrob Agents Chemother. 2000;44(11):3079–3086. doi: 10.1128/aac.44.11.3079-3086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elufisan TO, Rodríguez-Luna IC, Oyedara OO, Sánchez-Varela A, Hernández-Mendoza A, Dantán Gonzalez E, Paz-González AD, Muhammad K, Rivera G, Villalobos-Lopez MA, Guo X. The Polycyclic Aromatic Hydrocarbon (PAH) degradation activities and genome analysis of a novel strain Stenotrophomonas sp. Pemsol isolated from Mexico. Peer J. 2020;8:e8102. doi: 10.7717/peerj.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]